Ig VH somatic hypermutation has recently emerged as a novel prognostic factor in chronic lymphocytic leukemia (CLL), unmutated Ig VH genes predicting adverse prognosis.1-4 However, the clinical usefulness of Ig VH gene mutation analysis in predicting survival early in the course of the disease is offset by the high cost and level of expertise required for this technique. The report of Damle et al2 that CD38 expression can predict the Ig VHgene mutational status of CLL cells (positivity indicating unmutated and negativity, mutated, Ig VH genes) offered the possibility of a simple and inexpensive substitute for Ig VH gene analysis. However, the association between CD38 expression and Ig VH gene mutation could not be confirmed by Hamblin et al.5 This discrepancy prompted us to undertake a survey of our own CLL patients to examine further the relationship between CD38 expression, Ig VH gene mutation, and survival, and thereby determine to what extent measurement of CD38 can indeed be used as surrogate for Ig VH gene sequencing.
We included 40 CLL patients (27 men, 13 women) with known clinical history in the study. Of these, 6 were selected on the basis of their tumor cells expressing CD38. The mean age was 65 (men 61.5, women 72.3) years, and the mean follow-up time 5.4 (1-23) years. Ig VHgene sequence analysis was performed using a protocol based on that of Fais et al.6 Unmutated cases were defined as those with less than 2% mismatch to the most similar germline Ig VHgene in the V BASE directory.7 Immunophenotyping was performed on a Becton Dickinson FACS scan analyzer (San Jose, CA) using thawed samples of previously cryopreserved cells that had been double stained with CD38 PE and CD19 FITC (both from Becton Dickinson). The proportion of CD19+ cells coexpressing CD38 was determined; values more than 30% were considered positive. Survival analyses were performed by the method of Kaplan and Meier, and significance was analyzed using the log-rank test.
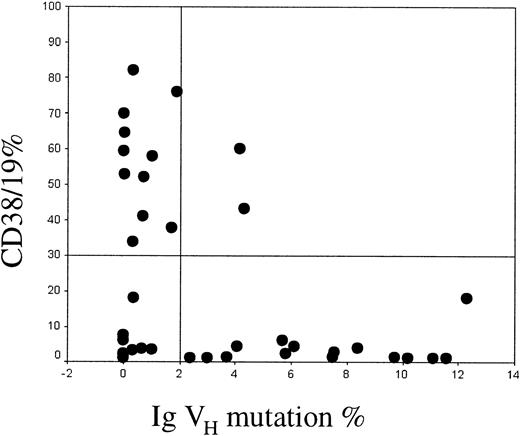
Twenty-four cases were unmutated and 16 mutated, with a male-female ratio of 1.6 in the mutated and 2.4 in the unmutated groups. These findings therefore support the observation made by Damle et al that there is an unequal sex distribution between the mutated and unmutated groups. CD38 was expressed in 2 (12.5%) of the 16 mutated cases and in 11 (46%) of the 24 unmutated cases. Among the 13 CD38+cases, 2 (15%) were mutated and 11 (85%) unmutated. Twenty-seven cases were CD38−, of which 13 (48%) were unmutated and 14 (52%) mutated. These results are summarized in Figure1.
Relationship between CD38 expression and Ig VH mutation.
The proportion of CD19+ cells coexpressing CD38 is shown on the y-axis. Values greater than 30% were considered positive. The percent mismatch with the closest Ig VH germline gene is shown on the x-axis. Cases with more than 2% mismatch were defined as being mutated.
Relationship between CD38 expression and Ig VH mutation.
The proportion of CD19+ cells coexpressing CD38 is shown on the y-axis. Values greater than 30% were considered positive. The percent mismatch with the closest Ig VH germline gene is shown on the x-axis. Cases with more than 2% mismatch were defined as being mutated.
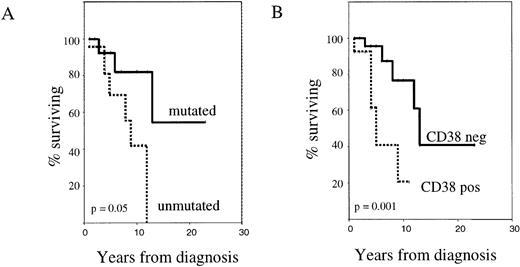
Survival analysis of mutated versus unmutated patients (Figure2A) showed that the absence of Ig VH gene somatic mutation was associated with a worse outcome (median survival 9 years, versus not reached;P = .05). Similarly, CD38+ patients had a shorter median survival than the CD38− group (5 years versus 13 years; P = .001) (Figure 2B).
CD38 expression and Ig VH gene mutational status as independent prognostic factors in B-CLL.
The figure shows Kaplan-Meier survival curves comparing cases with mutated versus unmutated Ig VH genes (A) and CD38+ versus CD38− cases (B). Deaths not attributable to CLL were censored.
CD38 expression and Ig VH gene mutational status as independent prognostic factors in B-CLL.
The figure shows Kaplan-Meier survival curves comparing cases with mutated versus unmutated Ig VH genes (A) and CD38+ versus CD38− cases (B). Deaths not attributable to CLL were censored.
Our results regarding the adverse prognostic significance of unmutated Ig VH genes and CD38 expression are entirely in keeping with Damle et al's and Hamblin et al's. Furthermore, in agreement with Damle et al, we also found that CD38 expression is, indeed, able to identify unmutated cases. Our confirmation that CD38 positivity is informative, both as an independent prognostic parameter and as a predictor of unmutated Ig VH status, therefore supports the idea that measurement of CD38 should form part of the routine diagnostic work-up of CLL.
Importantly, in our series only half of the CLL patients with unmutated Ig VH genes expressed CD38. This novel observation raises the interesting possibility that unmutated CLL tumors may be clonal expansions of 2 different types of pre–germinal center B cells: one being CD38+ and the other, CD38−.
Supported by grants from the Leukaemia Research Fund (UK) and North West Cancer Research Fund

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal