Abstract
The hemophagocytic lymphohistiocytoses (HLH) comprise a heterogeneous group of disorders characterized by dysregulated activation of T cells and macrophages. Although some patients with HLH harbor perforin gene mutations, the cause of the remaining cases is not known. The phenotype of HLH bears a strong resemblance to X-linked lymphoproliferative disease (XLP), an Epstein-Barr virus (EBV)-associated immunodeficiency resulting from defects in SH2D1A, a small SH2 domain-containing protein expressed in T lymphocytes and natural killer cells. Here it is shown that 4 of 25 male patients with HLH who were examined harbored germline SH2D1A mutations. Among these 4 patients, only 2 had family histories consistent with XLP. On the basis of these findings, it is suggested that all male patients with EBV-associated hemophagocytosis be screened for mutations in SH2D1A. Patients identified as having XLP should undergo genetic counseling, and be followed long-term for development of lymphoma and hypogammaglobulinemia.
Introduction
Patients affected by the hemophagocytic lymphohistiocytoses (HLH) experience recurring episodes of life-threatening, often infection-induced, lymphohistiocytic activation. HLH is genetically heterogeneous with both sporadic and familial forms described.1,2 The genetic defects underlying HLH are only partially defined3-5 and include germline mutations in PRF1, the gene-encoding perforin,6 in a subset of patients. Perforin is secreted by activated natural killer (NK) cells and T lymphocytes and mediates cytolysis by creating pores in target cell membranes. The discovery of these mutations establishes a critical role for perforin during negative regulation of antiviral immunity.
It is also possible that patients with HLH harbor mutations in genes cooperating with perforin during granule-mediated cytolysis, or participating in distinct biochemical pathways that control the activation of T cells and macrophages. To explore the latter possibility, we analyzed patients with HLH for germline mutations inSH2D1A7 (also known asSAP,8,DSHP9), the gene defective in X-linked lymphoproliferative disease (XLP). XLP is an immunodeficiency disorder characterized by increased susceptibility to Epstein-Barr virus (EBV)-induced fulminant infectious mononucleosis (FIM), malignant lymphoma, and hypogammaglobulinemia.10 We chose SH2D1A because the clinical and laboratory manifestations of XLP-associated FIM, including fever, hepatosplenomegaly, coagulopathy, cytopenias, cytokinemias, and NK cell dysfunction, closely resemble features observed in HLH.1,2 11-13 Among a cohort of 25 male patients with HLH, we found 4 (16%) carrying a previously unrecognized diagnosis of XLP. Two patients harbored hemizygous deletions encompassingSH2D1A exon 1, and 2 patients had nonsense mutations predicted to prematurely truncate the SH2D1A protein. Among these 4 patients, only 2 had family histories consistent with XLP.
Study design
Patients
Twenty-five male patients with HLH were identified at the Children's Research Hospital, Kyoto (n = 10), or the IRCCS Policlinico San Matteo, Italy (n = 15). All patients were given a diagnosis of HLH based on previously established guidelines.1 Pedigrees were generated through interviews with patients and parents. Clinical features are as described previously.13-16
SH2D1A mutational analysis
DNA was extracted from peripheral blood mononuclear cells or paraffin-embedded tissues according to standard protocols. TheSH2D1A coding sequence was polymerase chain reaction (PCR) amplified using Expand Taq polymerase (Roche) and primers flanking each of the 4 SH2D1A exons. To establish the centromeric boundaries for genomic deletions identified in patients HLH-PZ and HLH-OT, we used primers derived from the 5′ end of PAC contig dJ1052M9 (GenBank accession number AL022718) containing theSH2D1A gene. Conditions for PCR and primer sequences are available on request.
Results and discussion
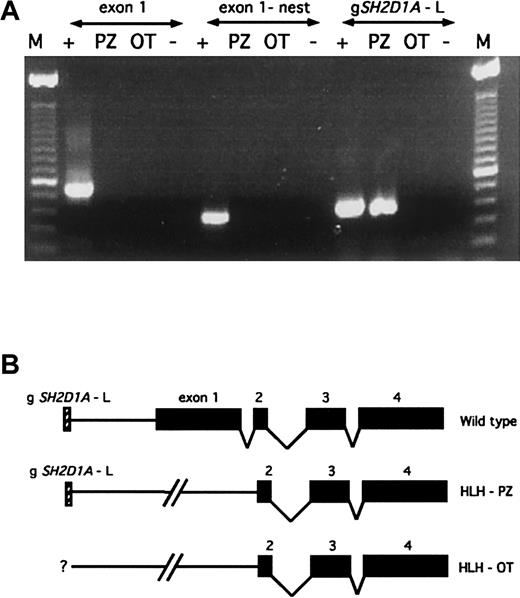
Nested PCR reactions failed to produce products forSH2D1A exon 1 in patients HLH-PZ and HLH-OT, strongly suggesting the presence of genomic deletions (Figure1A). Because confirmatory Southern blot analysis could not be performed due to limited quantities of patient DNA, we used PCR to perform a more detailed mapping. The centromeric boundaries for these deletions were estimated by using primers derived from genomic sequences proximal to SH2D1A exon 1. In HLH-PZ, this genomic region (gSH2D1A-L) is present, indicating a deletion smaller than 58 kilobase (kb), the distance between gSH2D1A-L and SH2D1A exon 2 (Figure 1B). The gSH2D1A-L was absent in HLH-OT, suggesting a deletion that is larger than 58 kb; however, no further mapping was performed (Figure1B). Automated sequence analysis of SH2D1A exons 2-4 revealed wild-type sequences in both patients. These deletions of exon 1 are predicted to be deleterious because of the removal of the start codon and 5′ regulatory sequences, which would interfere with the transcription of the SH2D1A message and its translation into protein.
Interstitial deletions encompassing SH2D1Aexon 1 are present in patients HLH-PZ and HLH-OT.
(A) Nested PCR analysis of SH2D1A exon 1 reveals no detectable product in patients HLH-PZ or HLH-OT, in comparison to a normal patient (denoted as “+”), indicating the presence of interstitial genomic deletions. Amplification of the centromeric sequence gSH2D1A-L reveals a product in patient HLH-PZ, establishing an approximate centromeric boundary for the deletion in this patient. The gSH2D1A-L is absent in HLH-OT, indicating the presence of a larger deletion, the centromeric boundary of which was not further mapped. “M” denotes the 123-base pair (bp) ladder, and “−” the negative control (no input DNA). (B) Schematic representation of the wild-type SH2D1A genomic locus and the constitutional deletions in these patients. SH2D1A exons are shown as black boxes, and the gSH2D1A-L marker as a hatched box.
Interstitial deletions encompassing SH2D1Aexon 1 are present in patients HLH-PZ and HLH-OT.
(A) Nested PCR analysis of SH2D1A exon 1 reveals no detectable product in patients HLH-PZ or HLH-OT, in comparison to a normal patient (denoted as “+”), indicating the presence of interstitial genomic deletions. Amplification of the centromeric sequence gSH2D1A-L reveals a product in patient HLH-PZ, establishing an approximate centromeric boundary for the deletion in this patient. The gSH2D1A-L is absent in HLH-OT, indicating the presence of a larger deletion, the centromeric boundary of which was not further mapped. “M” denotes the 123-base pair (bp) ladder, and “−” the negative control (no input DNA). (B) Schematic representation of the wild-type SH2D1A genomic locus and the constitutional deletions in these patients. SH2D1A exons are shown as black boxes, and the gSH2D1A-L marker as a hatched box.
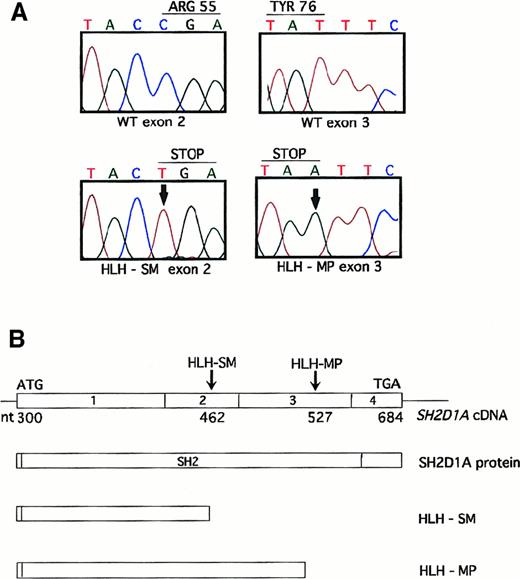
Automated sequence analysis of SH2D1A exons 1 to 4 from the remainder of the cohort identified 2 patients with nonsense mutations. Patient HLH-SM harbored a C to T transition at nucleotide position 462, introducing a stop codon at arginine 55 (Arg55) (Figure2A,B). This substitution has been observed previously7,9,17,18 and may represent a hot spot for mutational inactivation.9,17 Crystallographic analyses suggest that Arg55 is required for the binding of tyrosine by the SH2D1A SH2 domain19; therefore, termination at this residue is likely to generate a nonfunctional protein. Patient HLH-MP harbored a novel T to A change at nucleotide position 527 (Figure 2A,B). This mutation is predicted to truncate SH2D1A at tyrosine 76 (Tyr76), which resides within the βF pleated sheet, a conserved motif among SH2 domains. Termination at this residue is also anticipated to interfere with normal function.
Unique nonsense mutations are identified in patients HLH-SM and HLH-MP.
(A) Automated sequence analysis of exon 2 reveals a C462T nucleotide substitution (shown with an arrow) in patient HLH-SM (lower left-hand panel), that is predicted to substitute a stop codon for Arg55. Similarly, sequence analysis of exon 3 reveals a T527A nucleotide change in patient HLH-MP (lower right-hand panel), changing Tyr76 to a stop codon. (B) Schematic representation of the full-length SH2D1A complementary DNA and protein, and the location of both point mutations that are predicted to result in premature truncation within the SH2D1A SH2 domain.
Unique nonsense mutations are identified in patients HLH-SM and HLH-MP.
(A) Automated sequence analysis of exon 2 reveals a C462T nucleotide substitution (shown with an arrow) in patient HLH-SM (lower left-hand panel), that is predicted to substitute a stop codon for Arg55. Similarly, sequence analysis of exon 3 reveals a T527A nucleotide change in patient HLH-MP (lower right-hand panel), changing Tyr76 to a stop codon. (B) Schematic representation of the full-length SH2D1A complementary DNA and protein, and the location of both point mutations that are predicted to result in premature truncation within the SH2D1A SH2 domain.
With the exception that the 4 patients with SH2D1A mutations were of Italian ethnicity, there were no clinical or laboratory features at presentation distinguishing these patients from the rest of the cohort. The absence of mutations among the Japanese patients is unexplained, but may be due to the limited number of individuals available for analysis. Three patients were young, with an age at diagnosis of younger than 3 years (median 28 months, range 12 months to 18 years). Three patients died, despite the use of multiagent chemotherapy and, in one patient, bone marrow transplantation. The fourth patient survived his hemophagocytosis, but subsequently developed hypogammaglobulinemia develop. These outcomes are consistent with previous descriptions of XLP.11 Only 2 patients with mutations were known to have a history of EBV infection. Unfortunately, many samples included in this analysis were collected previously, and tissue was no longer available to assess for the presence of EBV.
Pedigrees were analyzed for the patients harboring germlineSH2D1A mutations. For HLH-PZ and HLH-MP, there were no additional family members with hemophagocytosis, immunodeficiency, or lymphoma. The pedigree of HLH-OT was notable for a brother with hemophagocytosis at 11 months, suggesting a diagnosis of familial HLH or XLP. The pedigree of patient HLH-SM was consistent with XLP as revealed by one brother with fever and cytopenia during infancy, and a second male sibling with Burkitt's lymphoma at 3 years. The identification of 2 patients with affected male relatives demonstrates the importance of obtaining family history information when caring for patients with hemophagocytosis. The presence of a typical family history supports a presumptive XLP diagnosis and should prompt molecular testing for SH2D1A mutations.18However, the absence of a positive family history, as seen in 2 of the 4 patients reported herein, does not rule out the possibility of XLP.
In summary, we have identified 4 boys with XLP among 25 carrying a clinical diagnosis of HLH. This observation supports the use ofSH2D1A mutation analysis to discern between these often clinically indistinguishable disorders and highlights several other important points. First, patients with hemophagocytosis and germlineSH2D1A mutations are at an increased risk for the development of other manifestations of XLP. Therefore, it is critical that clinicians monitor these patients over time for the development of lymphoma and hypogammaglobulinemia. Second, assignment of an XLP diagnosis necessitates consideration of genetic testing and counseling for family members at risk for XLP or to transmit the disease to their offspring. Third, the identification of 2 patients with “sporadic” XLP suggests that all male patients with EBV-associated hemophagocytosis, not just those with positive family histories, be considered for SH2D1A mutation analysis. Fortunately, both DNA and protein-based assays have been developed for this purpose.20 Ultimately, identification of additional genes underlying HLH, and improved understanding of their roles during the regulation of host antiviral immune responses, will facilitate development of novel therapies for these rare but devastating disorders.
Acknowledgments
We are grateful to the patients who participated by donating blood for these studies. In addition, we thank Mitchell Weiss, Michael Hogarty, John Maris, and Carolyn Felix for critically reviewing this manuscript.
Supported in part by grants K11AI01331-05 (K.E.N.); Telethon Italy, Grant C30 (C.D.) and E755 (M.A.); Ricerca Corrente 390RCR97/01 (M.A.); 16488/GEN 1999, IRCCS Policlinico San Matteo (M.A.); and The Histiocytosis Association of America (K.E.N.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kim E. Nichols, Division of Pediatric Oncology, Children's Hospital of Philadelphia, 34th St and Civic Center Blvd, Philadelphia, PA 19104; e-mail: nicholsk@emailchop.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal