Abstract
To understand the molecular basis for multidrug-resistant (MDR) cancer cells in vivo, this study analyzed molecular changes of the mdr1a gene region in leukemia cells in mice during continuous treatment with vincristine. An inverse insertion of murine leukemia retrovirus (MuLV) into the 5′-flanking region of the mdr1a gene was found. This insertion was concomitantly accompanied by up-regulation of themdr1a gene and the loss of chemosensitivity. Deletion of long-terminal repeat (LTR) sequences dramatically decreased themdr1a promoter-driven reporter activity. The MuLV LTR insertion appears to exert its enhancer activity onmdr1a transcription during the appearance of MDR leukemia cells. Two mechanisms were postulated to explain the mdr1agene activation by retrovirus insertion during in vivo chemotreatment: de novo insertion of MuLV induced by vincristine treatment and selection of a small fraction of pre-existing cells carrying MuLV insertion during vincristine treatment. No rearranged sequence was detected by polymerase chain reaction in parental cells. This result argued for the first mechanism. The randomly altered distribution of MuLV during repetitive chemotreatment might also be consistent with this hypothesis. On the other hand, the retrovirus insertion was detected at the same site of the mdr1a promoter region in 2 independent experiments, which suggests the second mechanism. It should be noted that in vivo chemotreatment using vincristine could generate the mdr1a-overexpressing cells through retrovirus insertion and the enhancer effect of the LTR.
Introduction
The acquisition of a multidrug-resistant (MDR) phenotype in cancer cells is often associated with increased expression of the cellular surface P-glycoprotein (P-gp), which functions as an energy-dependent drug efflux pump, thereby resulting in a decrease of drug concentrations within cells.1-3 Expression of P-gp in various malignancies might become one of the major obstacles to chemotherapy, especially in hematopoietic malignancies.4-6Mammalian P-gps are encoded by small families of genes known as linkedMDR genes, of which there are 2 known members in humans (PGY1 and PGY3, also known as MDR1 andMDR3, respectively)3,7,8 and 3 known members in mice (mdr1a, mdr1b, andmdr2).9-11 The transfection of complementary DNA (cDNA) derived from MDR1, mdr1a, andmdr1b confers multidrug resistance, but the transfection of cDNA from MDR3 and mdr2 does not.9,10 12-16
The MDR1 gene is expressed in both normal tissues17 and malignancies.18,19 It is thus important to understand how P-gp is specifically expressed in cancer cells in order to use P-gp as a diagnostic marker for MDR in human malignancies. Various mechanisms for MDR1 overexpression have been reported in cultured cancer cell lines or tumor samples from clinical patients. Gene amplification units of the MDR1 gene have often been detected in cancer cells selected for their drug resistance to multiple anticancer agents,20-24 but amplification has not appeared in clinical samples. The MDR1gene is activated through transcription factors NF-Y25 and Y-box binding protein (YB-1)26 in the presence of exogenous stimuli including ultraviolet light, irradiation, and carcinogens. Cellular translocation of YB-1 from cytoplasm to nuclei has been shown to be closely associated with P-gp expression in several cancers.27-29 The alteration of methylation status at CpG sites on MDR1 gene promoters has also been shown to be involved in MDR1 gene expression in cultured cancer cell lines.22,30 Demethylation at CpG sites on MDR1 gene promoters has also been closely associated with P-gp expression in patients with acute myeloid leukemias.31
Mickley et al32 have reported that gene rearrangement at the 5′-flanking region of the MDR1 gene was responsible for transcriptional activation of the MDR1 gene in drug-resistant cell lines derived from colon and breast cancers. The authors also showed rearrangement at the 5′-flanking region ofMDR1 in clinical samples of acute lymphoblastic leukemias. This activation might be induced by read-through transcription from the promoter of a fused, upstream-positioned gene. Determination of the rearranged sequence indicated that the gene rearrangements occurred from homologous recombination between Alu repeats.33
As an alternative approach to understanding how the initial onset of drug resistance in cancer cells appears during anticancer chemotreatment in vivo, an animal model could be useful to clarify the mechanism of overexpression of MDR genes. Chemotherapeutic models consisting of animals carrying transplanted cancer cells are expected to avoid the potential heterogeneity of tumor cells in human clinical samples.34-36 In this study we inoculated mouse leukemia cells into the intraperitoneal cavity and treated the mice with vincristine. We then examined the mechanisms involved in the appearance of leukemia cells that were refractory to chemotherapeutic agents. Based on these results, we discuss the appearance of P-gp–expressing leukemia cells in association with a novel type of gene rearrangement of the MDR gene by retroviral insertion.
Materials and methods
Oligonucleotides
The sequences of oligonucleotides that were used in this study are as follows: pr-1aF: 5′-GTAATGCCAGTGCTAGAGGC-3′; pr-1aR: 5′-TAATGTGTGCGTGTGTGTGC-3′; pr-mgF: 5′-GACCGGCTTGTATGCTATCC-3′; pr-mgR: 5′-TCATGCTTAACTCTGCAGGC-3′; ts-oligo1: 5′-CGAAGGAGAGGACGCTGTCTGTCGAAGGTAAGGAACGG- ACGAGAGAAGGGAGAG-3′; ts-oligo2: 5′-CATGAAGGAGAGGACGCTGTCTGTCGAAGGTAAGGAACGGACGAGAGAAGGGAGAG-3′; bs-oligo: 5′CTCTCCCTTCTCGAATCGTAACCGTTCGTACGAGAATCGC- TGTCCTCTCCTT-3′; uvp: 5′-CGAATCGTAACCGTTCGTACGAGA- ATCGCT-3′; msp: 5′-TGTAGATGAAATGTGCAGTCTGGTTTCCC-3′; musp1: 5′-ATGCCGACCACACAGGATTGGTACG-3′; musp2: 5′-CAATCAGCTCGCTTCTCGCTTCTGTACCCG-3′; pr-ltr/1aF: 5′-GCGTTACTGCAGTTAGCTGG-3′; pr-ltr/1aR: 5′-TGCAGTCTGGTTTCCCTCTC-3′; pr-1a3′F: 5′-GCATCTGTGGACCACATGAC-3′; pr-1a3′R: 5′-CTTTCACTTGAGCAGCATCG-3′; pr-luciF: 5′-CCATGGAGGAGCCGCAGTCAGATCC-3′; and pr-luciR: 5′-GAAGTGGAGAATGTCAGTCTGAGTCAGGCC-3′.
Cell lines and cell culture
Mouse fibroblast L cells and their vincristine-selected clone, LV500, were used for negative and positive control of P-gp, respectively. Mouse NIH 3T3 fibroblasts were used for transfection and luciferase reporter assay. They were cultured with Dulbecco modified Eagle medium containing 10% fetal bovine serum.
Animals and in vivo chemotherapeutic models
We used 5- to 6-week-old female CD2F1 mice (Charles River, Yokohama, Japan) and P388 murine leukemia cell lines (Human Science Research Resources Bank, Osaka, Japan). CD2F1 mice were inoculated in the abdomen with 1.0 × 106 P388 cells and derivatives.34 36 Mice in the vincristine treatment group were innoculated twice with 1 mg/kg interperitoneal vincristine, and mice in the control group were given saline. Survival of the mice was observed during the experimental period. The effectiveness of vincristine treatment was evaluated based on the mean survival days, which were analyzed by Student t test. P388 cells and derivatives were harvested from the tumor-carrying mice just before their death. Cells were suspended again in medium containing 1.0 × 106 cells and then transplanted to the next generation of mice. This cycle was repeated with 4 generations of mice.
Isolation of DNA and RNA
Semiquantitative reverse transcriptase–polymerase chain reaction
The semiquantitative reverse transcriptase–polymerase chain reaction (RT-PCR) method was previously described.31 38The template cDNA was diluted serially in water from 500 to 0.2 ng/μL. Each cycle of PCR included 30 seconds of denaturation at 94°C, 30 seconds of primer annealing at 55°C, and 30 seconds of polymerization at 72°C. We performed PCR with 35 cycles formdr1a and 25 cycles for β2-microglobulin. The PCR products were separated by electrophoresis on 3% agarose gels, stained with CYBR green (Molecular Probes, Eugene, OR), and image-analyzed by FAS 2000 (Fuji Photo Film, Tokyo, Japan).
Immunohistochemistry
Immunohistochemistry was performed using ImmunoCruz Staining Systems (Santa Cruz Biotechnology, Santa Cruz, CA) including goat polyclonal anti–P-gp antibody (C-19) and biotinylated antigoat secondary antibody (both from Santa Cruz Biotechnology) according to the manufacturer's instruction.
Southern blot analysis
The genomic DNA was digested by either a methylation-insensitive or methylation-sensitive restriction enzyme, MspI or HpaII, respectively. The digested DNA was separated on a 2% agarose gel and transferred to nylon filters. We used 370-bp (base pair) PstI fragments of the mouse mdr1a promoter region located 100 bp upstream from the transcriptional start of mdr1a as probes.
Vectorette PCR method
The vectorette linkers were prepared by annealing 2 oligonucleotides, ts-oligo1 and bs-oligo. The ts-oligo was not complementary to the bs-oligo in the middle 30-bp region.MspI-digested genomic DNA from vincristine-resistant P-V4 cells were separated on agarose gel, andMspI fragments approximately 500 bp in length were isolated and ligated by the vectorette linkers. When PCR is performed with the primer uvp, which is complementary to bs-oligo but not to ts-oligo, and the mdr1a-specific primer, msp, the sequence between primers uvp and msp is amplified, but it is not amplified between 2 uvp primers. Additional details about the vectorette PCR method were described previously.39 40 The vectorette linkers were also used to analyze the MuLV proviral distribution in the host genome. The genomic DNA derived from P388 and vincristine-selected derivatives (P-V1, P-V2, P-V3, and P-V4), were digested by restriction enzymesNheI, SpeI, and XbaI. Although each of these 3 enzymes recognizes the different sites, they generate the same 5′-overhanged cohesive end, 5′-CTAG-3′. The vectorette linkers prepared by annealing ts-oligo2 and bs-oligo are compatible to this cohesive end. PCR was performed by using the primer uvp and theMuLV-specific primer musp1 at first, which was then nested by primer uvp and the second MuLV-specific primer musp2. The nested PCR products were analyzed by separation on 10% acrylamide gel electrophoresis.
Luciferase reporter assay
The MuLV long-terminal repeat (LTR)/mdr1a promoter sequence was amplified from genomic DNA of P-V4 cells by PCR using the primer pair pr-luciF and pr-luciR. The MuLV-mdr1a promoter hybrid fragments were ligated upstream of luciferase cDNA of pGL3-basic vector (Promega, Madison, WI) and designated as pGL3-L/m1a. We next made an LTR-deleted construct, named pGL3-m1a, by PstI digestion. ThePstI-digested LTR fragment was cloned by SalI linker into the SalI site of pGL3-m1a, which is located downstream of luciferase, thereby generating pGL3-m1a/L. For transient transfection, NIH3T3 cells were cotransfected with 1 μg luciferase plasmid DNA and 20 ng pRL-CMV vector (Promega) by using LipofectAMINE 2000 (Gibco BRL Life Technologies, Rockville, MD) according to the manufacturer's instructions. The measurement of luciferase activity of transient transfectants was performed using a dual-luciferase assay protocol (Promega).
PCR to detect the existence of rearranged genomes
We used 10 μg genomic DNA, which was equivalent to 1.0 × 106 cells, as templates to amplify the rearranged MuLV LTR/mdr1a sequences by PCR with primers pr-ltr/1aF and pr-ltr/1aR. The PCR product was subcloned (pCR-L/m1a) and used as a template as follows. In the control experiment, 10 μg genomic DNA of mouse fibroblast L cells containing the serially diluted pCR-L/m1a from 3.3 pg (106 copies) to 3.3 × 10−6 pg (one copy) was used as template. PCR was performed with AmpliTaq Gold DNA polymerase (PE Applied Biosystems, Urayasu, Japan). Each cycle of PCR included 30 seconds of denaturation at 94°C, 30 seconds of primer annealing at 58°C, and 45 seconds of polymerization at 72°C. PCR was performed with 45 cycles and repeated again using the first PCR product as template.
Results
Appearance of leukemia cells refractory to chemotherapeutic treatment with vincristine
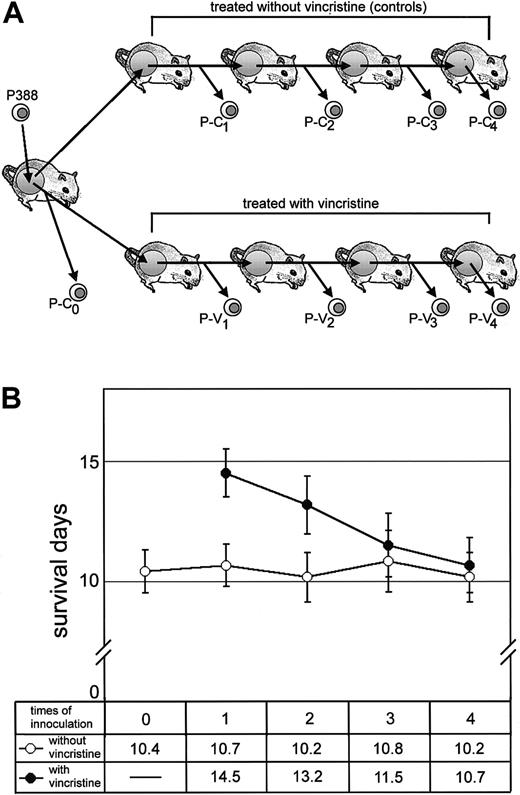
We used the mice carrying P388 murine leukemia cells as a chemotherapeutic model to investigate the molecular basis for the acquisition of MDR in vivo. We transplanted leukemia cells into the abdominal cavity of mice and continuously treated the mice with vincristine at each transplantation step (Figure1A). To assess the therapeutic efficacy of vincristine, we examined the life span of mice at each transplanted step (Figure 1B). The life span of mice treated with vincristine was prolonged by 13-15 days up to the second serial transplantation (early-transplantation mice); mice without any treatment died within 10-12 days after transplantation of leukemia cells (Figure 1B). By contrast, the life spans of mice became shorter with each sequential transplantation and repetition of vincristine treatment. After 3 or 4 transplantations (later-transplantation mice), the life spans plateaued at 10 days, which corresponds to the life spans of untreated control mice, in spite of vincristine treatment. This result suggested the appearance of drug-resistant leukemia cells in later-transplantation mice. It is also noted that this result was highly reproducible in the second experiment (data not shown).
Effect of vincristine treatment on the life span of mice carrying leukemias.
(A) Experimental design: CD2F1 mice were inoculated with 1.0 × 106 P388 cells, and the same number of cells was transplanted sequentially. Mice in the vincristine treatment group received a 1-mg/kg intraperitoneal dose of vincristine twice, and control mice received saline. We designated the P388 derivatives collected from the abdominal cavity of each successive group of mice as P-C0, P-C1, P-C2, P-C3, and P-C4 (untreated cells) or P-V1, P-V2, P-V3, and P-V4(vincristine-treated cells). These derivatives were used in the following experiments. (B) The life span of mice with (closed circles) or without (open circles) vincristine treatment. Arabic numbers (n = 0, 1, 2, 3, and 4) indicate the number of times that the P388 derivatives were inoculated. Mean survival days are also indicated in the lower box.
Effect of vincristine treatment on the life span of mice carrying leukemias.
(A) Experimental design: CD2F1 mice were inoculated with 1.0 × 106 P388 cells, and the same number of cells was transplanted sequentially. Mice in the vincristine treatment group received a 1-mg/kg intraperitoneal dose of vincristine twice, and control mice received saline. We designated the P388 derivatives collected from the abdominal cavity of each successive group of mice as P-C0, P-C1, P-C2, P-C3, and P-C4 (untreated cells) or P-V1, P-V2, P-V3, and P-V4(vincristine-treated cells). These derivatives were used in the following experiments. (B) The life span of mice with (closed circles) or without (open circles) vincristine treatment. Arabic numbers (n = 0, 1, 2, 3, and 4) indicate the number of times that the P388 derivatives were inoculated. Mean survival days are also indicated in the lower box.
Overexpression of the mdr1a gene in vincristine-treated leukemia cells
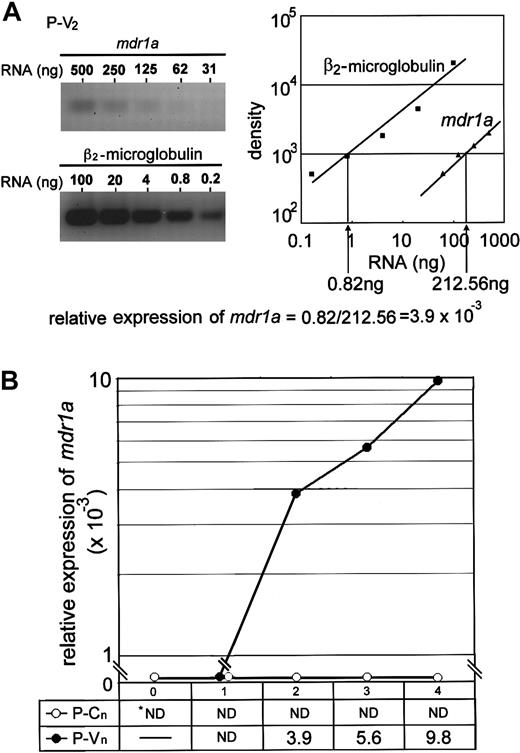
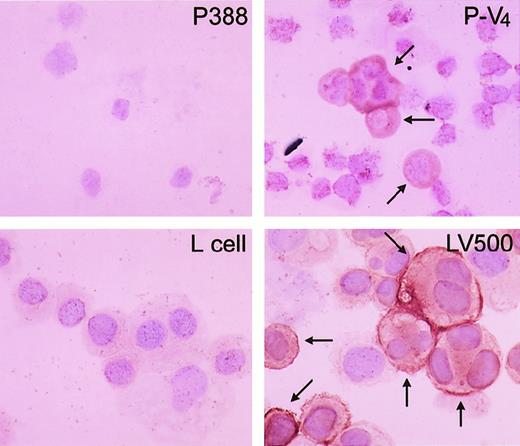
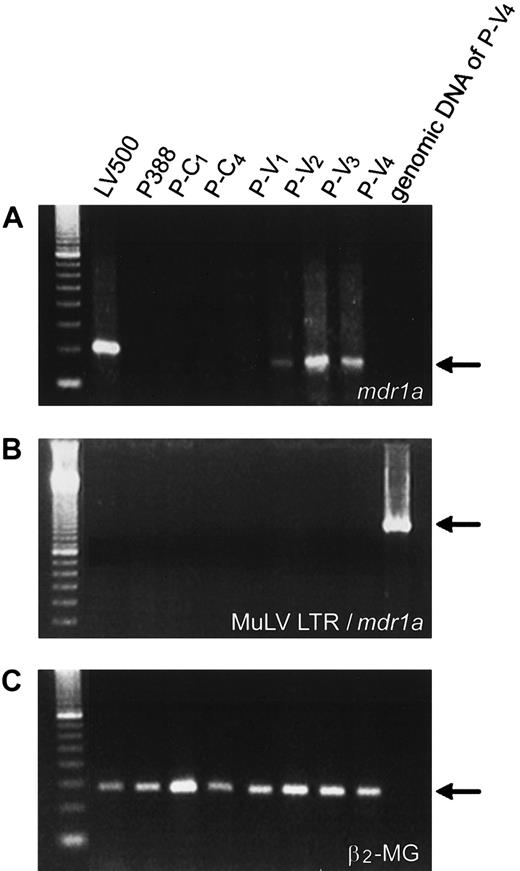
Because the mdr1a, mdr1b, andmrp41 genes are often highly expressed in cancer cells exposed to Vinca alkaloids, we next examined the expression of these genes by semiquantitative RT-PCR method (Figure2A). We could not find any significant increase in the level of mdr1b or mrp messenger RNA (mRNA) of the leukemia cells during repeated treatments with vincristine (data not shown). Only the level of mdr1a mRNA was elevated in vincristine-resistant cells (P-V2, P-V3, and P-V4) in comparison to controls (Figure 2B). Parental P388 cells did not show any expression ofmdr1a. P-V2 cells collected after the second transplantation with vincristine treatment showed a significant increase in mdr1a expression compared with parental cells. After later transplantations, P-V4 cells showed further increases in mdr1a expression, and a clear correlation between the increase of mdr1a expression and the decrease of the life span of mice was observed. Overexpression of themdr1a gene was also detected by immunohistochemistry during transplantations in P-V2, P-V3 (data not shown), and P-V4 cells (Figure3).
Expression of the
mdr1a gene in vincristine-treated leukemia cells. (A) Semiquantitative RT-PCR assay. The left panel shows a representative result of CYBR green staining after electrophoresis of PCR product amplified from P-V2 RT product withmdr1a-specific primers (left panel, upper) pr-1aF and pr-1aR, or β2-microglobulin–specific primers (left panel, lower) pr-mgF and pr-mgR, in a 3% agarose gel. The gel was analyzed with FAS200 and plotted as a graph (right panel: closed squares, β2-microglobulin, and closed triangles,mdr1a). The relative mdr1a expression was obtained from the dilution rate at the exponential range in the right panel and normalized by dividing by the dilution rate of β2-microglobulin. The relative expression ofmdr1a is calculated, for example, as a formula at the bottom. (B) Relative expression of mdr1a in mice. The value of relative expression is presented in the box and plotted as a graph. Closed and open circles indicate the P-V and P-C series, respectively. ND indicates not detected.
Expression of the
mdr1a gene in vincristine-treated leukemia cells. (A) Semiquantitative RT-PCR assay. The left panel shows a representative result of CYBR green staining after electrophoresis of PCR product amplified from P-V2 RT product withmdr1a-specific primers (left panel, upper) pr-1aF and pr-1aR, or β2-microglobulin–specific primers (left panel, lower) pr-mgF and pr-mgR, in a 3% agarose gel. The gel was analyzed with FAS200 and plotted as a graph (right panel: closed squares, β2-microglobulin, and closed triangles,mdr1a). The relative mdr1a expression was obtained from the dilution rate at the exponential range in the right panel and normalized by dividing by the dilution rate of β2-microglobulin. The relative expression ofmdr1a is calculated, for example, as a formula at the bottom. (B) Relative expression of mdr1a in mice. The value of relative expression is presented in the box and plotted as a graph. Closed and open circles indicate the P-V and P-C series, respectively. ND indicates not detected.
Immunohistochemical staining of P-gp in parental P388 and the vincristine-treated derivative, P-V4.
Representative results of immunohistochemical staining of P388 (upper left) and P-V4 (upper right). Lower panels show positive (LV500, right) and negative controls (L cell, left). Arrows indicate stained P-gp on the cell surface.
Immunohistochemical staining of P-gp in parental P388 and the vincristine-treated derivative, P-V4.
Representative results of immunohistochemical staining of P388 (upper left) and P-V4 (upper right). Lower panels show positive (LV500, right) and negative controls (L cell, left). Arrows indicate stained P-gp on the cell surface.
Gene rearrangement at the mdr1a promoter region in leukemia cells that are refractory to vincristine
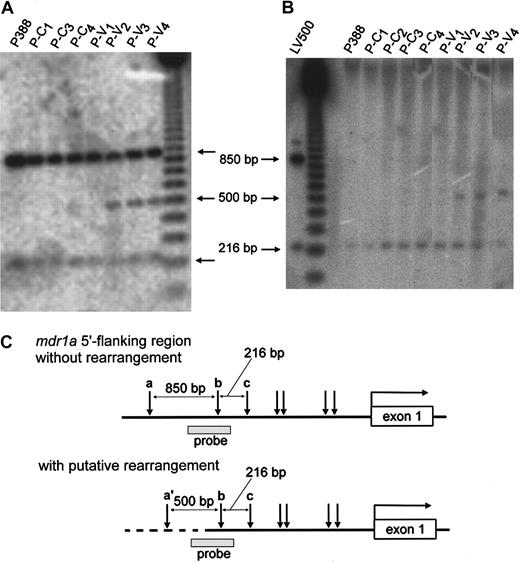
To identify the molecular mechanisms involved in up-regulation ofmdr1a expression, we first examined the gene amplification and methylation status of the promoter region of mdr1a by Southern blot analysis. As shown in Figure4A, there was no change in the intensity of 2 850- and 216-bp (base pair)–long bands among all samples, which suggests that the mdr1a gene was not amplified during vincristine treatment. We next examined the methylation status by digestion with a methylation-sensitive restriction endonuclease,HpaII. We detected 216-bp fragments and bands with higher molecular weight in all samples, but we did not detect change in intensity of those bands (Figure 4B), suggesting hypermethylation at the HpaII site a and hypomethylation at both site b and c (Figure 4C). This suggested that there was no alteration of methylation status during vincristine treatment. On the other hand, we detected extra fragments of 500 bp in the lanes derived from P-V2, P-V3, and P-V4 when digested with either MspI or HpaII (Figure 4A,B). We hypothesized that the extra fragments representing the gene rearrangement occurred upstream of the mdr1a promoter region in these cells. It should be noted that these cells also showed a relatively high mdr1a expression (Figure 2), suggesting that the rearrangement might be responsible for overexpression of themdr1a gene.
Southern blot analysis in the promoter region of
mdr1a. Genomic DNA of each P388 derivative was digested with MspI (A) or HpaII (B) and analyzed by Southern blotting using the PstI-digested 370-bp fragment as a probe (░, C). The physical maps of the mdr1a5′-flanking region without rearrangement (upper) and with putative rearrangement (lower) are presented in panel C. The open box and horizontal arrow correspond to mdr1a exon-1 and its transcriptional direction, respectively. Vertical arrows indicateMspI/HpaII sites. In the lower map the broken line represents the putative rearranged DNA, and the expectedMspI/HpaII site is indicated as site a′. The restriction patterns of HpaII suggested that site a was hypermethylated, and both sites b and c were hypomethylated.
Southern blot analysis in the promoter region of
mdr1a. Genomic DNA of each P388 derivative was digested with MspI (A) or HpaII (B) and analyzed by Southern blotting using the PstI-digested 370-bp fragment as a probe (░, C). The physical maps of the mdr1a5′-flanking region without rearrangement (upper) and with putative rearrangement (lower) are presented in panel C. The open box and horizontal arrow correspond to mdr1a exon-1 and its transcriptional direction, respectively. Vertical arrows indicateMspI/HpaII sites. In the lower map the broken line represents the putative rearranged DNA, and the expectedMspI/HpaII site is indicated as site a′. The restriction patterns of HpaII suggested that site a was hypermethylated, and both sites b and c were hypomethylated.
Characterization of the rearranged sequence
We next determined the junction sequence and molecular nature of the mdr1a gene rearrangement. We used a vectorette construct for selective amplification of the 500-bp MspI fragment. Subcloning and nucleotide sequence analysis revealed that the extra fragment actually consisted of 484 bp, including 355 bp of themdr1a sequence and a 129-bp sequence identical to the minus strand of the 5′ LTR of MuLV (Figure 5). The nucleotide sequence corresponding to theMspI/HpaII recognition site at the LTR is consistent with the generation of the 500-bp fragment byMspI/HpaII digestion from genomic DNA isolated from P-V2 to V4. These data suggested that the rearrangement resulted from the insertion of the MuLV genome into the promoter region of the mdr1a gene. Southern blot analysis and sequencing of the junction between the region further upstream of the mdr1a and the 3′ LTR confirmed this possibility (data not shown). It is noted that we detected exactly the same junction sequence when we repeated the in vivo chemotreatment protocol.
Junction sequence of MuLV–
mdr1a rearrangement. The sequence of the top row (plain capital letters) represents the complementary sequence of MuLV LTR. The middle row (bold capital letters) and bottom row (small letters) represent the rearranged sequence in this study and themdr1a promoter region, respectively. Vertical lines indicate the alignments between sequences.
Junction sequence of MuLV–
mdr1a rearrangement. The sequence of the top row (plain capital letters) represents the complementary sequence of MuLV LTR. The middle row (bold capital letters) and bottom row (small letters) represent the rearranged sequence in this study and themdr1a promoter region, respectively. Vertical lines indicate the alignments between sequences.
Characterization of the mdr1a mRNA product overexpressed in leukemia cells refractory to vincristine
We analyzed the mdr1a mRNA by RT-PCR methods using 2 pairs of primers—pr-ltr/1aF and pr-ltr/1aR, and pr-1a3′F and pr-1a3′R—that amplify the sequence derived from possible LTR/mdr1a hybrid mRNA and the normal transcript ofmdr1a, respectively. The mdr1a transcriptional products could be detected in LV500, P-V2, P-V3, and P-V4 cells but not in parental P388 cells (Figure 6A). However, only normal transcripts existed in these cells. Hybrid mRNA, which was expected to be generated if the LTR would act as an upstream-located promoter ofmdr1a transcription, could not be detected in any of the above cells (Figure 6B).
Characterization of
mdr1a transcripts by RT-PCR. The mdr1a(A), MuLV-mdr1a hybrid (B), and β2-microglobulin mRNA (C) were amplified by RT-PCR using primer pairs pr-1a3′F and pr-1a3′R, pr-lyr/1aF and pr-ltr/1aR, and pr-mgF and pr-mgR, respectively.
Characterization of
mdr1a transcripts by RT-PCR. The mdr1a(A), MuLV-mdr1a hybrid (B), and β2-microglobulin mRNA (C) were amplified by RT-PCR using primer pairs pr-1a3′F and pr-1a3′R, pr-lyr/1aF and pr-ltr/1aR, and pr-mgF and pr-mgR, respectively.
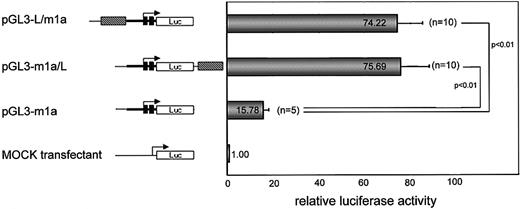
The MuLV LTR sequence shows positive enhancer activity tomdr1a promoter
We isolated the rearranged promoter region and examined whether the MuLV LTR sequence could activate the mdr1a gene by luciferase-reporter assay (Figure 7). We found that the LTR sequence increased the luciferase activity when it was positioned either upstream (pGL3-L/m1a) or downstream (pGL3-m1a/L) of mdr1a promoter-luciferase. Transfection of either pGL3-L/m1a or pGL3-m1a/L to NIH3T3 cells resulted in a 70- to 80-fold increase in luciferase activity relative to that resulting from transfection of the pGL3-basic vector. Next, we demonstrated that deletion of the LTR sequence drastically decreased the level of luciferase activity. The LTR-deleted construct pGL-m1a showed less luciferase activity, about one-fifth of the activity of pGL3-L/m1a and pGL3-m1a/L. These results strongly supported the idea that the opposite-directed MuLV LTR up-regulates the transcription of themdr1a gene as a positive enhancer.
The effect of MuLV LTR on
mdr1a promoter (luciferase-reporter assay). The blank, filled, and shadowed boxes correspond to dragonfly luciferase cDNA, exon-1 and exon-2 of mdr1a, and the MuLV LTR sequence, respectively. The filled arrow shows the transcriptional start and direction of the mdr1a gene. The average value of relative luciferase activity is indicated in the shaded column, and the number of assays performed is indicated in parenthesis.
The effect of MuLV LTR on
mdr1a promoter (luciferase-reporter assay). The blank, filled, and shadowed boxes correspond to dragonfly luciferase cDNA, exon-1 and exon-2 of mdr1a, and the MuLV LTR sequence, respectively. The filled arrow shows the transcriptional start and direction of the mdr1a gene. The average value of relative luciferase activity is indicated in the shaded column, and the number of assays performed is indicated in parenthesis.
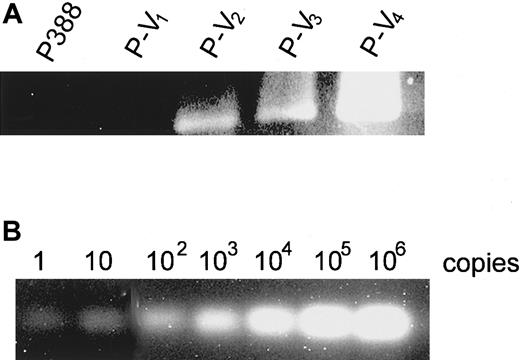
Detection of an MuLV LTR/mdrla rearranged genomic sequence in leukemia cells
It remains unclear whether the cell populations that primarily contain the rearranged sequence were selected by repeated vincristine treatment or the gene rearrangement was induced by MuLV insertion during the treatment. We thus examined whether the rearranged sequence existed primarily in the initial inoculums by PCR assay using genomic DNA of P388 and its derivatives as templates and LTR- andmdr1a-specific primers (Figure8A). We used 10 μg genomic DNA as a template, which was equivalent to 1.0 × 106 cells and to the initial inoculum. We could detect the amplified products in vincristine-treated derivatives P-V2, P-V3, and P-V4, but not in parental P388 or P-V1 cells (Figure 8A). In the control experiment, 10 μg genomic DNA of mouse fibroblast L cells containing the rearranged genomic DNA, which had been cloned into plasmid and serially diluted, was used as a template. The PCR product was detected even when one copy of the rearranged sequence was contained in the template (Figure 8B), suggesting that the PCR assay was sensitive enough to detect at least one copy of the rearranged sequence among genomic DNA equivalent to 1 × 106 cells.
Detection of rearranged sequence among P388 derivatives.
PCR was performed using 10 μg genomic DNA of (A) P388 derivatives or (B) L cells containing serially diluted rearranged sequence from 106 copies to one copy. Primer pairs corresponding to the MuLV-mdr1a rearranged sequence (pr-ltr/m1aF and pr-ltr/m1aR) were used.
Detection of rearranged sequence among P388 derivatives.
PCR was performed using 10 μg genomic DNA of (A) P388 derivatives or (B) L cells containing serially diluted rearranged sequence from 106 copies to one copy. Primer pairs corresponding to the MuLV-mdr1a rearranged sequence (pr-ltr/m1aF and pr-ltr/m1aR) were used.
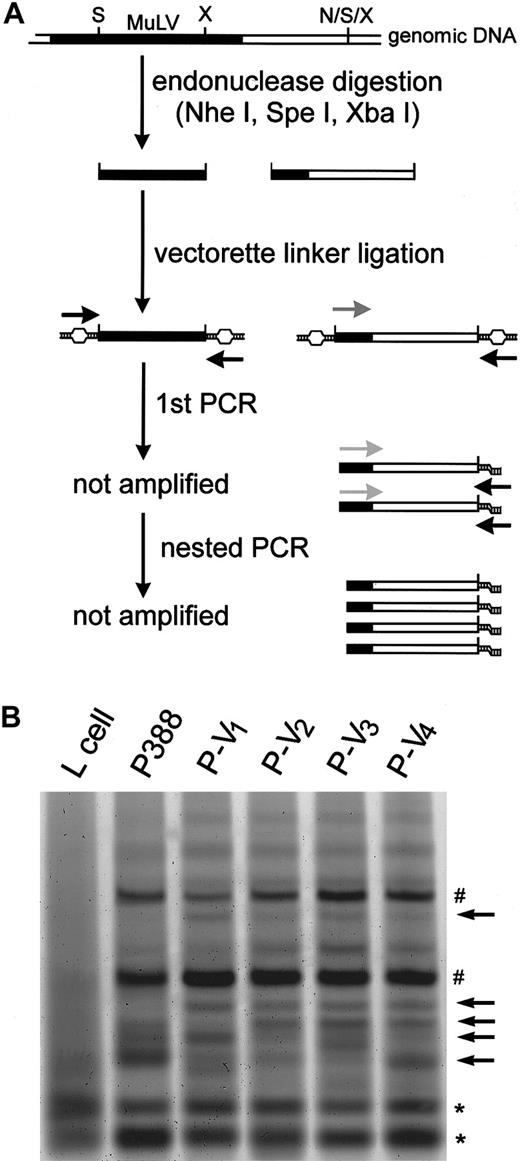
Alteration of the distribution of proviral MuLV during vincristine treatment
We hypothesized that the proviral MuLV was excised and integrated into the promoter region of the mdr1a gene during chemotreatment, probably by vincristine. We then examined whether the vincristine treatment could alter the distribution of the MuLV proviral genome. The experiment using a short-term culture of P388 in vitro failed, probably because the P388 cells, which had been maintained in vivo, have less viability in vitro. We then analyzed the MuLV distribution in the P388 derivatives by vectorette PCR (Figure9A). Although the number of bands detected was similar among the parental P388, P-V1, P-V2, P-V3, and P-V4 cells (Figure 9B, represented by sharp signs and asterisks), novel specific bands (arrows) were detected in P-V1, P-V2, P-V3, and P-V4 cells but not in parental P388 cells. This suggests that vincristine treatment might induce the excision and integration of proviral MuLV. By contrast, only a few bands were generated in mouse fibroblast L cells (Figure 9B, asterisks), suggesting that L cells might have fewer copies of MuLV than P388 leukemia cells. Similar but distinct patterns of the altered MuLV distribution were observed in the second independent experiment (data not shown). This result further suggested that the random integration of MuLV might be caused during these 2 independent experiments of vincristine chemotreatment.
Detection of the altered distribution of proviral MuLV during vincristine treatment.
(A) Schematic representation of the vectorette PCR strategy. The filled and open bars represent the MuLV proviral genome and mouse genomic DNA, respectively. The black, gray, and light gray arrows correspond to uvp, musp1, and musp2, respectively. The capital letters N, S and X correspond to restriction sites for NheI, SpeI, and XbaI, respectively. Further details are described in “Materials and methods.” (B) Vectorette PCR products separated by 10% acrylamide gel. The arrows indicate P-V–derivative specific bands; the sharp signs, P388- and P-V–derivative specific bands; and the asterisks, P388- and P-V–derivative non-specific bands.
Detection of the altered distribution of proviral MuLV during vincristine treatment.
(A) Schematic representation of the vectorette PCR strategy. The filled and open bars represent the MuLV proviral genome and mouse genomic DNA, respectively. The black, gray, and light gray arrows correspond to uvp, musp1, and musp2, respectively. The capital letters N, S and X correspond to restriction sites for NheI, SpeI, and XbaI, respectively. Further details are described in “Materials and methods.” (B) Vectorette PCR products separated by 10% acrylamide gel. The arrows indicate P-V–derivative specific bands; the sharp signs, P388- and P-V–derivative specific bands; and the asterisks, P388- and P-V–derivative non-specific bands.
Discussion
We examined the molecular basis for mdr1aoverexpression in leukemia cells during continuous treatment with vincristine. The therapeutic model we used has 2 advantages over analysis of human clinical samples: (1) the inoculated tumor is basically monoclonal, and (2) samples are available at every step during the clinical course. The therapeutic effect by vincristine on mice carrying the leukemia cells was abolished during repeated treatment, accompanying overexpression of mdr1amRNA. Previous studies have reported that demethylation of the CpG sites at the promoter region was inversely correlated with humanMDR1 overexpression both in vitro and in vivo.30,31 42 In the present study, however, no alteration in the methylation status of CpG sites occurred in any of the P388 derivatives, irrespective of whether or not mdr1a expression was observed. CpG methylation thus may not be responsible for the regulation of mdr1a expression in our experimental system.
Integration of MuLV was observed at the mdr1a promoter region in leukemia cells when expression of mdr1a was enhanced. This raises the question of whether or not the MuLV integration is associated with mdr1a overexpression. Overexpression of mdr1a mRNA could be induced by transcription that started from the LTR promoter through themdr1a gene. However, the transcriptional direction of the LTR promoter is opposite that of mdr1a, and no chimerical mRNA of mdr1a fused to the MuLV LTR was found. Payne et al43 have previously reported that retroviral 5′ LTR, which is inserted inverse to the transcriptional direction of thec-myc gene located downstream, enhances the transcription of the gene. Using a luciferase assay, we here demonstrated that insertion of the LTR of MuLV into themdr1a promoter region enhances the gene transcription irrespective of the position of the LTR. MuLV insertion could act as an enhancer element to activate the mdr1a gene.
Although MuLV insertion at the mdr1a gene in vivo has not been reported so far, a similar observation in vitro has been reported. Lepage et al44 reported the insertion of a murine mammary tumor virus and intracisternal A-particle at the mdr1apromoter region in vincristine- and adriamycin-resistant P388 leukemia cells, respectively. Both of the integration sites and viruses reported by the authors44 are different from those identified in our study. Infection of mouse bone marrow cells with retroviral vectors, followed by treatment with taxol, a P-gp–targeting agent, also efficiently enhanced integration of the retrovirus intomdr1a and mdr1b gene promoters (Y. Sugimoto and T. Tsuruo, personal communication, October 1999). In our present study, however, we could not observe any apparent increase inmdr1b expression. Treatment with P-gp–targeting drugs appears to rather specifically induce up-regulation of MDR-related genes through the insertion of retroviruses. These previous reports and our present in vivo study suggest that retroviral transposons may prefer to integrate into the mdr1a promoter region. Although there is no obvious sequence similarity among the integration sites, some type of preferential DNA or chromatin structure for retroviral integration in these promoter regions may exist.
The association between the virus integration and mdr1aoverexpression during chemotreatment may be attributable to either of 2 mechanisms: (1) de novo insertion of MuLV induced by vincristine treatment or (2) selection of a small fraction of pre-existing cells carrying MuLV insertion during vincristine treatment. We could not detect the rearranged genomic DNA in the parental cells by PCR assay, and we found the alteration of proviral MuLV distribution during chemotreatment. These data might suggest the possibility of de novo insertion. Alternatively, the same integration sites of the virus at the mdr1a promoter were identified in 2 independent experiments with the same therapeutic protocol. This finding would seem to suggest the second mechanism. The alteration of MuLV distribution might reflect the selection of leukemia cells, which intrinsically carried the altered pattern of MuLV distribution. Thus, based on these results, we were unable to determine which of the above mechanisms was more likely. Whatever the precise mechanism is, it should be noted that in vivo chemotreatment using vincristine could induce generation ofmdr1a-overexpressing cells through retrovirus insertion and the enhancer effect of LTR.
Concerning the plausible mechanism for MuLV insertion into themdr1a gene, DNA damaging agents often activate transposable elements in genomes and induce gene rearrangement. For example, Ty retrotransposons can be activated by ultraviolet light in yeast.45 Tanaka and Ishihara46 also reported radiation-induced intracisternal A-particle integration in mouse myeloid leukemia cell strains. Vincristine treatment is reported to generate oxiradicals,47 and enhancement of the viral integration by oxidative stress has also been reported.48The retroviral integration might be enhanced by vincristine-induced oxidative stress, but the details have not yet been clarified. Gene rearrangements, associated with human MDR1 gene overexpression, have been observed in cultured multidrug-resistant cell lines and in clinical samples.32,49 Such drug-resistant human cancer cell lines express hybrid mRNAs containing sequences from both MDR1 and unrelated genes located upstream. We have recently reported Alu repeats near the junction sequence in these MDR cancer cell lines.33 The genomic instability intrinsic in malignant cancer cells also appears to modulate MDR gene expression. Further studies are now in progress to determine whether the human MDR1 gene can be activated by retroviral integration in clinical specimens of human hematopoietic malignancies that are refractory to chemotherapeutic agents.
Acknowledgments
We thank Dr Kathleen W. Scotto (Memorial Sloan-Kettering Cancer Center, New York, NY), Dr Igor B. Roninson (University of Illinois, Urbana), and Dr Kimitoshi Kohno (University of Occupational and Environmental Health, Kitakyushu, Japan) for fruitful discussion. We also thank Dr Yoshikazu Sugimoto (Japanese Foundation for Cancer Research) and Dr Takashi Tsuruo (The University of Tokyo, Japan) for providing important information.
Supported by a grant from the Ministry of Education, Science, Sports, and Culture of Japan (11694286); a grant from the Second-Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health and Welfare (H12gan011); and a grant from Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Corporation (JST), Tokyo.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Morimasa Wada, Department of Medical Biochemistry, Graduate School of Medical Sciences, Kyushu University, 3-1-1 Maidashi Higashi-ku, Fukuoka, 812-8582, Japan; e-mail:wada@biochem1.med.kyushu-u.ac.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal