Abstract
The transcription factor encoded by the mi locus (MITF) is a transcription factor of the basic-helix-loop-helix zipper protein family. Mice of mi/mi genotype express a normal amount of abnormal MITF, whereas mice oftg/tg genotype do not express any MITFs due to the transgene insertional mutation. The effect of normal (+) and mutant (mi) MITFs on the expression of mouse mast cell protease (MMCP) 6 and 7 was examined. Both MMCP-6 and MMCP-7 are tryptases, and their coding regions with high homology are closely located on chromosome 17. Both MMCP-6 and MMCP-7 genes are expressed in normal cultured mast cells (+/+ CMCs). Although the transcription of MMCP-6 gene was severely suppressed in bothmi/mi and tg/tg CMCs, that of MMCP-7 gene was severely suppressed only in mi/mi CMCs. The study identified the most significant segment for the transcription in the 5′ flanking region of MMCP-7 gene. Unexpectedly, no CANNTG motifs were found that are recognized and bound by +-MITF in this segment. Instead, there was an AP-1 binding motif, and binding of c-Jun to the AP-1 motif significantly enhanced the transcription of MMCP-7 gene. The complex formation of c-Jun with either +-MITF ormi-MITF was demonstrated. The binding of +-MITF to c-Jun enhanced the transactivation of MMCP-7 gene, and that ofmi-MITF suppressed the transactivation. Although the former complex was located only in the nucleus, the latter complex was predominantly found in the cytoplasm. The negative effect ofmi-MITF on the transcription of MMCP-7 gene appeared to be executed through the interaction with c-Jun.
Introduction
We have been studying the involvement of the transcription factor encoded by the mi locus (MITF) in the transcription of genes expressed by mast cells. MITF is a member of the basic-helix-loop-helix–leucine zipper (bHLH-Zip) protein of transcription factors.1,2 The mutant mice ofmi/mi genotype were found by Hertwig among the offspring of an X-irradiated male mouse.3,4 The mutant gene was introduced into C57BL/6 background and has been kept in the Jackson Laboratory (Bar Harbor, ME). The MITF encoded by the mutantmi allele (hereafter mi-MITF) deletes 1 of 4 consecutive arginines in the basic domain.1,5,6 Themi-MITF is defective in the DNA-binding activity7 and the nuclear localization potential.8
A VGA-9-tg/tg transgenic mouse possessing the transgene-insertional mutation at the 5′ flanking region of themi gene was produced.1,9 The expression of MITF was undetectable in various cells of VGA-9-tg/tg mice, including mast cells.1 We introduced the tgtransgene into C57BL/6 background and compared messenger RNA (mRNA) expression between cultured mast cells (CMCs) derived from C57BL/6-mi/mi mice and those of C57BL/6-tg/tgmice.10 The transcription of 3 genes encoding mouse mast cell proteases (MMCPs), MMCP-4, MMCP-5, and MMCP-6, was impaired in CMCs of both mice. However, the transcription of the gene encoding the c-kit receptor, tryptophan hydroxylase, or granzyme B was remarkably reduced in CMCs of C57BL/6-mi/mi mice, but the reduction was significantly smaller in CMCs of C57BL/6-tg/tg mice, suggesting the inhibitory effect ofmi-MITF on the transactivation of particular genes in CMCs.10
MMCPs are serine proteases and are useful for studying the mechanism of gene expression in mast cells because of their abundant expression. Nine MMCPs have been known.11-18MMCP-1,11 MMCP-2,12 MMCP-4,11,13MMCP-5,11,14 and MMCP-918 are chymases, and the genes encoding each of them reside on chromosome 14. Although the expression of both MMCP-4 and MMCP-5 genes were impaired inmi/mi CMCs, the mechanism of the impaired expression was different.19 20
MMCP-615 and MMCP-716 are tryptases, and the genes encoding each of them reside on the chromosome 17. The coding region of the MMCP-6 gene is highly homologous with that of MMCP-7.16 Both MMCP-6 and MMCP-7 mRNAs are present in mast cells of the skin but not in mast cells of the intestinal mucosa.16 However, MMCP-6 and MMCP-7 showed some different biological characteristics. Mast cells in the peritoneal cavity express MMCP-6 mRNA but not MMCP-7 mRNA.16 A considerable amount of MMCP-6 mRNA is expressed in CMCs throughout the culture period, but the amount of MMCP-7 mRNA decreases markedly 3 weeks after starting the culture of CMCs.16
We attempted to examine the effect of MITF on the expression of the MMCP-7 gene. However, the MMCP-7 gene is not expressed in mast cells of the C57BL/6 strain due to a point mutation at the first nucleotide of exon 2/intron 2 splice site.21 22 Because the MMCP-7 gene is normally expressed in the WB strain, we crossed WB-+/+ mice to C57BL/6-mi/+ or C57BL/6-tg/+ mice and finally obtained the mice possessing the WB strain-derived MMCP-7 gene and double gene dose of either mi or tg mutant allele. The expression of the WB strain-derived MMCP-7 gene remarkably decreased in CMCs of the mi/mi mice but decreased only slightly in CMCs of the tg/tg mice. The regulation of MMCP-7 gene expression appeared to be different from that of MMCP-6 gene. We examined the mechanism and found that either +-MITF ormi-MITF interacted with c-Jun and affected the expression of the MMCP-7 gene through the c-Jun.
Materials and methods
Mice
The original stock of C57BL/6J-mi/+ mice was purchased from the Jackson Laboratory (Bar Harbor, ME) and was maintained in our laboratory by consecutive backcrosses with C57BL/6 mice of our own inbred colony. The original stock of VGA-9-tg/tg mice,1 in which the mouse vasopressin–Escherichia coli–galactosidase transgene was integrated at the 5′ flanking region of the mi (MITF) gene, was kindly given by Dr H. Arnheiter (National Institutes of Health, Bethesda, MD). The integrated transgene was maintained by repeated backcrosses to C57BL/6 mice of our own inbred C57BL/6 colony.
The mutant allele of MMCP-7 possessed by the C57BL/6 strain was described as MMCP-7B6, and the normal allele of the WB strain as MMCP-7WB. Mice of (MMCP-7WB/MMCP-7WB,mi/mi), (MMCP-7WB/MMCP-7WB,tg/tg), and control (MMCP-7WB/MMCP-7WB, +/+) genotype were necessary in the present experiment. First, we crossed WB-(MMCP-7WB/MMCP-7WB, +/+) mice with C57BL/6-(MMCP-7B6/MMCP-7B6,mi/+) mice or with C57BL/6-(MMCP-7B6/MMCP-7B6,tg/+) mice. Then we selected (MMCP-7WB/MMCP-7B6,mi/+) mice by the presence of a white belly spot3,4 and (MMCP-7WB/MMCP-7B6,tg/+) mice by the presence of a tg transgene. Then we backcrossed these F1 hybrid mice to WB-(MMCP-7WB/MMCP-7WB, +/+) mice and selected (MMCP-7WB/MMCP-7WB,mi/+) and (MMCP-7WB/MMCP-7WB,tg/+) mice by examining the MMCP-7 genotype.22We crossed together mice of each genotype and finally selected mice of (MMCP-7WB/MMCP-7WB,mi/mi) and of (MMCP-7WB/MMCP-7WB,tg/tg) with white coat and mice of (MMCP-7WB/MMCP-7WB, +/+) that had completely black coat and no tg transgene. Because all mice were homozygous at the MMCP-7WB allele, we describe them hereafter just as mi/mi, tg/tg, and +/+ mice.
Examination of genomic DNA
Tail tip was cut under ether anesthesia, and DNA was extracted as described by Blin and Stafford.23 The presence or absence of tg transgene was examined by polymerase chain reaction (PCR) with sense (5′-CATTAAATGTGAGCGAGTAACAACCCGTCG-3′) and antisense (5′-CTTTCGCCAGCTGGCGTAATAGCGAAGAGG-3′) primers.22
The region, including the exon 2/intron 2 splice site, of the MMCP-7 gene was amplified by PCR with the use of sense (5′-GCCCTGGCAGGTGAGCCTGC-3′) and antisense (5′-TTGTTGGGGTCAGCAACATC-3′) primers.22 The correctly sized PCR products were purified by QIAquick PCR Purification Kit (Qiagen, Santa Clarita, CA). The purified PCR products were directly sequenced with standard dideoxy sequencing techniques by Model 373A DNA sequencer (Applied Biosystems, Foster City, CA).
Promoter region of MMCP-7 gene
The isolation of the promoter region of MMCP-7 gene was performed with the Mouse Genome Walker Kit (Clonetech Laboratories, Palo Alto, CA) according to the manufacturer's instructions. The isolated promoter region was cloned into Bluescript KS (−) plasmids (pBS; Stratagene, La Jolla, CA).
Cells
Pokeweed mitogen-stimulated spleen cell condition medium (PWM-SCM) was prepared according to the method described by Nakahata et al.24 Mice of mi/mi,tg/tg, and +/+ genotype were used at an age of 2 to 3 weeks to obtain CMCs. Mice were killed by decapitation after ether anesthesia, and the spleens were removed. Spleen cells of each genotype were cultured in α-minimal essential medium (α-MEM; ICN Biomedicals, Costa Mesa, CA) supplemented with 10% PWM-SCM and 10% fetal calf serum (FCS; Nippon Biosupp Center, Tokyo, Japan). Half of the medium was replaced every 7 days. Three weeks after initiation of the culture, more than 95% of cells were CMCs.25 These CMCs were harvested and used for experiments.
FMA/3, mouse mastocytoma cell line, was maintained in α-MEM supplemented with 10% FCS and antibiotics. MST,26 the stable heparin-producing cell line derived from the Furth murine masytoma, was also maintained in α-MEM supplemented with 10% FCS and antibiotics. Jurkat, human T-cell line, was maintained in RPMI 1640 supplemented with 10% FCS and antibiotics. Monkey COS-7 cells were maintained in Dulbecco modification of Eagle medium (DMEM) supplemented with 10% FCS and antibiotics.
Construction of expression and reporter plasmids
The luciferase gene subcloned into pSP72 (pSPLuc) was kindly given by Dr K. Nakajima (Osaka City University Medical School, Osaka, Japan).27 To construct reporter plasmids, a DNA fragment containing a promoter region and the first exon (noncoding region) of the MMCP-7 gene (−1892 to +20) was cloned into the upstream of luciferase gene in pSPLuc. The deletion of the MMCP-7 promoter was done by using the appropriate restriction enzyme or PCR. The mutation in the AP-1 binding consensus motif was introduced by PCR with mismatch sense (5′-TGGAGTCTGAGTTGCCCATT-3′) and antisense (5′-AATGGGCAACTCAGACTCCA-3′) primers. Deleted or mutated products were verified by DNA sequencing.
pEF-BOS expression vector was kindly given by Dr S. Nagata (Osaka University Medical School).28 pEF-BOS containing the whole coding region of +-MITF or mi-MITF had been constructed in our laboratory.29,30 The Myc-tagged +- ormi-MITF constructs has been described.8 pBS containing the whole coding region of c-Jun complementary DNA (cDNA) (pBS-c-Jun) was constructed in our laboratory. c-Jun cDNA was subcloned into pEF-BOS and pcDNA3-FLAG. pEF-BOS containing the dominant negative c-Jun, TAM-67,31 was kindly given by Dr K. Shimozaki and Dr S. Nagata (Osaka University Medical School).
Northern blot analysis
Total RNAs (20 μg) were isolated with the lithium chloride–urea method.32 Northern blot analyses were performed using the fragments of MMCP-7, MMCP-6, MC-CPA, and GAPDH cDNAs as probes after being labeled with [32P]-α-dCTP (DuPont/NEN Research Products, Boston, MA; 10mCi/mL) by random oligonucleotide priming. The probe of MMCP-7 cDNA was a 247–base pair (bp) MMCP-7 gene specific fragment that corresponds to the nucleotides 2071 to 2317 of the gene. The probes of MMCP-6, MC-CPA, and GAPDH have been described.15 33 After hybridization at 45°C, blots were washed to a final stringency of 0.2 × SSC (1 × SSC is 150 mM NaCl, 15 mM trisodium citrate, pH 7.4) at 55°C and subjected to autoradiography.
Luciferase assay
FMA/3 or Jurkat cells (5 × 106 cells in 300 μL of the culture medium in a 0.4-cm cuvette) were transfected with a luciferase-reporter plasmid (10 μg), effector plasmids (each 1 μg), and an expression vector containing β-galactosidase gene (2 μg) by electroporation (Gene Pulser; Bio-Rad Laboratories, Hercules, CA) at a setting of 975 μF/300V and at room temperature. The expression vector containing β-galactosidase gene was used to normalize the varying transfection efficiencies. The compositions of transfected expression plasmids are described in figure legends. The total amount of transfected DNA was always kept at 20 μg, using a backbone plasmid, pEF-BOS. After 24 to 36 hours, cells were harvested, and luciferase activity was assayed with a luminometer LB96P (Berthold, GmbH, Wildbad, Germany). By determining the β-galactosidase activity and the total protein concentration, the luciferase activity was normalized as described previously.10
Overexpression of dominant negative c-Jun, TAM-67, in MST cells
MST cells (2 × 107 cells) were transfected with 10 μg pEF-BOS containing dominant negative c-Jun, TAM-67, or empty pEF-BOS with the electroporation, as mentioned above. After 4 days of culture, total RNAs (20 μg) were extracted and subjected to Northern blot analysis.
Electrophoretic mobility shift assay
The production and purification of glutathione-S-transferase (GST) fusion protein has been described.7 The wild-type oligonucleotide of the MMCP-7 promoter sequence (5′-AGCTGTCCCTGGAGTCTGAGTCACCCATTTAGCT-3′) was labeled with [32P]-α-dCTP by filling 5′-overhangs and was used as a probe of Electrophoretic mobility shift assay (EMSA). The DNA binding reaction and electrophoresis were done as described previously.7 For the competition assay, unlabeled wild-type oligonucleotide or mutated oligonucleotide (5′-AGCTGTCCCTGGAGTCTGAGTTGCCCATTTAGCT-3′) was added.
In vitro binding assays
The 35S-labeled c-Jun protein was synthesized, using the reticulocyte lysate system (TNT system, Promega, Madison, WI). c-Jun cDNA in pBluescript was transcribed with T7 RNA polymerase and translated in the presence of 35S-methionine. For the binding assays, the 35S-labeled c-Jun protein was incubated for 1 hour at room temperature, with GST-+-MITF, GST-mi-MITF, or GST alone immobilized on glutathione-agarose beads. The beads were washed 4 times. Proteins retained on the beads were subsequently analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. As controls, purified GST-+-MITF, GST-mi-MITF, or GST was subjected to SDS-PAGE, and the gel was stained with Coomassie brilliant blue.
Preparation of nuclear and cytoplasmic extracts
Nuclear and cytoplasmic extracts were prepared as described before.8 COS-7 cells were transfected with 5 μg each of the expression plasmids by calcium phosphate method. Forty-eight hours after transfection, the cells were collected and washed with PBS. Then the cells were resuspended in 100 μL buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 15 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). The cells were allowed to swell on ice for 3 minutes, and Nonidet P-40 was added to a final concentration of 0.1%. After vigorous vortexing for 10 seconds, the homogenate was centrifuged at 3000 rpm for 3 minutes. The supernatant was used as cytoplasmic fraction. The pellet was resuspended in 100 μL buffer B (20 mM HEPES [pH 7.9], 400 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 1 mM PMSF). The homogenate was allowed to swell on ice for 30 minutes, subsequently diluted to 200 mM NaCl in the same buffer lacking NaCl, and centrifuged at 15 000 rpm for 20 minutes at 4°C. The supernatant was used as the nuclear extract.
Immunoprecipitation
The nuclear or cytoplasmic extracts prepared as described above were incubated with LIP buffer (10 mM HEPES, 250 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA, and 1 mM PMSF) and Protein G Sepharose (Amersham Pharmacia Biotech, Bucks, United Kingdom) for 1 hour with gentle rocking and centrifuged at 3000 rpm for 3 minutes. The supernatants were transferred into new tubes and incubated with protein G Sepharose and anti-FLAG monoclonal antibody (mAb) (M2, Sigma, St. Louis, MO) or anti-Myc mAb (9E10, Pharmingen, San Diego, CA) for 1 hour in LIP buffer. Immunocomplexes were washed 4 times with LIP buffer, resuspended in loading buffer, boiled, and analyzed by Western blot with anti-Myc or anti-FLAG mAb.
Results
Expression of MMCP-7
The expression of MMCP-7 gene is abnormal in the C57BL/6 strain because of the point mutation of an exon/intron splice site.22 However, the MMCP-7 gene is normally expressed in the WB strain.21 We crossed WB-mice (MMCP-7WB/MMCP-7WB, +/+) with C57BL/6-mice (MMCP-7B6/MMCP-7B6,mi/+) or with C57BL/6-mice (MMCP-7B6/MMCP-7B6,tg/+) and ultimately obtained mice of (MMCP-7WB/MMCP-7WB,mi/mi), (MMCP-7WB/MMCP-7WB,tg/tg), or (MMCP-7WB/MMCP-7WB, +/+) genotype. Because all mice used in the present experiment hadMMCP-7WB/MMCP-7WB genotype, we hereafter describe them just as mi/mi, tg/tg, or +/+ mice.
CMCs were obtained using spleens of mi/mi, tg/tg, or +/+ mice. Total RNA was extracted from CMCs of each genotype, and the expression of MMCP-7 gene was examined by Northern blot analysis. As controls, the expression of MMCP-6 and MC-CPA genes was also examined. The expression of MMCP-6 gene was severely impaired in bothmi/mi and tg/tg CMCs; that of MC-CPA gene was normal in both mi/mi and tg/tg CMCs. However, the expression of MMCP-7 gene was different between mi/mi andtg/tg CMCs. It was severely reduced in mi/mi CMCs but only slightly reduced in tg/tg CMCs (Figure1).
Expression of MMCP-7, MMCP-6, and MC-CPA mRNA in +/+,
tg/tg, and mi/mi CMCs.Twenty micrograms of total RNA prepared from +/+, tg/tg, ormi/mi CMCs was loaded in each lane. The membranes were hybridized with specific DNA probes. Reprobing with GAPDH allowed verification that an equal amount of mRNA was loaded per lane.
Expression of MMCP-7, MMCP-6, and MC-CPA mRNA in +/+,
tg/tg, and mi/mi CMCs.Twenty micrograms of total RNA prepared from +/+, tg/tg, ormi/mi CMCs was loaded in each lane. The membranes were hybridized with specific DNA probes. Reprobing with GAPDH allowed verification that an equal amount of mRNA was loaded per lane.
The promoter region of the MMCP-7 gene
McNeil et al16 reported the sequence of MMCP-7 gene 80 bp upstream of the transcription initiation site. We cloned further 1811 bp upstream region to examine the regulation of transcription (Figure 2). Although the coding regions of MMCP-7 and MMCP-6 genes showed a remarkable homology (73%),16 the promoter regions of these genes showed less homology (51%) (Figure 2).
The nucleotide sequence of 5′ flanking region of MMCP-7 gene.
Each CANNTG motif was boxed. The AP-1 binding consensus motif was underlined. A part of the first exon is shown by capitals, and 5′ flanking region is shown by lower case. The transcription initiation site was numbered as +1.
The nucleotide sequence of 5′ flanking region of MMCP-7 gene.
Each CANNTG motif was boxed. The AP-1 binding consensus motif was underlined. A part of the first exon is shown by capitals, and 5′ flanking region is shown by lower case. The transcription initiation site was numbered as +1.
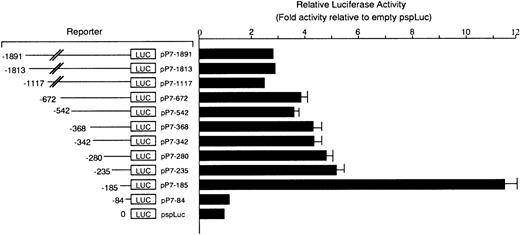
We constructed the reporter plasmid that contained the luciferase gene under the control of the MMCP-7 promoter starting from nt −1891 (+1 shows transcription initiation site, hereafter referred to as pP7-1891). We also constructed the deleted reporter plasmids containing the MMCP-7 promoter starting from nt −1813, −1117, −672, −542, −368, −342, −280, −235, −185, or −84. Each reporter plasmid was transfected to FMA/3 mastocytoma cells, which weakly expressed the MMCP-7 mRNA. When the reporter plasmid deleted up to position −235 (pP7-235) was transfected, a small but significant luciferase activity was found when compared to the basal activity. The reporter plasmid deleted to position −185 (pP7-185) showed a high luciferase activity, and pP7-84 showed the basal luciferase activity. Thus, the promoter region between nt −185 and −85 appeared important for the expression of the MMCP-7 gene (Figure 3).
Luciferase activity under the control of sequentially deleted MMCP-7 promoter.
The luciferase gene under the control of intact or deleted MMCP-7 promoter was transfected to FMA/3 cells. The value of the luciferase activity was divided by the value obtained with backbone vector pSPLuc and was shown as the relative luciferase activity. The data represent the mean ± standard error (SE) of 3 experiments. In some cases, the SE was too small to be shown by bars.
Luciferase activity under the control of sequentially deleted MMCP-7 promoter.
The luciferase gene under the control of intact or deleted MMCP-7 promoter was transfected to FMA/3 cells. The value of the luciferase activity was divided by the value obtained with backbone vector pSPLuc and was shown as the relative luciferase activity. The data represent the mean ± standard error (SE) of 3 experiments. In some cases, the SE was too small to be shown by bars.
Role of AP-1 binding motif and c-Jun
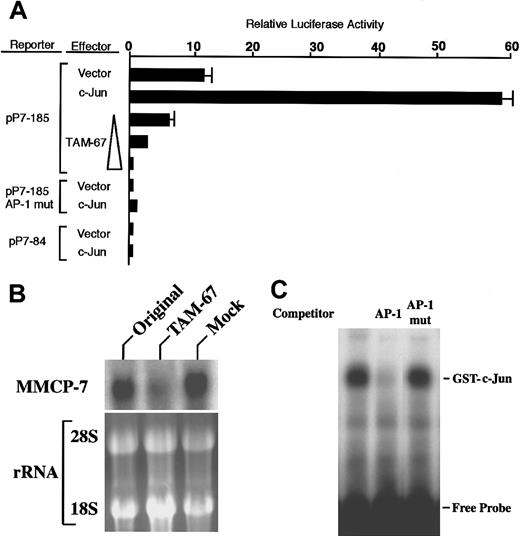
Between nt −185 and − 85, no CANNTG motif that is recognized and bound by MITF was present. However, an AP-1 binding consensus motif was found (Figure 2). The reporter plasmid pP7-185 was mutated at the AP-1 binding motif (hereafter called pP7-185-AP-1-mut) and transfected into FMA/3 cells. The mutation abolished the luciferase activity of pP7-185 (Figure 4A), suggesting that the AP-1 binding motif was used for the transactivation of MMCP-7 gene.
Involvement of c-Jun in transactivation of MMCP-7 promoter.
(A) The reporter plasmid containing the intact, mutated, or deleted AP-1 motif (pP7-185, pP7-185-AP-1-mut, or pP7-84, respectively) was cotransfected with effector plasmids to FMA/3 cells. The value of luciferase activity was divided by that obtained with the cotransfection of pP7-84 and pEF-BOS and was shown as relative luciferase activity. (B) Reduced expression of MMCP-7 gene by the overexpression of dominant negative form of c-Jun, TAM-67. Total RNA was extracted from MST cells (indicated as Original), MST cells overexpressing TAM-67, or MST cells overexpressing empty expression vector (indicated as Mock). The expression of MMCP-7 gene was examined with Northern blot analysis. (C) Binding of c-Jun to the AP-1 motif in the MMCP-7 promoter. The 5′-AGCTGTCCCTGGAGTCTGAGTCACCCATTTAGCT, designated as AP-1, was used as a probe (the AP-1 motif is underlined). The oligonucleotide mutated in the AP-1 motif (TGAGTCA to TGAGTTG) was designated as AP-1-mut (the mutated nucleotides are underlined). The excess amount of nonlabeled AP-1 or AP-1-mut was added as a competitor.
Involvement of c-Jun in transactivation of MMCP-7 promoter.
(A) The reporter plasmid containing the intact, mutated, or deleted AP-1 motif (pP7-185, pP7-185-AP-1-mut, or pP7-84, respectively) was cotransfected with effector plasmids to FMA/3 cells. The value of luciferase activity was divided by that obtained with the cotransfection of pP7-84 and pEF-BOS and was shown as relative luciferase activity. (B) Reduced expression of MMCP-7 gene by the overexpression of dominant negative form of c-Jun, TAM-67. Total RNA was extracted from MST cells (indicated as Original), MST cells overexpressing TAM-67, or MST cells overexpressing empty expression vector (indicated as Mock). The expression of MMCP-7 gene was examined with Northern blot analysis. (C) Binding of c-Jun to the AP-1 motif in the MMCP-7 promoter. The 5′-AGCTGTCCCTGGAGTCTGAGTCACCCATTTAGCT, designated as AP-1, was used as a probe (the AP-1 motif is underlined). The oligonucleotide mutated in the AP-1 motif (TGAGTCA to TGAGTTG) was designated as AP-1-mut (the mutated nucleotides are underlined). The excess amount of nonlabeled AP-1 or AP-1-mut was added as a competitor.
c-Jun cDNA was cotransfected with pP7-185 to FMA/3 cells as an effector molecule. The cotransfection increased the luciferase activity of pP7-185 58-fold when compared to the basal level (4.8-fold as compared with pP7-185 alone) (Figure 4A). However, when c-Jun cDNA and pP7-185-AP-1-mut were cotransfected, the transactivating effect of c-Jun was not detectable (Figure 4A).
A dominant negative c-Jun, TAM-67, was used to examine whether endogenous c-Jun contributed to the promoter activity. TAM-67 is a variant of c-Jun in which amino acids 3 to 122 are deleted. TAM-67 may dimerize and bind DNA but lacks the transactivating potential.31 When various amounts of TAM-67 cDNA were cotransfected with pP7-185 into FMA/3 cells, the luciferase activity of pP7-185 decreased in a dose-dependent manner (Figure 4A).
In the next experiment, TAM-67 was transfected into the MST cells, because they express MMCP-7 mRNA at an easily detectable level. After 4 days of culture, the expression of MMCP-7 gene was examined by Northern blot analysis. The overexpression of TAM-67, which dominantly inhibits the DNA binding of endogenous c-Jun, significantly reduced the amount of MMCP-7 mRNA (Figure 4B). This suggested that the endogenous c-Jun was involved in the transactivation of MMCP-7 gene.
The binding of c-Jun with the MMCP-7 promoter was examined by EMSA. GST–c-Jun fusion protein bound the oligonucleotide probe containing the AP-1 binding motif (Figure 4C). The binding of c-Jun was specifically abolished by the excess amount of the normal (5′-AGCTGTCCCTGGAGTCTGAGTCACCCATTTAGCT-3′) nonlabeled oligonucleotide but not by the mutated (5′-AGCTGTCCCTGGAGTCTGAGTTGCCCATTTAGCT-3′) nonlabeled oligonucleotide.
Functional cooperation between c-Jun and MITF
The expression of MMCP-7 gene was enhanced through cis elements located between nt −185 and −85. An AP-1 binding consensus motif but no CANNTG motif was present between nt −185 and −85. Therefore, c-Jun was considered to be involved in the transactivation of MMCP-7 through the AP-1 binding sequence. Then we examined whether MITF affected the transactivation activity of c-Jun. When cDNA encoding +-MITF was cotransfected with pP-185 to FMA/3 cells, the luciferase activity increased 38-fold when compared to the basal level (3.2-fold when compared to pP7-185 alone) (Figure5A). However, the promoter activity of pP-185 was reduced by the cotransfection of mi-MITF in a dose-dependent manner (Figure 5A).
Functional cooperation between +-MITF and c-Jun in the promoter activation of MMCP-7 gene.
(A) The reporter plasmids were transfected to FMA/3 cells with the expression plasmid containing +-MITF or mi-MITF cDNA or the expression vector alone. (B) The reporter plasmids were transfected to Jurkat cells with the effector plasmids in various combinations. Control vector was used to adjust the total amount of DNA used in each transfection.
Functional cooperation between +-MITF and c-Jun in the promoter activation of MMCP-7 gene.
(A) The reporter plasmids were transfected to FMA/3 cells with the expression plasmid containing +-MITF or mi-MITF cDNA or the expression vector alone. (B) The reporter plasmids were transfected to Jurkat cells with the effector plasmids in various combinations. Control vector was used to adjust the total amount of DNA used in each transfection.
To remove the possible involvement of the endogenous MITF, we used the Jurkat human T-cell line that does not express MITF. The reporter plasmid pP7-185 was not transactivated by the endogenous factors in Jurkat cells (Figure 5B). The transfection of either c-Jun cDNA alone or +-MITF cDNA alone moderately increased the luciferase activity of pP7-185 (2.1-fold and 3.5- fold, respectively). The cotransfection of both c-Jun and +-MITF cDNAs remarkably increased the luciferase activity of pP7-185 (9.5-fold).
Physical interaction between c-Jun and MITF
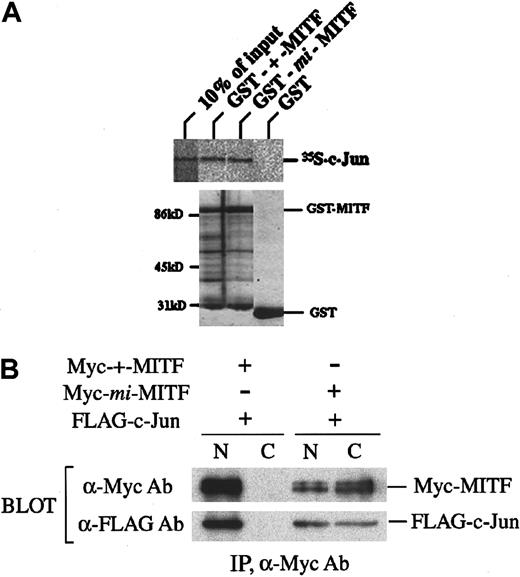
Physical interaction between c-Jun and MITF was examined. The coprecipitation of 35S-labeled c-Jun with GST-+-MITF or with GST-mi-MITF was examined by SDS-PAGE. The complex of c-Jun and GST-+-MITF and that of c-Jun and GST-mi-MITF were detected, but the complex of c-Jun and GST was not (Figure6A).
Physical interaction between MITF and c-Jun.
(A) In vitro binding of c-Jun to MITF. 35S-labeled c-Jun was subjected to coprecipitation with GST-+-MITF, GST-mi-MITF, or GST-coated beads, and the protein complex was analyzed by SDS-PAGE. Coomassie brilliant blue staining for each GST fusion protein was also shown. (B) Coimmunoprecipitation of MITF and c-Jun. COS-7 cells were transfected with FLAG-tagged–c-Jun and Myc-tagged–+MITF or Myc-tagged–mi-MITF. Nuclear or cytoplasmic extract was subjected to immunoprecipitation with anti-Myc antibody. Precipitates were separated by SDS-PAGE and immunoblotted with anti-Myc or anti-FLAG antibody.
Physical interaction between MITF and c-Jun.
(A) In vitro binding of c-Jun to MITF. 35S-labeled c-Jun was subjected to coprecipitation with GST-+-MITF, GST-mi-MITF, or GST-coated beads, and the protein complex was analyzed by SDS-PAGE. Coomassie brilliant blue staining for each GST fusion protein was also shown. (B) Coimmunoprecipitation of MITF and c-Jun. COS-7 cells were transfected with FLAG-tagged–c-Jun and Myc-tagged–+MITF or Myc-tagged–mi-MITF. Nuclear or cytoplasmic extract was subjected to immunoprecipitation with anti-Myc antibody. Precipitates were separated by SDS-PAGE and immunoblotted with anti-Myc or anti-FLAG antibody.
Inhibitory effect of mi-MITF on nuclear translocation of c-Jun
Myc-tagged-+-MITF or Myc-tagged-mi-MITF cDNA was cotransfected with FLAG-tagged-c-Jun cDNA into COS-7 cells. Lysate of transfected cells was divided into nuclear and cytoplasmic fractions. Each fraction was subjected to immunoprecipitation with anti-Myc antibody and Western blot analysis with anti-Myc or anti-FLAG antibody. When Myc-tagged–+-MITF and FLAG-tagged–c-Jun were transfected, Myc-tagged–+-MITF and coprecipitated FLAG-tagged–c-Jun were detected only in the nuclear fraction (Figure 6B). However, when Myc-tagged–mi-MITF and FLAG-tagged–c-Jun were cotransfected, one third of Myc-tagged–mi-MITF and FLAG-tagged–c-Jun were detected in the nuclear fraction, and the remaining Myc-tagged–mi-MITF and FLAG-tagged–c-Jun were in the cytoplasmic fraction (Figure 6B).
Discussion
Both MMCP-7 and MMCP-6 are tryptases,15,16 and genes encoding each of them reside on chromosome 17.34,35 The coding region of MMCP-7 gene is highly homologous with that of MMCP-6 gene,16 but the 5′ flanking region of both genes showed a remarkable difference. Three MITF-binding consensus sequences are present in the promoter region of MMCP-6 gene, all of which are recognized and bound by +-MITF.30 In fact, +-MITF appeared to play important roles for the transcription of MMCP-6 gene.30 However, we could not find any MITF-binding consensus sequences in the most significant segment (pP7-185) of the MMCP-7 promoter. Although the transcription of MMCP-6 gene was severely impaired in both mi/mi and tg/tg CMCs, that of MMCP-7 gene was severely impaired only in mi/mi CMCs. The abnormal mi-MITF was expressed in mi/mi CMCs but practically no MITFs were expressed in tg/tg CMCs. Therefore, the presence of mi-MITF rather than the absence of +-MITF appeared to have a negative effect on the transcription of MMCP-7 gene.
We investigated the mechanism of the negative effect ofmi-MITF. In spite of the lack of the MITF-binding consensus sequence, presence of +-MITF enhanced the promoter activity of pP7-185 and that of mi-MITF suppressed it. Both +-MITF andmi-MITF appeared to influence the promoter function through binding to other transcription factors. There is an AP-1 binding consensus motif in the pP-185 promoter. The mutation of the AP-1 binding motif abolished the promoter activity. The coexpression of normal c-Jun enhanced the promoter activity, and that of dominant negative mutant of c-Jun suppressed it. The binding of c-Jun to the AP-1 binding motif activated the promoter, and binding of +-MITF to the c-Jun further enhanced the promoter and that ofmi-MITF suppressed it. In fact, we demonstrated the physical interaction of c-Jun with either +-MITF or mi-MITF.
The complex of c-Jun and +-MITF was detectable only in the nuclear fraction, but that of c-Jun and mi-MITF was observed in both nuclear and cytoplasmic fractions. A possible explanation of suppressing potential of mi-MITF may be its deficient nuclear localization potential. Not only mi-MITF by itself but also the complex of mi-MITF and c-Jun appeared to have deficient nuclear translocation potential. Hemesath et al5 showed a deficient DNA-binding ability of the heterodimer of mi-MITF and TEFB. Because TFEB is another bHLH-Zip–type transcription factor, this mechanism may be understandable. However, because there was no CANNTG consensus sequence that may be recognized and bound by bHLH-Zip–type transcription factors in the pP-185 promoter, it is difficult to explain the negative effect of mi-MITF by the deficient DNA binding potential.
We produced a cDNA library from +/+ CMCs.34 By subtracting mRNAs expressed by mi/mi CMCs ortg/tg CMCs, we obtained (+/+-mi/mi) and (+/+-tg/tg) subtraction libraries.10,36 The number of clones that may hybridize cDNAs contained by the cDNA library of +/+ CMCs was significantly greater in the (+/+-mi/mi) library than in the (+/+-tg/tg) library.10 This is consistent with the present result. In addition to the loss of transactivation potential, the presence of mi-MITF suppressed the transactivation potential through other transcription factors. The transcription of c-kit, granzyme B, and tryptophan hydroxylase genes was also suppressed by the presence ofmi-MITF.10 Probably a similar suppressing mechanism may play a role, but counterpart transcription factors that make a complex with mi-MITF may be different in each case.
Comparison of +/+, mi/mi, and tg/tg CMCs may be a convenient method to examine the influence of MITF on the expression of genes that are expressed in CMCs. By screening the (+/+-mi/mi) subtraction library, the maximum number of possible target genes of MITF may be obtained. The following 2 types of genes are included: (1) genes for which transcription +-MITF is essential and (2) genes of which transcription was suppressed bymi-MITF. When the expression of type 2 genes was compared between +/+ CMCs and tg/tg CMCs, the expression intg/tg CMCs was slightly but significantly lower than that of +/+ CMCs. Even if such genes were not detected in the (+/+-tg/tg) subtraction library, +-MITF may enhance their expression. Taken together, the mechanism of the transcription regulation by MITF was different between MMCP-6 and MMCP-7 genes, and the comparison of expression among +/+, mi/mi, andtg/tg CMCs may be useful for understanding the mechanism.
Acknowledgments
The authors thank Dr J. D. Esko of University of California, San Diego for MST cells, Dr H. Arnheiter of National Institutes of Health for tg/tg mice, Dr K. Shimozaki and Dr S. Nagata of Osaka University for TAM-67 in pEF-BOS, Dr S. Nagata of Osaka University for pEF-BOS, Dr I. Matsumura of Osaka University for pcDNA3-FLAG, and T. Jippo and S. Adachi of Osaka University and T. Sawado of Fred Hutchinson Cancer Research Center for helpful discussion.
Supported by grants from the Ministry of Education, Science and Culture, the Ministry of Health and Welfare, the Organization for Pharmaceutical Safety and Research, and the Welfide Medicinal Research Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Yukihiko Kitamura, Department of Pathology, Osaka University Medical School, Yamada-oka 2-2, Suita 565-0871, Japan; e-mail: kitamura@patho.med.osaka-u.ac.jp.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal