This study examined the effectiveness of 3 leukocyte-reduction (LR) methods in depleting the residual level of cytomegalovirus (CMV) in blood products measured by quantitative polymerase chain reaction (QA-PCR). At 2 locations over 3 allergy seasons, apheresis platelets and whole blood were collected from 52 healthy CMV seropositive subjects having an elevated titer of CMV DNA (median = 2400 genome equivalents [GE]/mL) resulting in 32 evaluable LR apheresis platelets, 31 filtered platelets from whole blood, and 31 filtered red blood cells (RBCs) from whole blood. Leukoreduction by apheresis and filtration resulted in substantial reduction of detectable CMV DNA levels with 99.9% of the LR products expected to have less than 500 GE/mL of CMV DNA. No difference was found between methods (P = .52). CMV genomic leukocyte subset localization was determined by QA-PCR of fluorescence-activated cell sorter (FACS)-sorted peripheral blood from 20 seropositive subjects (n = 10 > 100 GE/mL, n = 10 QA-PCR negative). CMV was detected in monocyte (13 of 20) and granulocyte (3 of 20) fractions. Presence of competent virus in QA-PCR positive (> 100 GE/mL) peripheral blood samples was verified with 4 of 19 subjects positive in shell vial assay, and 8 of 18 positive for CMV gene products (messenger RNA). We observed a seasonal DNAemia variation in seropositive subjects. CMV seropositive subjects (n = 45) entered into longitudinal monitoring in March/April 1999 were QA-PCR negative at baseline. Subjects converted to a positive QA-PCR coincident with increased seasonal allergen levels (Norfolk 15 of 18 evaluable in 43.4 ± 9.48 days; Denver, 16 of 23 evaluable in 96 ± 26.3 days). These data demonstrate effective reduction of CMV load by LR during periods of DNAemia in CMV seropositive subjects.
Introduction
Transfusion-transmitted cytomegalovirus (CMV) infections cause only mild illness in individuals with normal immune systems, but can cause serious and even fatal illness (including retinitis, gastroenteritis, and interstitial pneumonitis) in the seronegative, immunocompromised recipient.1,2 To avoid these serious complications, it is recommended that seronegative immunocompromised patients receive blood components from seronegative donors.3 Typically, the majority of the blood donors in a geographic region are seropositive for CMV, which makes providing CMV seronegative blood components at times a difficult logistical task. Because it is believed that latent CMV resides in the white blood cells (WBCs), an alternative to using seronegative donors has been to remove most of the leukocytes from transfused products.4 A number of studies have suggested that reduction in the total number of leukocytes in blood products (leukocyte-reduction [LR]) below 1 × 107 to 1 × 106 would reduce the risk of transmission of CMV to a level comparable to that of seronegative blood products.5,6 Such levels of LR have been achieved either by filtration7 or during collection of apheresis platelets using a cell separator.8
Some investigators have suggested that various methods of LR result in different distributions of WBC subsets in the final product. Based on the assumption that latent CMV is only in a particular cell type, they further extrapolate these data to suggest that different methods of preparation will result in varying effectiveness for reduction of the viral load presented by the blood donor.9 Residual WBCs in leukocyte-reduced products are rare events with typically fewer than 100 cells/mL. The seemingly simple process of obtaining an accurate WBC count in these products has historically been difficult. Further isolation and subtyping of these cells presents many technical challenges, such as isolating WBCs without cell loss and observing enough events to make accurate estimates. Inferences regarding CMV safety from these data require additional biologic assumptions regarding the exclusive location of latent or active virus. We wished to determine if there is a difference in the effectiveness of LR methods in rendering blood components CMV safe, and believed quantitative polymerase chain reaction (QA-PCR) offered the most sensitive measure of low levels of virus without the pitfalls of the previously mentioned assumptions.
Patients, materials, and methods
Study subjects
Men and women at least 18 years of age were selected from eligible blood donors at the American Red Cross Mid-Atlantic Region (Norfolk, VA) and Bonfils Memorial Blood Center (Denver, CO). All subjects were positive for anti-CMV antibodies by microhemagglutination assay in Norfolk (Olympus, Melville, NY) or by a passive latex agglutination assay in Denver (CVM Scan, Becton Dickinson, Cockeysville, MD). Subjects were screened at 1- to 2-week intervals for detectable levels of CMV genome in peripheral blood using QA-PCR. Those with elevated CMV levels were enrolled to donate platelets by apheresis and whole blood on 2 separate occasions. The sequence of donation was mixed by the collection staffs but not subjected to a planned randomization scheme. Subject complete blood counts (CBCs) with automated differentials were performed in the local clinical hematology laboratory. This protocol was approved by the Institutional Review Board of the University of Colorado and the Eastern Virginia Medical School.
Blood products
Whole blood was collected using the Leukotrap RC-PL (Medsep Corporation, Covina CA) with CP2D anticoagulant. Filtered platelet-rich plasma (PRP) and AS3 containing red blood cells (RBCs) were prepared according the manufacturer's instructions within 4 hours of collection.10 The RC-PL system results in one PRP and one RBC with additive. These products are then individually filtered through 2 separate filters in the LR step. Platelet apheresis was performed using the COBE Spectra LRS or LRS TURBO (Gambro BCT, Lakewood, CO).11 Residual WBCs in leukocyte-reduced blood products were determined at Gambro BCT by a previously described flow cytometry protocol.12
Products collected in Denver were shipped overnight with 22°C gel packs from Lakewood to Norfolk for CMV QA-PCR. Subject peripheral blood samples used for CMV QA-PCR screening and reverse transcription–PCR (RT-PCR) were collected in EDTA tubes and shipped overnight at 4°C. Blood products and samples were held at 4°C 0 to 3 days before DNA extraction.
WBC subset isolation and testing for CMV genome
To determine which WBC subsets carried the CMV genome, PCR+ and PCR− subject samples from Norfolk during the spring of 1998 were separated by density gradient separation using OptiPrep density gradient media (Nycomed Pharma AS, Oslo, Norway) into monocyte, lymphocyte, and granulocyte fractions. The separation was done according to the protocol recommended by the manufacturer. The enriched monocytes (present in the top fraction of the gradient) were further purified by FACS (BD, San Jose, CA) based on CD14 markers to remove the small amount of contaminating lymphocytes. The lymphocyte fractions (present in the middle part of the gradient) were further purified by FACS using CD3, and CD4 markers to isolate T cells, and CD19 marker to isolate B cells (BD; Diatec AS, Oslo, Norway). Granulocytes present in the pellet fraction were used directly for further analysis. Analysis of the latter fraction indicated that contamination with CD14+ cells was less than 0.5% and contamination with lymphocytes (CD3, CD4, CD19 combined) was less than 3%. One million (1 × 106) cells were used from the isolated granulocytes, monocytes, B lymphocytes, and T lymphocytes for detection of the presence of CMV genome by QA-PCR and by RT-PCR for expression of viral gene products p72 and viral DNA polymerase.
QA-PCR
DNA preparation from blood or blood products.
DNA was extracted from 100μL peripheral blood or 1 × 106 cells from the various cell fractions by lysing cells, solubilizing proteins with 1 mL DNAzol (Gibco-BRL, Rockville, MD) and precipitating with 0.5 mL ethanol according to the manufacturer's recommendation. DNA was washed twice with 70% ethanol, dried, and resuspended in water for use in PCR reactions.
Leukocyte-reduced platelets and RBCs were prepared for QA-PCR by fully extracting DNA from 60 mL using a silica-based method. Briefly, to 60 mL product was added 32.0 g guanidine-HCl, 1.2 mL 2M Tris-HCl, pH 7.4, 0.6 mL 0.5M EDTA, and 120 μL 2-mercaptoethanol. After dissolution of the guanidine-HCl, the mixture was incubated for 10 minutes at 37°C to solubilize membrane proteins. Prewashed silica (100 μL of a 30% mix in water) was added and mixed for 15 minutes. The silica was separated from the solution by centrifugation, washed 3 times in 4 M guanidine-HCl, washed twice in ice-cold Tris-HCl, pH 7.4/50% ethanol (pH 7.4, 5 mM EDTA), and dried. Water (100 μL) was added to the silica and the bound DNA was eluted by incubation at 57°C for 5 minutes. The silica was removed by centrifugation and the DNA was precipitated with sodium acetate and isopropanol, washed with 70% ethanol and dried, and finally resuspended in 100 μL water for PCR.
CMV QA-PCR assay.
Quantitative PCR was performed by amplifying a 240–base pair (bp) region of CMV DNA-polymerase gene using primers 5′-GCT ATG TTT CAG ATG TCG CCG CC (5′-biotin), 5′-CCC ACC TCG GGC TCA AAC AC (Midland Certified Reagent, Midland, TX). One microgram of DNA per reaction tube was amplified using the AmpliTaq Gold enzyme (PerkinElmer, Norwalk, CT). The conditions were: initial melt 94° for 6 minutes, followed by 39 cycles melting at 94°C for 20 seconds and annealing at 58°C for 30 seconds and extension at 72°C for 40 seconds. Amplification products were resolved by acrylamide gel electrophoresis, developed with ethidium bromide, and visualized with an imaging system (Stratagen Eagle Eye II Camera System, La Jolla, CA). The image was stored as a TIFF file and the intensity of the reaction products were analyzed using SigmaGel (SPSS Science, Chicago, IL). CMV+ or CMV− genomic controls were run in each batch of PCR reaction. Calibration curves were prepared by amplification of known copy numbers (0, 10, 100, 1000, 10 000, 100 000) of cloned CMV polymerase gene (pCMPol 2B) diluted in 500 ng human CMV genome-negative genomic DNA. This standard was included in each batch of PCR runs using the same 240-bp amplicon for detection (Figure1).13 Some samples were retested using enzyme-linked immunosorbent assay (ELISA)-based hybridization assay to confirm the accuracy of QA-PCR. Briefly, the PCR products from the samples and also the standards were hybridized to a DNA-bind plate, which contained a covalently coupled probe corresponding to a 27-bp region of the amplified PCR product (5′-ACG CCG CCA CTA CTG CCG GAG CCG ACG). The hybridized DNA was detected by incubation with alkaline phosphatase-labeled streptavidin and subsequent color development. The color reaction was analyzed by an ELISA reader and the values were compared to the standard curve derived from the known copy numbers.
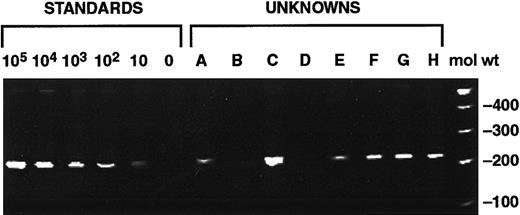
QA-PCR calibration curve of known copy numbers of cloned CMV polymerase gene.
Lanes 1 to 6 show 240-bp amplification product of pCMPol2B at 105, 104, 103, 102, 10, 0 GE/500 ng human CMV− genomic DNA. Lanes 7 to 14 are test samples (A-H) from this study. Molecular weight markers are in lane 15 (mol wt).
QA-PCR calibration curve of known copy numbers of cloned CMV polymerase gene.
Lanes 1 to 6 show 240-bp amplification product of pCMPol2B at 105, 104, 103, 102, 10, 0 GE/500 ng human CMV− genomic DNA. Lanes 7 to 14 are test samples (A-H) from this study. Molecular weight markers are in lane 15 (mol wt).
Because of the very low levels of residual WBCs in the blood products (typically < 3 WBC/μL), a positive control for DNA amplification was run with each sample. A 430-bp region of the hemachromatosis gene was amplified as above using primers 5′-TCC TGG CAA GGG TAA ACA GAT CC and 5′-CTC CTC AGG CAC TCC TCT CAA CC (5′-biotin). Resolved amplification products were identified using the probe 5′-CAA GGA GTT CGA ACC TAA AGA CGT ATT GCC CA. The assay was completed as described for CMV genome detection. The sensitivity of detection of the hemachromatosis gene was 50 copies per PCR reaction.
Determination of the detection limit of QA-PCR.
Because no cell lines containing latent CMV genomes are in existence, and independent quantitation of CMV genomes in infected cells is not possible, the detection limits for viral genomes in these preparation and assay methods were determined using blood products spiked with Namalva cell line containing 2 copies of Epstein-Barr virus (EBV) per cell. The primers used for this assay were 5′ TAT GAC AAA GCC CGC TCC TAC CT and 5′ GGG AAT ACA CGG CTT TTA ATA CG and the size of the PCR product was 236 bp. The sensitivity of EBV PCR was similar to the CMV PCR as was determined in previous studies. This method allowed us not only to determine the lowest viral copy numbers to be detected but also indicated that we can isolate DNA from fewer than 500 cells diluted in 60 mL blood product. Spiked RBCs and platelets were extracted using the silica protocol followed by QA-PCR using primers for EBV. The lower detection limits of 100 GE/mL (genomic equivalent) for peripheral blood and 4 GE/mL for the blood products were observed (Figure2). However, because detection of CMV genome and EBV genome is done with different primers, the possibility exists that the sensitivity corresponding to the EBV genome cannot be directly adapted to the detection of CMV genome. In a recent multilaboratory blinded study our laboratory could detect and accurately quantitate 10 genome copies of CMV in duplicate samples indicating that the QA-PCR adapted in our laboratory can reproducibly detect low copy numbers of CMV (manuscript submitted for publication).
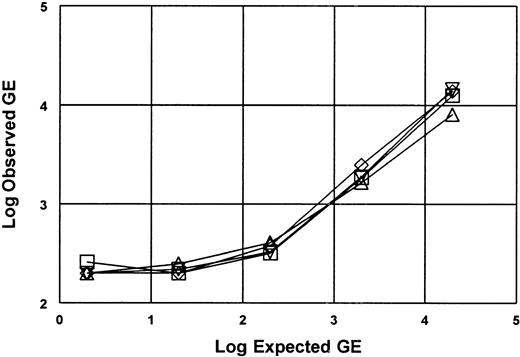
Lower limit of detection for DNA extraction and QA-PCR amplification was determined using blood products spiked with EBV containing Namalva cell line.
Namalva cell line containing 2 copies per cell of EBV was spiked into RBC and platelets to known concentrations. Spiked RBC and platelets (50 mL) were extracted using silica protocol followed by QA-PCR. The plot shows recovered copies (observed) versus expected copies for 2 RBC (■ ⋄) and 2 platelet experiments (▵ ▿). Signal was not detectable above a background of 200 GE, thus the limit of detection is 4 GE/mL.
Lower limit of detection for DNA extraction and QA-PCR amplification was determined using blood products spiked with EBV containing Namalva cell line.
Namalva cell line containing 2 copies per cell of EBV was spiked into RBC and platelets to known concentrations. Spiked RBC and platelets (50 mL) were extracted using silica protocol followed by QA-PCR. The plot shows recovered copies (observed) versus expected copies for 2 RBC (■ ⋄) and 2 platelet experiments (▵ ▿). Signal was not detectable above a background of 200 GE, thus the limit of detection is 4 GE/mL.
RT-PCR
Cell pellets (1 × 106 to 5 × 106) from whole blood or cell fractions were resuspended in Trizol reagent (Gibco-BRL) and total RNA was purified according to the manufacturer's directions. Polyadenylated messenger RNA (mRNA) was isolated from the purified RNA by binding the mRNA to Instant mRNA Capture Disc (Trevigen, Gaithersburg, MD) according to the manufacturer's recommendation. Bound mRNAs were eluted with 50 μL Rnase-free water for use in the RT-PCR assay. The assay was performed with 10 μL of the mRNA preparation (corresponding to 1 × 106 cells) using the EZ-RT-PCR kit (PerkinElmer). The tubes were incubated at 62°C for 20 minutes to convert RNA to DNA; then amplification was performed by cycling 36 times to 94°C for 0.3 minutes, annealing at 58°C for 0.4 minutes, and elongating at 72°C for 0.5 minute. Amplification products were resolved by acrylamide gel electrophoresis, developed with ethidium bromide, and visualized with an imaging system (Stratagen Eagle Eye II Camera System).
CMV culture
To determine if the detection of CMV genome was indicative of the presence of infectious virus, samples from positive donors during the spring of 1999 were evaluated via shell vial assay. Although primary cell line cultures of various foreskin fibroblast are frequently used in some cell vial assays, we found that MRC-5 fibroblast cells can be equally well infected with CMV strains from various patients to be used in this assay. Therefore, the fibroblast cell line MRC-5 (American Tissue Culture Collection, Rockville, MD) was used in our shell vial assays. Cells were overlaid in triplicate with buffy coat purified from 1 mL of subject peripheral blood sample and incubated at 37°C, 5% CO2 for 10 days. Cultures were checked for infectivity at days 3 and 10 by staining the cells with a monoclonal antibody against the CMV p72 antigen and detection of p72 mRNA expression by RT-PCR.
Cytokine assays
Plasma was separated within 30 minutes from peripheral blood samples collected in EDTA and frozen at −70°C. Quantitative sandwich enzyme immunoassays were performed on thawed plasma for interleukin (IL)-2, IL-4, and IL-10 and tumor necrosis factor-α (TNF-α) (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions.
Pollen data
Local pollen data were kindly provided by Dr Alpha A. Diallo, Department Public Health, Norfolk, and obtained from the American Academy of Allergy, Asthma, and Immunology Web site for Denver (data collected at National Jewish Medical and Research Center).
Statistical methods
The primary end point, CMV genome residual in leukocyte-reduced blood products, was evaluated as a dichotomous outcome variable (detectable or nondetectable at 4 GE/mL) for those procedures where there was detectable CMV genome in the subject before collection. Matched subject effects for the 3 preparation methods were evaluated by the McNemar χ2 test. Logistic regression of outcome variable on preparation methods was performed with global testing of equivalence of the preparation methods. As secondary analyses, estimates of the probability of detection an global equivalency tests were conducted using logistic regression at detection thresholds of 50 and 80 GE/mL in the products.
As additional secondary analyses, residual WBCs in the products were evaluated as a log-normal distribution as described by Dumont and colleagues.14 The effect of residual WBCs on final genome concentration in the end products was determined by logistic analysis that included preparation methods as predictor variables. The association of subjects' reported allergy condition with the level of peripheral blood CMV genome was evaluated with a general linear regression model of the logarithm of peripheral blood CMV regressed on questionnaire responses. The Fisher exact test was applied to the differences in questionnaire responses between sites. All tests were performed at an alpha of 0.05.
Results
During February and March 1998, 148 CMV-seropositive blood donors at the American Red Cross, Norfolk. Zero versus an anticipated 35% to 50% of these donors were positive for CMV DNA by QA-PCR. Because of previous local observations of season association with CMV DNAemia, the screening was suspended until April, whereupon the CMV positivity increased to 39 of 40 subjects (95%, median = 4200 GE/mL, range = < 100-1 000 000 GE/mL). Nine of these subjects with median screening levels of 15 800 GE/mL were then entered into phase 2 (blood product collection). Logistical delays resulted in up to a 30-day interval from the time of screening until the first collection, whereupon many of the subjects' levels had dropped to below detection level. Noticing the apparent seasonal association of this reactivation, we reinitiated screening during the fall of 1998 in Denver and Norfolk. Although reactivation was observed during the fall mold seasons at both locations, the absolute peak levels of CMV DNA were lower than observed in the spring with a rapid falloff over 2 weeks. Therefore, in the spring of 1999 enrollment was increased in both locations, gathering longitudinal data on subjects over a long calendar period. In Norfolk, 20 CMV-seropositive subjects were entered into weekly monitoring (phase 1) between March 2 and 30, 1999. Two subjects withdrew from the study and 15 of 18 converted to a positive CMV DNAemia in a mean of 43.4 days (SD = 9.48, range = 22-57 days). Twelve positive subjects were entered into the collection phase (phase 2). Subjects were followed for a maximum of 65 days, through May 5. In Denver, 25 CMV-seropositive subjects were entered into phase 1 between March 15 and April 23, 1999, with initial monitoring every other week changing to weekly as the Denver tree pollen season approached. Of these, 2 subjects withdrew and 16 of 23 converted to a positive CMV DNAemia in a mean of 96 days (SD = 26.3, range = 0-120 days). Thirteen positive subjects were entered into phase 2. Subjects were followed for a maximum of 120 days, through July 7. Figure3 shows the maximum CMV DNA titers observed for subjects throughout the monitoring and collection phases. Norfolk subjects reactivated during April and Denver subjects during June/July, both following the local tree pollen rise.
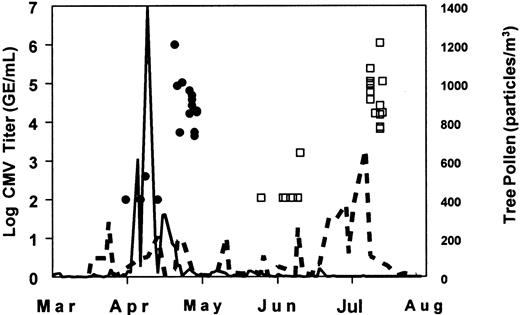
Subject maximum CMV titer and local tree pollen levels for spring 1999 demonstrate association of CMV reactivation and local pollen level.
The maximum CMV DNA levels observed for each subject throughout the monitoring and collection/treatment phases and local tree pollen counts are shown for Norfolk and Denver (each subject shown once—minimum detection level of 100 GE/mL). Twenty Norfolk CMV seropositive subjects were entered into the monitoring phase March 2 to 30 and followed for a maximum of 65 days, through May 5. Twenty-five Denver CMV seropositive subjects were entered into the monitoring phase March 15 to April 23 and followed for a maximum of 120 days, through July 7. Norfolk subjects reactivated during April and Denver subjects during June/July. Denver had several short-lived increases in tree pollen throughout the spring, but the largest, sustained period immediately preceded the subject CMV titer rise. Key: Norfolk subjects, (●); Norfolk tree pollen, (—) ; Denver subjects (■); Denver tree pollen, (– – –).
Subject maximum CMV titer and local tree pollen levels for spring 1999 demonstrate association of CMV reactivation and local pollen level.
The maximum CMV DNA levels observed for each subject throughout the monitoring and collection/treatment phases and local tree pollen counts are shown for Norfolk and Denver (each subject shown once—minimum detection level of 100 GE/mL). Twenty Norfolk CMV seropositive subjects were entered into the monitoring phase March 2 to 30 and followed for a maximum of 65 days, through May 5. Twenty-five Denver CMV seropositive subjects were entered into the monitoring phase March 15 to April 23 and followed for a maximum of 120 days, through July 7. Norfolk subjects reactivated during April and Denver subjects during June/July. Denver had several short-lived increases in tree pollen throughout the spring, but the largest, sustained period immediately preceded the subject CMV titer rise. Key: Norfolk subjects, (●); Norfolk tree pollen, (—) ; Denver subjects (■); Denver tree pollen, (– – –).
To determine the WBC subtype localization of the CMV, we identified 10 CMV DNA positive and 10 negative subjects from the April-May 1998 Norfolk screening. During a subsequent visit, a second sample was taken and WBC subsets isolated as described in “Patients, materials, and methods.” The CMV was detected in monocytes (CD14) in 13 of 20 samples; even for those samples that had an original whole blood level of less than 100 GE/mL. CMV was detected in the density gradient granulocyte fraction for 3 subjects (Table1).
Location of cytomegalovirus in white blood cell subsets
| Subject . | Peripheral blood . | FACS analysis (sample 2) . | |||||
|---|---|---|---|---|---|---|---|
| GE/mL . | GE/mL . | ||||||
| Sample 1 . | Sample 2 . | CD14 . | CD4 . | CD3 . | CD19 . | GRAN . | |
| 1 | 2 100 | — | POS | — | — | — | — |
| 2 | 25 200 | 4 800 | POS | — | — | — | — |
| 3 | — | 24 000 | POS | — | — | — | POS |
| 4 | — | — | — | — | — | — | — |
| 5 | — | — | — | — | — | — | — |
| 6 | 43 600 | — | POS | — | — | — | — |
| 7 | 17 400 | — | POS | — | — | — | — |
| 8 | 15 800 | — | POS | — | — | — | — |
| 9 | — | — | — | — | — | — | — |
| 10 | 2 600 | — | POS | — | — | — | — |
| 11 | — | — | — | — | — | — | — |
| 12 | — | — | POS | — | — | — | — |
| 13 | 300 | — | — | — | — | — | — |
| 14 | 3 100 | — | POS | — | — | — | — |
| 15 | 1 000 000 | 720 000 | POS | — | — | — | POS |
| 16 | — | — | — | — | — | — | — |
| 17 | — | — | POS | — | — | — | — |
| 18 | — | — | — | — | — | — | — |
| 19 | — | — | POS | — | — | — | — |
| 20 | 3 800 | — | POS | — | — | — | POS |
| Subject . | Peripheral blood . | FACS analysis (sample 2) . | |||||
|---|---|---|---|---|---|---|---|
| GE/mL . | GE/mL . | ||||||
| Sample 1 . | Sample 2 . | CD14 . | CD4 . | CD3 . | CD19 . | GRAN . | |
| 1 | 2 100 | — | POS | — | — | — | — |
| 2 | 25 200 | 4 800 | POS | — | — | — | — |
| 3 | — | 24 000 | POS | — | — | — | POS |
| 4 | — | — | — | — | — | — | — |
| 5 | — | — | — | — | — | — | — |
| 6 | 43 600 | — | POS | — | — | — | — |
| 7 | 17 400 | — | POS | — | — | — | — |
| 8 | 15 800 | — | POS | — | — | — | — |
| 9 | — | — | — | — | — | — | — |
| 10 | 2 600 | — | POS | — | — | — | — |
| 11 | — | — | — | — | — | — | — |
| 12 | — | — | POS | — | — | — | — |
| 13 | 300 | — | — | — | — | — | — |
| 14 | 3 100 | — | POS | — | — | — | — |
| 15 | 1 000 000 | 720 000 | POS | — | — | — | POS |
| 16 | — | — | — | — | — | — | — |
| 17 | — | — | POS | — | — | — | — |
| 18 | — | — | — | — | — | — | — |
| 19 | — | — | POS | — | — | — | — |
| 20 | 3 800 | — | POS | — | — | — | POS |
From the spring 1998 cohort, 10 DNAemia positive and 10 negative subjects were identified from the initial screening (sample 1) of peripheral blood samples (limit of detection 100 GE/mL). During a subsequent visit, a second sample (sample 2) was obtained. White blood cells (WBCs) were isolated and sorted by surface antigen phenotype. Isolated subpopulations were then assayed by QA-PCR. The CMV was detected in monocytes (CD14) in 13 of 20 samples, even for those samples that had a titer of less than or equal to 100 GE/mL. Cytomegalovirus (CMV) was detected in the granulocyte fraction for 3 subjects.
GE indicates genome equivalents; FACS, fluorescence-activated cell sorter; POS, positive; GRAN, granulocyte.
Aggregate CMV QA-PCR levels are shown for phase 2 treatments in Figure4. Reactivated subjects in the spring of 1999 were entered into phase 2 within 7 days of a positive screening result. Subject preprocedure CMV DNA levels in phase 2 were 50 to 100 times above the limit of detection of the assay. Blood products collected were within the typical ranges of volume and cellular content (Table 2). Residual WBC in the products were: filtered RBC median = 7.1 × 104, 98.8% less than 1 × 106 (n = 51); filtered platelets median = 7.9 × 103, 99.9% less than 1 × 106 (n = 51); apheresis platelets median = 4.0 × 104, 85% less than 1 × 106 (n = 52). Seven of the apheresis platelet procedures were flagged as nonleukocyte reduced by the equipment. Note that flagged procedures in clinical practice would not be regarded as CMV safe. Procedures indicated by the apheresis equipment as leukocyte reduced resulted in apheresis platelet WBC residuals median = 3.1 × 104, 99.8% less than 1 × 106 (n = 45). However, all apheresis procedures (n = 52) were included for the CMV analysis.
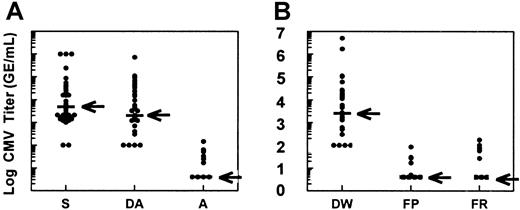
Phase 2 treatment results demonstrate reduction in CMV burden by all LR methods.
CMV DNA titer by QA-PCR (GE/mL). (A) S indicates positive screening test immediately before entry into phase 2; DA, donor titer immediately before apheresis donation; A, apheresis product titer. (B) DW indicates donor titer immediately before whole blood donation; FP, titer in filtered platelets prepared from whole blood; FR, titer in filtered RBCs prepared from whole blood. Median values are indicated by horizontal bar and arrow. Minimum detection level 100 GE/mL from peripheral blood (S, DA, DW) and 4 GE/mL for final products (A, FP, FR).
Phase 2 treatment results demonstrate reduction in CMV burden by all LR methods.
CMV DNA titer by QA-PCR (GE/mL). (A) S indicates positive screening test immediately before entry into phase 2; DA, donor titer immediately before apheresis donation; A, apheresis product titer. (B) DW indicates donor titer immediately before whole blood donation; FP, titer in filtered platelets prepared from whole blood; FR, titer in filtered RBCs prepared from whole blood. Median values are indicated by horizontal bar and arrow. Minimum detection level 100 GE/mL from peripheral blood (S, DA, DW) and 4 GE/mL for final products (A, FP, FR).
Description of collected blood products by city and season
| . | . | Apheresis volume (mL) . | Apheresis yield (plt × 1011) . | WB volume (mL) . | WB total WBC (109) . | Prefiltration RBC volume (mL) . | Postfiltration RBC volume (mL) . | Filtered PRP volume (mL) . | Filtered PRP yield (Plt × 1010) . |
|---|---|---|---|---|---|---|---|---|---|
| D-Spring99 | Mean | 321 | 5.2 | 428 | 2.5 | 235 | 217 | 217 | 8.0 |
| SD | 56 | 0.9 | 18 | 0.3 | 32 | 28 | 34 | 2.3 | |
| Min | 238 | 3.9 | 390 | 2.1 | 177 | 173 | 166 | 3.9 | |
| Max | 429 | 7.1 | 450 | 3.3 | 294 | 267 | 272 | 10.6 | |
| N | 13 | 13 | 13 | 13 | 12* | 12* | 12* | 11† | |
| N-Spring99 | Mean | 286 | 3.2 | 489 | 3.0 | 300 | 299 | 265 | 7.0 |
| SD | 35 | 1.2 | 16 | 0.7 | 26 | 22 | 11 | 2.6 | |
| Min | 212 | 1.3 | 442 | 2.2 | 252 | 247 | 250 | 4.2 | |
| Max | 334 | 5.2 | 503 | 4.3 | 331 | 320 | 283 | 11.9 | |
| N | 12 | 12 | 11‡ | 11‡ | 11‡ | 11‡ | 11‡ | 11‡ | |
| D-Fall98 | Mean | 412 | 7.1 | 450 | ND | 272 | 232 | 199 | ND |
| SD | 92 | 0.9 | 22 | 29 | 31 | 41 | |||
| Min | 268 | 5.7 | 427 | 225 | 180 | 148 | |||
| Max | 532 | 8.2 | 478 | 299 | 259 | 249 | |||
| N | 6 | 6.0 | 6 | 6 | 6 | 6 | |||
| N-Fall98 | Mean | 280 | 3.2 | 519 | 3.3 | 320 | 336 | 232 | 7.3 |
| SD | 44 | 1.5 | 28 | 0.7 | 50 | 41 | 30 | 3.0 | |
| Min | 224 | 0.7 | 441 | 2.3 | 226 | 244 | 171 | 3.5 | |
| Max | 368 | 6.4 | 544 | 4.4 | 399 | 405 | 279 | 14.9 | |
| N | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | |
| N-Spring98 | Mean | 290 | 3.5 | 521 | 2.9 | 355 | 358 | 213 | 6.7 |
| SD | 55 | 0.8 | 16 | 0.8 | 41 | 44 | 41 | 1.8 | |
| Min | 234 | 2.3 | 492 | 2.0 | 299 | 305 | 140 | 3.9 | |
| Max | 401 | 4.8 | 545 | 4.3 | 439 | 449 | 278 | 9.7 | |
| N | 9 | 9 | δ 8 | δ 8 | δ 8 | δ 8 | δ 8 | δ 8 |
| . | . | Apheresis volume (mL) . | Apheresis yield (plt × 1011) . | WB volume (mL) . | WB total WBC (109) . | Prefiltration RBC volume (mL) . | Postfiltration RBC volume (mL) . | Filtered PRP volume (mL) . | Filtered PRP yield (Plt × 1010) . |
|---|---|---|---|---|---|---|---|---|---|
| D-Spring99 | Mean | 321 | 5.2 | 428 | 2.5 | 235 | 217 | 217 | 8.0 |
| SD | 56 | 0.9 | 18 | 0.3 | 32 | 28 | 34 | 2.3 | |
| Min | 238 | 3.9 | 390 | 2.1 | 177 | 173 | 166 | 3.9 | |
| Max | 429 | 7.1 | 450 | 3.3 | 294 | 267 | 272 | 10.6 | |
| N | 13 | 13 | 13 | 13 | 12* | 12* | 12* | 11† | |
| N-Spring99 | Mean | 286 | 3.2 | 489 | 3.0 | 300 | 299 | 265 | 7.0 |
| SD | 35 | 1.2 | 16 | 0.7 | 26 | 22 | 11 | 2.6 | |
| Min | 212 | 1.3 | 442 | 2.2 | 252 | 247 | 250 | 4.2 | |
| Max | 334 | 5.2 | 503 | 4.3 | 331 | 320 | 283 | 11.9 | |
| N | 12 | 12 | 11‡ | 11‡ | 11‡ | 11‡ | 11‡ | 11‡ | |
| D-Fall98 | Mean | 412 | 7.1 | 450 | ND | 272 | 232 | 199 | ND |
| SD | 92 | 0.9 | 22 | 29 | 31 | 41 | |||
| Min | 268 | 5.7 | 427 | 225 | 180 | 148 | |||
| Max | 532 | 8.2 | 478 | 299 | 259 | 249 | |||
| N | 6 | 6.0 | 6 | 6 | 6 | 6 | |||
| N-Fall98 | Mean | 280 | 3.2 | 519 | 3.3 | 320 | 336 | 232 | 7.3 |
| SD | 44 | 1.5 | 28 | 0.7 | 50 | 41 | 30 | 3.0 | |
| Min | 224 | 0.7 | 441 | 2.3 | 226 | 244 | 171 | 3.5 | |
| Max | 368 | 6.4 | 544 | 4.4 | 399 | 405 | 279 | 14.9 | |
| N | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | |
| N-Spring98 | Mean | 290 | 3.5 | 521 | 2.9 | 355 | 358 | 213 | 6.7 |
| SD | 55 | 0.8 | 16 | 0.8 | 41 | 44 | 41 | 1.8 | |
| Min | 234 | 2.3 | 492 | 2.0 | 299 | 305 | 140 | 3.9 | |
| Max | 401 | 4.8 | 545 | 4.3 | 439 | 449 | 278 | 9.7 | |
| N | 9 | 9 | δ 8 | δ 8 | δ 8 | δ 8 | δ 8 | δ 8 |
WB indicates whole blood; RBC, red blood cell; PRP, platelet-rich plasma; Plt, platelet; ND, not determined.
n = 1 not counted.
n = 2 not counted.
n = 1 subject dropped from WB arm.
Phase 2 collections with the subject predonation CMV DNA level more than 100 GE/mL were selected for preparation method effect analyses resulting in 32 of 52 evaluable apheresis products, 32 of 51 evaluable filtered platelets, and 32 of 51 evaluable filtered RBC. The effect of matched preparation methods with one subject undergoing all treatment arms was evaluated by the McNemar χ2 test and found to be insignificant (P = .257). The detectable presence of CMV in the leukocyte-reduced product was analyzed by logistic regression using apheresis, filtered platelet, and filtered RBC to determine the effect of methods on residual CMV, using detection thresholds of 4 (primary analysis) and 50 and 80 GE/mL (secondary analyses). Global testing of preparation method effect showed no difference between apheresis platelets, filtered platelets, and filtered RBC (P = .522; Table 3). As a secondary analysis, the effect of the logarithm of residual WBC was included in the model at the CMV threshold of 4 GE/mL and found to be an insignificant contributor (χ2 = 0.224, df = 1, P = 0.636).
Effect of preparation method on residual CMV in leukocyte-reduced blood products
| . | Observed frequency . | Probability . | 95% CI . | Odds ratio . | Test . | ||
|---|---|---|---|---|---|---|---|
| df . | χ2 . | Probability . | |||||
| Probability of GE > 4/mL | |||||||
| Apheresis | 7/32 | .22 | 0.04-0.68 | 1.0 | 2 | 1.30 | .522 |
| Filtered Platelet | 4/32 | .13 | 0.02-0.56 | 0.5 | |||
| Filtered RBC | 7/32 | .22 | 0.11-0.39 | 1.0 | |||
| Probability of GE > 50/mL | |||||||
| Apheresis | 4/32 | .13 | 0.01-0.58 | 0.6 | 2 | 4.45 | .108 |
| Filtered Platelet | 1/32 | .03 | 0.00-0.41 | 0.1 | |||
| Filtered RBC | 6/32 | .19 | 0.09-0.36 | 1.0 | |||
| Probability of GE > 80/mL | |||||||
| Apheresis | 1/32 | .03 | 0.00-0.52 | 0.3 | 2 | 1.57 | .456 |
| Filtered Platelet | 1/32 | .03 | 0.00-0.52 | 0.3 | |||
| Filtered RBC | 3/32 | .09 | 0.03-0.25 | 1.0 | |||
| . | Observed frequency . | Probability . | 95% CI . | Odds ratio . | Test . | ||
|---|---|---|---|---|---|---|---|
| df . | χ2 . | Probability . | |||||
| Probability of GE > 4/mL | |||||||
| Apheresis | 7/32 | .22 | 0.04-0.68 | 1.0 | 2 | 1.30 | .522 |
| Filtered Platelet | 4/32 | .13 | 0.02-0.56 | 0.5 | |||
| Filtered RBC | 7/32 | .22 | 0.11-0.39 | 1.0 | |||
| Probability of GE > 50/mL | |||||||
| Apheresis | 4/32 | .13 | 0.01-0.58 | 0.6 | 2 | 4.45 | .108 |
| Filtered Platelet | 1/32 | .03 | 0.00-0.41 | 0.1 | |||
| Filtered RBC | 6/32 | .19 | 0.09-0.36 | 1.0 | |||
| Probability of GE > 80/mL | |||||||
| Apheresis | 1/32 | .03 | 0.00-0.52 | 0.3 | 2 | 1.57 | .456 |
| Filtered Platelet | 1/32 | .03 | 0.00-0.52 | 0.3 | |||
| Filtered RBC | 3/32 | .09 | 0.03-0.25 | 1.0 | |||
The detectable presence of CMV in the leukocyte-reduced product was regressed on treatment type using logistic regression to determine the effect of apheresis, filtered platelet, and filtered RBC methods on residual CMV. The results are shown as the probability of CMV above the detection thresholds 4 (primary analysis), 50 and 80 GE/mL (secondary analyses). The results of global tests of the equivalence of treatment type are shown with the chi-squared statistic and P value (Test). There was no significant difference in leukocyte-reduction (LR) methods.
Pooled Norfolk and Denver subject CBC at baseline and immediately before phase 2 collections are shown in Table4. There were no significant changes in baseline numbers taken in February and those at the height of the CMV DNAemia. A small increase in mean lymphocyte count was observed (P = .037). Other changes, which may have been indicative of an allergic response, such as eosinophilia, were not seen.
Pooled Norfolk and Denver subject complete blood counts
| . | HCT (%) . | PLT (×103/μL) . | MPV (fL) . | Donor WBCs (×103/μL) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | |
| Mean | 43 | 42 | 40 | 244 | 247 | 210 | 9.3 | 8.9 | 9.3 | 6.4 | 6.4 | 6.2 |
| SD | 4 | 4 | 3 | 61 | 59 | 49 | 1.3 | 1.0 | 1.1 | 1.7 | 1.3 | 1.5 |
| Min | 36 | 35 | 34 | 141 | 151 | 135 | 7.6 | 7.4 | 7.6 | 3.4 | 4.4 | 3.7 |
| Max | 50 | 50 | 46 | 390 | 374 | 300 | 13.5 | 11.6 | 11.9 | 10.4 | 9.7 | 9.7 |
| . | HCT (%) . | PLT (×103/μL) . | MPV (fL) . | Donor WBCs (×103/μL) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | |
| Mean | 43 | 42 | 40 | 244 | 247 | 210 | 9.3 | 8.9 | 9.3 | 6.4 | 6.4 | 6.2 |
| SD | 4 | 4 | 3 | 61 | 59 | 49 | 1.3 | 1.0 | 1.1 | 1.7 | 1.3 | 1.5 |
| Min | 36 | 35 | 34 | 141 | 151 | 135 | 7.6 | 7.4 | 7.6 | 3.4 | 4.4 | 3.7 |
| Max | 50 | 50 | 46 | 390 | 374 | 300 | 13.5 | 11.6 | 11.9 | 10.4 | 9.7 | 9.7 |
| . | Segmented neutrophils (×103/μL) . | Lymphocytes (×103/μL) . | Monocytes (×103/μL) . | Eosinophils (×103/μL) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | |
| Mean | 3.6 | 3.7 | 3.4 | 1.9 | 2.1 | 2.1 | 0.5 | 0.5 | 0.4 | 0.1 | 0.2 | 0.1 |
| SD | 1.1 | 0.9 | 1.1 | 0.6 | 0.6 | 0.6 | 0.2 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 |
| Min | 1.6 | 2.2 | 1.9 | 1.0 | 1.1 | 1.3 | 0.2 | 0.2 | 0.2 | 0.0 | 0.0 | 0.0 |
| Max | 6.2 | 5.5 | 5.7 | 3.4 | 3.3 | 3.3 | 0.9 | 0.9 | 0.7 | 0.3 | 0.4 | 0.3 |
| . | Segmented neutrophils (×103/μL) . | Lymphocytes (×103/μL) . | Monocytes (×103/μL) . | Eosinophils (×103/μL) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | Baseline . | Proc1 . | Proc2 . | |
| Mean | 3.6 | 3.7 | 3.4 | 1.9 | 2.1 | 2.1 | 0.5 | 0.5 | 0.4 | 0.1 | 0.2 | 0.1 |
| SD | 1.1 | 0.9 | 1.1 | 0.6 | 0.6 | 0.6 | 0.2 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 |
| Min | 1.6 | 2.2 | 1.9 | 1.0 | 1.1 | 1.3 | 0.2 | 0.2 | 0.2 | 0.0 | 0.0 | 0.0 |
| Max | 6.2 | 5.5 | 5.7 | 3.4 | 3.3 | 3.3 | 0.9 | 0.9 | 0.7 | 0.3 | 0.4 | 0.3 |
| . | Basophils (×103/μL) . | . | . | . | . | . | . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline . | Proc1 . | Proc2 . | ||||||||||
| Mean | 0.0 | 0.0 | 0.0 | |||||||||
| SD | 0.0 | 0.0 | 0.0 | |||||||||
| Min | 0.0 | 0.0 | 0.0 | |||||||||
| Max | 0.1 | 0.1 | 0.1 | |||||||||
| . | Basophils (×103/μL) . | . | . | . | . | . | . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline . | Proc1 . | Proc2 . | ||||||||||
| Mean | 0.0 | 0.0 | 0.0 | |||||||||
| SD | 0.0 | 0.0 | 0.0 | |||||||||
| Min | 0.0 | 0.0 | 0.0 | |||||||||
| Max | 0.1 | 0.1 | 0.1 | |||||||||
Norfolk baseline 1 month before entrance to study (February 1999), Denver baseline from March 1999. Complete blood count (CBC) and differential performed on day of apheresis and whole blood collections (Proc1 and Proc2). No significant changes from baseline were observed, except for small increase in mean lymphocyte count (P= .037).
Subjects entering the collection phase in the spring of 1999 were presented with a questionnaire, which asked about various aspects of allergic status and treatment. There were no differences between the Norfolk and Denver groups for known allergies or receipt of allergy shots (P < .16). No Denver subject and 33% of Norfolk subjects were being seen by an allergist (P = .039). None of these categories were significantly correlated with the maximum level of CMV titer observed in the subject (P > .5).
To determine if reactivation may be caused or associated with increased systemic cytokine levels, a panel of cytokines (IL-2, IL-4, IL-10, and TNF-α) was run on retained subject plasma from baseline (before reactivation) and during reactivation periods for a subset of subjects who reactivated and a control cohort of non-reactivators from both locations. No increases in any of these cytokines were observed for either group, with all cytokine concentrations remaining within the normal ranges (data not shown).
Determination of competent virus via mRNA detection and tissue culture are shown in Table 5. Messenger RNA was detected in 8 of 19 subjects. Four of 19 subjects had positive tissue culture by day 10. Both transcription and positive culture observations were associated with higher levels of genome copies.
Determination of competent virus via messenger RNA detection and tissue culture
| Subject5-150 . | Location . | Day 0 . | Day 5 . | Tissue culture . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CMV (GE/mL) . | RNA IE . | RNA POL . | RNA late . | CMV (GE/mL) . | RNA IE . | RNA POL . | RNA late . | 3 d . | 10 d . | ||
| H1 | Denver | 1 000 000 | POS | 1 000 000 | POS | POS | POS | POS | |||
| N2 | Denver | 1 000 000 | POS | POS | POS | 1 000 000 | POS | POS | POS | POS | POS |
| O2 | Denver | 110 800 | POS | 100 | |||||||
| Q2 | Denver | 110 000 | POS | 351 400 | |||||||
| D | Norfolk | 105 000 | POS | 468 200 | POS | POS | |||||
| J1 | Denver | 97 300 | POS | 426 300 | |||||||
| M1 | Denver | 93 000 | 105 200 | ||||||||
| B1 | Norfolk | 64 900 | POS | POS | 506 200 | POS | POS | ||||
| R1 | Denver | 60 100 | 73 100 | ||||||||
| K1 | Norfolk | 49 000 | 52 800 | ||||||||
| O1 | Denver | 41 200 | 100 | ||||||||
| A1 | Norfolk | 36 100 | 53 300 | ||||||||
| F | Norfolk | 33 100 | 41 600 | ||||||||
| E | Norfolk | 26 800 | 69 100 | ||||||||
| A2 | Norfolk | 24 200 | 62 800 | ||||||||
| P1 | Denver | 16 700 | 56 100 | POS | |||||||
| C1 | Norfolk | 16 600 | 31 900 | ||||||||
| Q1 | Denver | 13 400 | 47 900 | ||||||||
| J2 | Denver | 12 300 | 106 100 | ||||||||
| C2 | Norfolk | 11 700 | 34 300 | ||||||||
| B2 | Norfolk | 11 200 | 24 700 | ||||||||
| R2 | Denver | 9 400 | 16 800 | ||||||||
| G | Denver | 5 700 | 21 800 | ||||||||
| N1 | Denver | 4 200 | POS | 31 300 | |||||||
| H2 | Denver | 4 100 | 105 500 | ||||||||
| L1 | Denver | 3 200 | 19 500 | ||||||||
| L2 | Denver | 2 700 | 23 200 | ||||||||
| I | Denver | 2 600 | 3 800 | ||||||||
| S | Denver | 2 000 | 10 200 | ||||||||
| M2 | Denver | 1 600 | 4 700 | ||||||||
| K2 | Norfolk | 1 382 | 10 600 | ||||||||
| P2 | Denver | 100 | 3 200 | ||||||||
| Subject5-150 . | Location . | Day 0 . | Day 5 . | Tissue culture . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CMV (GE/mL) . | RNA IE . | RNA POL . | RNA late . | CMV (GE/mL) . | RNA IE . | RNA POL . | RNA late . | 3 d . | 10 d . | ||
| H1 | Denver | 1 000 000 | POS | 1 000 000 | POS | POS | POS | POS | |||
| N2 | Denver | 1 000 000 | POS | POS | POS | 1 000 000 | POS | POS | POS | POS | POS |
| O2 | Denver | 110 800 | POS | 100 | |||||||
| Q2 | Denver | 110 000 | POS | 351 400 | |||||||
| D | Norfolk | 105 000 | POS | 468 200 | POS | POS | |||||
| J1 | Denver | 97 300 | POS | 426 300 | |||||||
| M1 | Denver | 93 000 | 105 200 | ||||||||
| B1 | Norfolk | 64 900 | POS | POS | 506 200 | POS | POS | ||||
| R1 | Denver | 60 100 | 73 100 | ||||||||
| K1 | Norfolk | 49 000 | 52 800 | ||||||||
| O1 | Denver | 41 200 | 100 | ||||||||
| A1 | Norfolk | 36 100 | 53 300 | ||||||||
| F | Norfolk | 33 100 | 41 600 | ||||||||
| E | Norfolk | 26 800 | 69 100 | ||||||||
| A2 | Norfolk | 24 200 | 62 800 | ||||||||
| P1 | Denver | 16 700 | 56 100 | POS | |||||||
| C1 | Norfolk | 16 600 | 31 900 | ||||||||
| Q1 | Denver | 13 400 | 47 900 | ||||||||
| J2 | Denver | 12 300 | 106 100 | ||||||||
| C2 | Norfolk | 11 700 | 34 300 | ||||||||
| B2 | Norfolk | 11 200 | 24 700 | ||||||||
| R2 | Denver | 9 400 | 16 800 | ||||||||
| G | Denver | 5 700 | 21 800 | ||||||||
| N1 | Denver | 4 200 | POS | 31 300 | |||||||
| H2 | Denver | 4 100 | 105 500 | ||||||||
| L1 | Denver | 3 200 | 19 500 | ||||||||
| L2 | Denver | 2 700 | 23 200 | ||||||||
| I | Denver | 2 600 | 3 800 | ||||||||
| S | Denver | 2 000 | 10 200 | ||||||||
| M2 | Denver | 1 600 | 4 700 | ||||||||
| K2 | Norfolk | 1 382 | 10 600 | ||||||||
| P2 | Denver | 100 | 3 200 | ||||||||
Peripheral blood samples from 7 Norfolk subjects and 12 Denver on the day of CMV-DNAemia were further evaluated by RT-PCR forIE, POL, and late gene products on days 0 and 5 of culture. Tissue culture was performed by incubation of fibroblast with peripheral blood white cells for 18 hours and staining of fibroblast with monoclonal antibodies to IE protein and RT-PCR for IE gene products at days 3 and 10 after incubation.
See Table 1 for other abbreviations.
Number indicates first or second sample for subjects having samples from 2 separate dates tested.
Discussion
In this study, we have observed a seasonal variation in the level of CMV DNAemia that is associated with increases in environmental allergens, most notably pine tree pollen. This effect has been previously observed in the Norfolk area, but in this study confirmed in 2 locations over 2 allergy seasons, with CMV DNAemia varying from 0% to 100% in normal healthy CMV seropositive subjects. Various investigators have reported a low frequency of CMV DNAemia in serologically positive blood donors (Bitsch and coworkers15 0 of 100 [0%], Urushibara and colleagues16 0 of 155 [0%], Smith and coworkers17 7 of 86 [8%], and Krajden and associates18 0-8 of 101 [0%-8%]). More recently, Larsson and coworkers19 observed 41.4% (60 of 145) CMV DNAemia in seropositive blood donors. In a small cohort followed over time, Larsson also found that seropositive donors initially negative for CMV DNA converted to positive. The variety of specific preparation and amplification methods used in these studies may explain some of the variations among the groups. Our data show latent CMV may be at very low levels (< 100 GE/mL) in the peripheral blood of seropositive, healthy individuals, then reactivated resulting in extremely high, transient titers of CMV DNAemia (> 1 000 000 GE/mL). The triggering event of reactivation is not known. Hahn and colleagues20demonstrated in vitro reactivation of latently infected granulocyte-macrophage progenitors in the presence of interferon-γ or TNF-α, Taylor-Wiedeman21 stimulated reactivation in monocytes in vitro by inducing differentiation to monocyte-derived macrophages, and Soderberg-Naucler22 in response to allogeneic stimulation. Ohsawa and colleagues23demonstrated accelerated CMV replication in rats in response to in vivo stimulation with allogeneic spleen cells. We did not see a systemic increase in cytokines in contrast to results obtained by Humar and coworkers24 in bone marrow transplant recipients and Asanuma and associates25 in infants. This observation does not preclude local tissue or transient increases in response to environmental allergens.
We have also demonstrated by viral DNA transcription and in vitro infectivity that the increase in DNAemia can produce competent virus in some CMV seropositive individuals. We have not investigated these effects in serologically negative subjects, and do not know if there could be a similar phenomenon in such a cohort. However, others have reported detectable CMV DNA in seronegative individuals: Larsson 19 of 140 (13.6%)26 and Taylor-Weideman 3 of 9 (33%).27 Reactivation with transient DNAemia associated with competent virus may suggest a variation in infectivity risk for blood products from both seropositive and seronegative blood donors. Antibody response to a reactivation event was not reported in these studies nor was it determined in the present work. If development of antibody is delayed in response to reactivation events, a window period of potential infectivity could thus be present before a detectable rise in antibody occurs. Therefore, LR may also add another level of protection for blood products from seronegative donors. This suggests an area for further investigation.
Although we have shown that some of the subjects in this study with high CMV levels had competent virus in vitro, we cannot generalize this observation. Further, we have not shown that the resultant levels present after LR are infective or not infective in vivo. To our knowledge, a minimal in vivo infective dose for CMV has not been determined. Our own in vitro data suggest high levels are required before any competency can be demonstrated. One sample at 4200 GE/mL with detectable IE-mRNA did not culture out after 10 days nor did the mRNA persist. The residual CMV levels for the 3 LR methods (apheresis, filtered platelets, and filtered red cells) were pooled and evaluated as a log-normal distribution. This analysis suggests more than 99.9% of leukocyte-reduced products would be expected to be less than 500 GE/mL. This substantial reduction of viral load in leukocyte-reduced blood products supports the clinical efficacy in avoidance of CMV transmission observed in several studies, reviewed by Preiksaitis.28 However, the data presented in this study are not conclusive evidence that LR provides absolute protection against CMV infection for reasons mentioned above: sample size constraints on the estimated proportion of products with detectable CMV DNA (Table 3) and unknown minimal in vivo infective dose. One potential source of bias in this study was that the laboratory performing QA-PCR was not fully blinded to the source or identification of the study samples. We do not feel this is a significant risk to the results, however, because of the objective nature of the measurements; negative and positive controls were run with each batch as described previously, and samples were encoded and processed by different individuals within the laboratory.
The primary objective of this study was to test the difference in the effectiveness of 3 LR methods in reducing CMV levels. These methods are used widely for preparation of blood products for indications such as avoidance of nonhemolytic febrile transfusion reactions, alloimmunization, and immunomodulation. Several groups have attempted to subtype the residual WBCs in these blood products and have suggested that there are substantial differences between preparation methods based on their observations. This line of thought has been extended to suggest that these differences are great enough to result in significant differences in clinical outcomes. Residual WBC in leukocyte-reduced blood products are rare events and present substantial challenges in simply enumerating the WBC. The next level of subtyping offers even greater technical challenges. This is evidenced by the lack of repeatability between groups that have attempted this arduous task. For example, in Amicus apheresis products, Tiulzi et al29 found 5 granulocytes/mL, 31 monocytes/mL, and 217 B cells/mL, whereas Johnson and colleagues30 observed 21 granulocytes/mL, 2 monocytes/mL, and 873 B cells/mL.
For CMV transmission, this trail of logic requires further assumptions regarding the location and activity of latent virus. Although substantial progress has been made over the past few years in localizing predominant cell types harboring the latent virion31 and the configuration of the genome,32 whether these cell types are the only site of latency or competent virus is still controversial. This study attempted to avoid issues of rare WBC event isolation and subtyping as well as assumptions regarding viral vector location. Instead, we looked specifically for CMV genome in the aggregate final product. Quantitative molecular biology methods were enhanced by extracting nucleic acids from large volumes of final products, with a resulting lower limit of detection of 4 GE/mL. We found a substantial reduction in viral genome load during periods of reactivation in the donor, with more than 99.9% of leukocyte-reduced products expected to have less than 500 GE/mL of CMV DNA. Further, we found apheresis platelets, filtered platelets prepared from whole blood, and filtered RBCs prepared from whole blood to be equivalent in residual CMV DNA in the final product.
The authors would like to thank the apheresis and research staffs of Bonfils Memorial Blood Center and the American Red Cross, Mid-Atlantic Region for their contribution to this work. Carol Afflerbach, Deanna McNeil, Sherrie Sawyer, Bonnie Zishka, and Yvonne Runyan made special contributions that made this logistically difficult study possible. Renee Louis performed the cytokine assays, and Art Hamstra performed WBC counting. Robert Schuyler made very helpful suggestions on the final manuscript.
L.J.D. and T.V.B. are employed by Gambro BCT.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Larry J. Dumont, 10811 Collins Ave, Lakewood, CO 80215; e-mail: larry.dumont@gambrobct.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal