Pyrimidine 5′ nucleotidase (P5′N-1) deficiency is an autosomal recessive condition causing hemolytic anemia characterized by marked basophilic stippling and the accumulation of high concentrations of pyrimidine nucleotides within the erythrocyte. It is implicated in the anemia of lead poisoning and is possibly associated with learning difficulties. Recently, a protein with P5′N-1 activity was analyzed and a provisional complementary DNA (cDNA) sequence published. This sequence was used to study 3 families with P5′N-1 deficiency. This approach generated a genomic DNA sequence that was used to search GenBank and identify the gene for P5′N-1. It is found on chromosome 7, consists of 10 exons with alternative splicing of exon 2, and produces proteins 286 and 297 amino acids long. Three homozygous mutations were identified in this gene in 4 subjects with P5′N-1 deficiency: codon 98 GAT→GTT, Asp→Val (linked to a silent polymorphism codon 92, TAC→TAT), codon 177, CAA→TAA, Gln→termination, and IVS9-1, G→T. The latter mutation results in the loss of exon 9 (201 bp) from the cDNA. None of these mutations was found in 100 normal controls. The DNA analysis was complicated by P5′N-1 pseudogenes found on chromosomes 4 and 7. This study is the first description of the structure and location of the P5′N-1 gene, and 3 mutations have been identified in affected patients from separate kindreds.
Introduction
Pyrimidine 5′ nucleotidase (P5′N-1, also known as uridine-5′-monophosphate hydrolase-1) catalyzes the dephosphorylation of the pyrimidine 5′ monophosphates UMP and CMP to the corresponding nucleosides. A deficiency of this enzymatic activity was first identified by Valentine et al1,2 in erythrocyte stroma while investigating patients with hemolytic anemia characterized by marked basophilic stippling. Initial studies showed very high concentrations of what were assumed to be adenine nucleotides in the erythrocytes, but these were later found to be pyrimidine nucleotides; the red blood cells (RBCs) also contained high levels of glutathione and reduced activity of ribose-phosphate pyrophosphokinase. Studies on 3 additional kindreds with hemolytic anemia and basophilic stippling demonstrated absent or markedly reduced pyrimidine 5′ nucleotidase activity in their RBCs.3 Reports of 40 patients with this condition have been published, with presumably large numbers undetected. However, because of the lack of a simple and reliable test for carriers, the exact prevalence of the condition is unknown. Reported numbers of homozygotes suggest that it is the third most common RBC enzymopathy—after glucose-6-phosphate dehydrogenase and pyruvate kinase deficiency—causing hemolysis.4 Although the first 6 patients reported were all female, a number of affected males have been reported since then, and the pattern of inheritance is typical of an autosomal recessive disorder.5
Additional studies have suggested that there are 2 isozymes of P5′N in RBCs, one with a preference for UMP and CMP, referred to as P5′N-1, and one able to hydrolyze deoxy-pyrimidine nucleotide monophosphates (P5′N-2).6,7 These are not separable by electrophoresis in humans but have distinct kinetic properties and genetics. P5′N-2 has been assigned to the long arm of chromosome 17 by studying human–mouse somatic cell hybrids.8 The most convincing evidence for the existence of 2 isozymes arises from studies of patients with hemolytic anemia, in whom P5′N-1 activity is greatly reduced but P5′N-2 is normal.7 Purification and partial protein sequencing of P5′N-1 from RBCs led to the identification and cloning of the complementary DNA (cDNA) and the expression of the recombinant enzyme in Escherichia coli.9,10 The protein consists of 286 amino acids and is identical to a previously identified lupus inclusion protein, p36, though the significance of this is unclear. A mammalian 5′(3′)-deoxyribonucleotidase with similar properties to P5′N-2, though lacking its apparent phosphotransferase activity, has also been cloned.11 12 Although P5′N-1 and P5′N-2 are not separable by electrophoresis in humans, the 2 proteins show no homology. We used the putative cDNA sequence for P5′N-1 to screen families known to have hemolytic anemia caused by pyrimidine 5′ nucleotidase deficiency for causative mutations.
Materials and methods
Case histories
Four patients with P5′N deficiency are included in this study, including one pair of siblings.
Norwegian family.
In the early 1980s, a brother and sister from Norway were found to have P5′N deficiency with typical hemolytic anemia and basophilic stippling.13 It was unusual that both siblings had intravascular hemolysis, urinary iron loss, iron deposition in the kidneys, and iron deficiency. This has not been noted in other published reports of P5′N deficiency. Their parents were hematologically normal and had a common ancestor 5 generations earlier. Three other children were found to be unaffected. Both affected siblings are now in their late twenties and maintain hemoglobin levels of approximately 10 g/dL with no significant complications from their chronic hemolysis.
South African 1.
A South African girl was examined in early childhood for failure to thrive and typical hemolytic anemia with basophilic stippling, and she was found to have P5′N-1 deficiency. She was adopted, and it is unknown whether her parents were related in any way. She appeared to be of mixed-race origin. After diagnosis, she was lost to follow-up.
South African 2.
A 55-year-old white farmer was referred with anemia, which was found to be of hemolytic origin and associated with low P5′N levels and intra-erythrocytic accumulation of pyrimidine nucleotides. He was most recently seen in his sixties and continues well without any significant problems related to P5′N-1 deficiency.
Hematologic analysis
Venous blood was collected with informed consent from patients and relatives. Automated full blood counts and manual reticulocyte counts on blood stained supravitally with brilliant cresyl blue were taken.
Enzyme and nucleotide studies
Pyrimidine 5′ nucleotidase in hemolysates was determined as P5′N-1 and P5′N-2 activities14 by a method using high-performance liquid chromatography with UMP and deoxy-UMP as respective substrates15 or as P5′N-1 by measuring inorganic phosphate release by a colorimetric method after incubation with CMP.16 RBC nucleotides were quantitated spectrophotometrically.17
DNA analysis
Epstein-Barr virus–transformed lymphoblastoid cell lines were established from fresh EDTA blood of all patients and from the 2 parents and 3 siblings of the Norwegian family. Genomic DNA was extracted from EDTA-anticoagulated blood or cultured lymphoblasts of patients, family members, and 100 anonymous healthy white controls (QIAamp, DNA blood kit; Qiagen, Crawley, United Kingdom). Control cell lines were set up from controls without hemolytic anemia. Messenger RNA (mRNA) was extracted from the cell lines using standard techniques, and first-strand cDNA was synthesized using oligo dT priming and MMLV reverse transcriptase (Gibco-BRL Life Technologies, Paisley, United Kingdom). The coding region of P5′N-1 cDNA was amplified using nested primers in 2 rounds of amplification (Table1). Polymerase chain reaction (PCR) products were gel purified (Qiaex II Gel Extraction Kit; Qiagen) and directly sequenced (dRhodamine Terminator cycle sequencing kit; PE Applied Biosystems, Warrington, United Kingdom) on an ABI377 automated fluorescent DNA sequencer (PE Applied Biosystems). Reticulocyte mRNA was extracted from control erythrocytes using standard methods and subjected to an identical nested PCR protocol.18Reticulocyte mRNA was unavailable from the affected families. To define partially the genomic structure of the P5′N-1 gene, combinations of cDNA-specific primers were used to amplify genomic DNA across introns. Gel-purified PCR products were sequenced, and primers were designed to amplify identified exons from genomic DNA (Table 1). When mutations were identified, they were screened for in 100 normal controls using restriction endonuclease digestion of PCR-amplified exons (enzymes from New England Biolabs, Hitchin, United Kingdom). The silent codon 92 TAC→TAT mutation was screened for by PCR amplification with a mismatched primer that creates a DdeI site in the presence of the TAC allele.
PCR primers used in this study
| Primer . | Sequence . | Comment . |
|---|---|---|
| cDNA 165 forward | GGCGGGGGTGGTGCTGGCTC | cDNA amplification outer primers |
| cDNA 1232 reverse | GGTGGAGAAAAGGAGCTTCCAG | |
| cDNA 180 forward | GGTGGAGAAAAGGAGCTTCCAG | cDNA amplification nested primers |
| cDNA 1174 reverse | GTTATACAAAGGGAACACTTCGAGAG | |
| cDNA 315 forward | TATCAAAGGAGGAGCTGCCAAAC | cDNA sequencing |
| cDNA 443 reverse | AATAGCGTAGTATTTTTCCTTTAGTTGC | cDNA sequencing |
| cDNA 452 forward | GAATGTAGAAAAAAGTTATTGCAACTAAAG | cDNA sequencing |
| cDNA 552 forward | TATACTAAATCACATGGTTTGCTTGTTC | cDNA sequencing |
| cDNA 705 forward | GAATCGGCGATGTACTAGAGGAAG | cDNA sequencing |
| cDNA 734 reverse | ATAAAATTGGACACAACTTTGACATTG | cDNA sequencing |
| cDNA 849 forward | ATGATGGTGCCTTGAGGAATACAG | cDNA sequencing |
| cDNA 928 reverse | ATGTGCTCAACATTGGCCACT | cDNA sequencing |
| Exon 10 forward (with cDNA 1232 reverse) | TTGCCCAAGAGATCTAACAACGAG | Genomic PCR exon 10 |
| Exon 9 forward | AATATGATGAAATAATAGTGACATCAATTAAG | Genomic PCR exon 9 |
| Exon 9 reverse | CATCACTACACTCTAGCCTCGGTAAC | |
| Exon 8 forward | TGAAATGAGAATATGAGTGAGCCATAG | Genomic PCR exon 8 |
| Exon 8 reverse | TATCGGCTTGGCCTAATTTCTG | |
| Exon 7 forward | ATGCTTTGGAATAAAGATAATAATTTTAG | Genomic PCR exon 7 |
| Exon 7 reverse | TTTAAAAAGTACAACTGACTACATAAATAGC | |
| Exon 6 forward | TTTATTGTTGGTTTTTACTCATCTGAAAG | Genomic PCR exon 6 |
| Exon 6 reverse | TTATTAATCTGTTTGTTTGCAATACAGG | |
| Codon 92 mismatch reverse (with exon 6 forward) | GTAAGAACAGGATCAACTTCAATAGCTTA | Mismatch primer creates a DdeI site in the presence of the codon 92 |
| (TAC) polymorphism. The mismatch is underlined. | ||
| Pseu 226 forward (with cDNA 1232 reverse) | CAAGAACCCTACAAGAGTAGAAGAAATG | PCR of pseudogene from genomic DNA |
| Pseu 902 reverse (with cDNA 165 forward) | CATCCACTCTATCATTTAGATATCCAAC | PCR of pseudogene from genomic DNA |
| Primer . | Sequence . | Comment . |
|---|---|---|
| cDNA 165 forward | GGCGGGGGTGGTGCTGGCTC | cDNA amplification outer primers |
| cDNA 1232 reverse | GGTGGAGAAAAGGAGCTTCCAG | |
| cDNA 180 forward | GGTGGAGAAAAGGAGCTTCCAG | cDNA amplification nested primers |
| cDNA 1174 reverse | GTTATACAAAGGGAACACTTCGAGAG | |
| cDNA 315 forward | TATCAAAGGAGGAGCTGCCAAAC | cDNA sequencing |
| cDNA 443 reverse | AATAGCGTAGTATTTTTCCTTTAGTTGC | cDNA sequencing |
| cDNA 452 forward | GAATGTAGAAAAAAGTTATTGCAACTAAAG | cDNA sequencing |
| cDNA 552 forward | TATACTAAATCACATGGTTTGCTTGTTC | cDNA sequencing |
| cDNA 705 forward | GAATCGGCGATGTACTAGAGGAAG | cDNA sequencing |
| cDNA 734 reverse | ATAAAATTGGACACAACTTTGACATTG | cDNA sequencing |
| cDNA 849 forward | ATGATGGTGCCTTGAGGAATACAG | cDNA sequencing |
| cDNA 928 reverse | ATGTGCTCAACATTGGCCACT | cDNA sequencing |
| Exon 10 forward (with cDNA 1232 reverse) | TTGCCCAAGAGATCTAACAACGAG | Genomic PCR exon 10 |
| Exon 9 forward | AATATGATGAAATAATAGTGACATCAATTAAG | Genomic PCR exon 9 |
| Exon 9 reverse | CATCACTACACTCTAGCCTCGGTAAC | |
| Exon 8 forward | TGAAATGAGAATATGAGTGAGCCATAG | Genomic PCR exon 8 |
| Exon 8 reverse | TATCGGCTTGGCCTAATTTCTG | |
| Exon 7 forward | ATGCTTTGGAATAAAGATAATAATTTTAG | Genomic PCR exon 7 |
| Exon 7 reverse | TTTAAAAAGTACAACTGACTACATAAATAGC | |
| Exon 6 forward | TTTATTGTTGGTTTTTACTCATCTGAAAG | Genomic PCR exon 6 |
| Exon 6 reverse | TTATTAATCTGTTTGTTTGCAATACAGG | |
| Codon 92 mismatch reverse (with exon 6 forward) | GTAAGAACAGGATCAACTTCAATAGCTTA | Mismatch primer creates a DdeI site in the presence of the codon 92 |
| (TAC) polymorphism. The mismatch is underlined. | ||
| Pseu 226 forward (with cDNA 1232 reverse) | CAAGAACCCTACAAGAGTAGAAGAAATG | PCR of pseudogene from genomic DNA |
| Pseu 902 reverse (with cDNA 165 forward) | CATCCACTCTATCATTTAGATATCCAAC | PCR of pseudogene from genomic DNA |
5′-ends of primers are numbered according to the complementary DNA (cDNA) sequence in Figure 2.
Results
Hematologic analysis
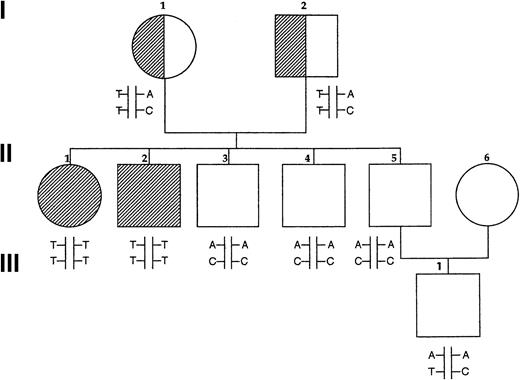
Basic clinical and hematologic data are summarized in Table2. Results of P5′N assays for the Norwegian family are shown in Figure 1and Table 3. Both affected siblings had very low levels of P5′N-1 levels with normal P5′N-2 levels. The parents and 3 healthy siblings had P5′N-1 levels varying upward from the lower end of the normal range, making it difficult to accurately identify carriers of the condition. P5′N-2 levels were all normal, and we have previously found that the P5′N-1/P5′N-2 ratio is a better way of detecting reduced P5′N-1 levels; a ratio of 0.7 or less is significant.19 This ratio shows that both parents are carriers and that the 3 healthy siblings have normal P5′N-1 activity. Analysis of a child of one of the healthy siblings also suggests that he is not a carrier. Both affected siblings were found to have a massive accumulation of intra-erythrocytic pyrimidine nucleotides at the time of diagnosis.13
Clinical features of subjects with P5′N-1 deficiency
| . | Sex . | Age at presentation . | Presenting features . | Bilirubin μM . | Hemoglobin g/dL . | Reticulocytes (%) . |
|---|---|---|---|---|---|---|
| Norway 1 | M | 4 d | Neonatal jaundice | 63 | 10.9 | 8 |
| Norway 2 | F | 7 y | Dark urine, anemia | 47 | 10.7 | 9 |
| S Africa 1 | F | 2 y | Pallor, chest infections | 47 | 8.1 | 6.7 |
| S Africa 2 | M | 55 y | Anemia | Unknown | Unknown | Unknown |
| Normal range | 0-15 | F 11-15 | < 1 | |||
| M 12-16 |
| . | Sex . | Age at presentation . | Presenting features . | Bilirubin μM . | Hemoglobin g/dL . | Reticulocytes (%) . |
|---|---|---|---|---|---|---|
| Norway 1 | M | 4 d | Neonatal jaundice | 63 | 10.9 | 8 |
| Norway 2 | F | 7 y | Dark urine, anemia | 47 | 10.7 | 9 |
| S Africa 1 | F | 2 y | Pallor, chest infections | 47 | 8.1 | 6.7 |
| S Africa 2 | M | 55 y | Anemia | Unknown | Unknown | Unknown |
| Normal range | 0-15 | F 11-15 | < 1 | |||
| M 12-16 |
Pedigree of Norwegian family.
Fully shaded areas show homozygotes for P5′N-1 deficiency, and half-shaded symbols show heterozygotes, based on P5′N-1/P5′N-2 ratios. Letters show the results of genotyping the P5′N-1. Upper letters refer to mutations of codon 98 (A, wild type; T, mutated) and lower letters refer to codon 92 (C, wild type; T, mutated). Reduced P5′N-1 activity segregates with the codon 98 T mutation and with the T/T haplotype.
Pedigree of Norwegian family.
Fully shaded areas show homozygotes for P5′N-1 deficiency, and half-shaded symbols show heterozygotes, based on P5′N-1/P5′N-2 ratios. Letters show the results of genotyping the P5′N-1. Upper letters refer to mutations of codon 98 (A, wild type; T, mutated) and lower letters refer to codon 92 (C, wild type; T, mutated). Reduced P5′N-1 activity segregates with the codon 98 T mutation and with the T/T haplotype.
Activities of P5′N-1 (NR 9-20 μmol/h per g Hb), P5′N-2 (NR 7-28 μmol/h per g Hb), and P5′N-1/P5′N-2 ratio in members of the Norwegian family
| . | I.1 . | I.2 . | II.1 . | II.2 . | II.3 . | II.4 . | II.5 . | III.1 . |
|---|---|---|---|---|---|---|---|---|
| P5′N-1 | 3.5 | 6.4 | 0.4 | 0.5 | 5.5 | 7.9 | 7.7 | 9.6 |
| P5′N-2 | 7.8 | 9.4 | 9.2 | 7.7 | 7.0 | 8.1 | 8.5 | 8.3 |
| P5′N-1/P5′N-2 | 0.45 | 0.68 | .04 | .06 | 0.79 | 0.98 | 0.91 | 1.1 |
| . | I.1 . | I.2 . | II.1 . | II.2 . | II.3 . | II.4 . | II.5 . | III.1 . |
|---|---|---|---|---|---|---|---|---|
| P5′N-1 | 3.5 | 6.4 | 0.4 | 0.5 | 5.5 | 7.9 | 7.7 | 9.6 |
| P5′N-2 | 7.8 | 9.4 | 9.2 | 7.7 | 7.0 | 8.1 | 8.5 | 8.3 |
| P5′N-1/P5′N-2 | 0.45 | 0.68 | .04 | .06 | 0.79 | 0.98 | 0.91 | 1.1 |
Interpretation of ratios: P5′N-1/P5′N-2 ≤ 0.1, homozygous deficiency; 0.1 < P5′N-1/P5′N-2 ≤ 0.7, carriers of P5′N-1 deficiency; P5′N-1/P5′N-2 > 0.7, normal.
Fresh blood was unavailable from both South African patients, but assays at the time of diagnosis showed P5′N activities of 1.8 and 0.4 μmol/h per g Hb, respectively (normal range, 6.9-10.7). These assays measured the generation of inorganic phosphate.
DNA analysis
Sequencing of P5′N-1 nested PCR product from controls and patients showed multiple heterozygous nucleotide substitutions and a 1-bp insertion compared to the published sequence. Most of the sequence changes are predicted to produce nonconservative amino acid changes, and the insertion produces a frame shift and a premature termination of the predicted protein. The presence of these enzyme-inactivating mutations in patients and controls is consistent with the coamplification of a processed P5′N-1 pseudogene from genomic DNA contaminating the RNA preparations. Pseudogene-specific primers were used to amplify and sequence the pseudogene in 2 overlapping segments from genomic DNA. A similarity search of the GenBank databases showed a match between the pseudogene and 169 bp of a contig derived from chromosome 7p14-p15 (NT_002053, locus AC007312, clone RP11-349E11), corresponding to the 5′ end of the P5′N-1 cDNA and consistent with partial genomic duplication of the P5′N-1 gene. The cDNA sequence showed extensive homology to elements of a working draft sequence of the chromosome 4 clone, RP11-778G8, and the absence of intervening sequences is characteristic of a processed pseudogene. Amplification of genomic DNA using cDNA primers yielded a single homozygous sequence corresponding to the chromosome 4 pseudogene. It is, therefore, likely that the complex sequence produced by the nested reverse transcription–PCR is produced by amplification of both the P5′N-1 mRNA and contaminating DNA from the processed pseudogene. The chromosome 4 pseudogene sequence contained 2 restriction sites not present in the P5′N-1 cDNA sequence, a PvuII site at nucleotide 353 and anHpaII site at nucleotide 606. Digestion of the nested PCR product with one of these before sequencing the pseudogene and produced a homozygous sequence corresponding to the published sequence for P5′N-1.
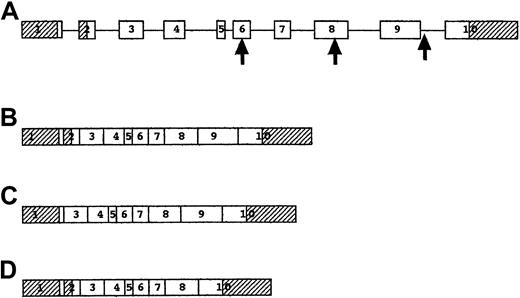
Two PCR products of slightly different sizes were amplified from control cell lines and reticulocytes. The smaller transcript was identical to the P5′N-1 cDNA published sequence,9 but the larger transcript contained a 55-bp insertion near the 5′ end. A selection of primers was used to generate intronic sequence from the 3′ end of the gene, and this information was used to search GenBank. Complete homology was found to a working draft of chromosome 7 clone RP11-162O1 (AC083863). Analysis of this revealed 10 exons with intron–exon boundaries adhering to the canonical AG-GT rule (Figure2). The 55-bp insertion noted in some transcripts corresponded to the second exon of the genomic DNA and was predicted to result in a protein with an additional 11 amino acids at the N-terminal (Figure 3).
Nucleotide sequence of the P5′N-1 gene.
The exonic sequence is shown in uppercase letters, and the intronic sequence is shown in lowercase letters. Predicted amino acid sequence is shown below the nucleotide sequence. The final codon of exon 1 acts as the initiator codon for the 286–amino acid form of P5′N-1, whereas the translation is initiated by the first codon of exon 2 in the longer 297–amino acid form of the protein.
Nucleotide sequence of the P5′N-1 gene.
The exonic sequence is shown in uppercase letters, and the intronic sequence is shown in lowercase letters. Predicted amino acid sequence is shown below the nucleotide sequence. The final codon of exon 1 acts as the initiator codon for the 286–amino acid form of P5′N-1, whereas the translation is initiated by the first codon of exon 2 in the longer 297–amino acid form of the protein.
Structure and splicing of the P5′N-1 gene.
(A) Genomic structure of P5′N-1 gene. Exons are shown as boxes and introns as lines. Shaded exons represent the 5′ and 3′ untranslated regions. The first arrow shows the site of the codon 98 mutation, the second arrow shows the codon 177 mutation, and the third arrow shows the IVS9-1 mutation. (B) mRNA for 297–amino acid form of P5′N-1, including all the exons, though the initiator codon in exon 1 is inactive. (C) mRNA for 286–amino acid form of P5′N-1, with translation initiated by the last codon of exon 1 and with exon 2 spliced out. (D) mRNA resulting from IVS9-1, G→T mutation, with skipping of exon 9.
Structure and splicing of the P5′N-1 gene.
(A) Genomic structure of P5′N-1 gene. Exons are shown as boxes and introns as lines. Shaded exons represent the 5′ and 3′ untranslated regions. The first arrow shows the site of the codon 98 mutation, the second arrow shows the codon 177 mutation, and the third arrow shows the IVS9-1 mutation. (B) mRNA for 297–amino acid form of P5′N-1, including all the exons, though the initiator codon in exon 1 is inactive. (C) mRNA for 286–amino acid form of P5′N-1, with translation initiated by the last codon of exon 1 and with exon 2 spliced out. (D) mRNA resulting from IVS9-1, G→T mutation, with skipping of exon 9.
Sequencing of the P5′N-1 transcript from the affected Norwegian siblings showed 2 homozygous mutations, codon 92 TAC→TAT and codon 98 GAT→GTT. The codon 92 polymorphism was silent, but the alteration of codon 98 produced the nonconservative amino change of aspartate to valine. Both parents are obligate carriers and are heterozygous for the same mutation. The codon 98 mutation destroyed an MboI site. A 273-bp fragment of genomic DNA containing codon 98 was amplified, andMboI restriction enzyme analysis confirmed that the codon 98 and codon 92 mutations were linked and homozygous in the affected siblings and heterozygous in the parents. All 3 unaffected siblings were found to be homozygous for the wild type. The son of one of the unaffected siblings was heterozygous for the silent codon 92 polymorphism, and it is most likely that he inherited the TAT allele from his mother (Figure 1). The codon 98 GAT→GTT mutation was not found in 100 normal European controls, but the silent codon 92 polymorphism was present at an allele frequency of 0.29 in the same population.
The cDNA from South African patient 1 with P5′N-1 deficiency was found to be homozygous for a mutation in codon 177 CAA→TAA (glutamine to stop codon). It was predicted that this would produce a protein truncated by 165 residues. The mutation created an MseI restriction site. MseI restriction of the PCR-amplified genomic DNA confirmed the homozygosity of the mutation in this patient. The mutation was not found in 200 alleles amplified from controls.
A single PCR product of reduced size was amplified from cDNA of South African patient 2. Sequencing showed a 201-bp deletion corresponding to exon 9 of the genomic sequence, removing 67 amino acids from the predicted protein. Genomic sequencing of a region including exon 9 showed a normal-sized product but a homozygous T → G mutation in the highly conserved GT of the splice donor site of intron 9 (GTAAGT to GGAAGT). No cryptic splice sites were found within 250 bp of the mutated splice donor site, and the mutation therefore resulted in exon skipping. The mutation created an MnlI site and was not found in 100 healthy controls.
Discussion
In this study we report 2 novel findings. First, we identified a genomic DNA sequence corresponding to the previously published cDNA for P5′N-1. Second, we identified significant mutations of this genomic sequence in 3 families with P5′N-1 deficiency. This gene for P5′N-1 is found on chromosome 7 and consists of 10 exons. Two alternatively spliced cDNAs were identified and sequenced, predicting the synthesis of proteins 286 and 297 amino acids long. The shorter protein has been shown to have characteristics of P5′N-1,9 but the longer form is yet to be characterized. Both forms are expressed in reticulocytes and lymphocytes.
The presence of 2 P5′N-1 cDNAs in lymphoblastoid cell lines and reticulocytes is a consequence of alternative splicing of exon 2. The poly-pyrimidine tract of the intron 1 splice acceptor site is displaced from the conserved AG dinucleotide, and no branch site with homology to the consensus branch site sequence YNYTRAY could be identified within 100 bases of this splice site. This is characteristic of splice acceptors flanking alternatively spliced exons.20 It is interesting that 2 electrophoretic forms of P5′N-1 have been demonstrated in nucleated cells and RBCs,14 which is consistent with our finding that alternative splicing predicts 2 P5′N-1 isoforms. The effect of the addition of 11 amino acids on catalytic activity requires further investigation. It is possible that the isozymes are differentially expressed in various tissues, as has been shown for pyruvate kinase, in which differentially spliced forms of the enzyme are expressed in liver and RBCs.21
We identified 3 different homozygous mutations in unrelated patients with the clinical and biochemical features of P5′N-1 deficiency. Two of the mutations resulted in large deletions of the protein and would be expected to result in severe disruption of enzyme structure and function. The point mutation in the Norwegian siblings involved a nonconservative amino acid change, segregated with P5′N-1 activity in this family, and was not found in 100 healthy controls. Our findings thus establish a direct causal relation between mutation of the P5′N-1 gene and hemolytic anemia with basophilic stippling from RBC P5′N-1 deficiency.
The 286–amino acid form of the P5′N-1 enzyme has a predicted mass of 32.7 kd. It contains 5 cysteine residues, some of which may be implicated in the acquired P5′N-1 deficiency associated with lead poisoning22 and the oxidative stress of β-thalassemia23; interestingly, these cysteine residues all occurred within 70 amino acids of the NH2-amino end of the protein. The tertiary structure of the protein is not yet known, but predictions of secondary structure suggest that it is a globular protein consisting of approximately 30% α helices and 26% extended strands.24 It is notable that this protein is unlike other nucleotidases, particularly the 5′(3′)-deoxyribonucleotidase thought to be responsible for P5′N-2 activity.11 As previously reported, the P5′N-1 protein is apparently identical to p36, a protein found in lupus inclusion bodies in response to interferon-α treatment. The function of p36 protein is obscure, and the relevance of the identity between the 2 apparently unrelated proteins is not yet known.9
Similarity searches of the GenBank EST database revealed matches with a number of partial mouse cDNA sequences. A consensus Mus musculus polypeptide derived from these sequences shows 95% homology with the human P5′N-1. Searches identified 3 other homologous proteins of unknown function (Figure 4). Interestingly, the Asp98, mutated in the Norwegian siblings, is conserved in the CG3362 gene product of Drosophila melanogaster.
Amino acid sequence homology of human P5′N-1 to similar proteins.
Dash indicates gaps in the aligned sequences, and x indicates amino acids identical to those in the equivalent position of the wild-type human protein. Residue Asp98 (boxed) is mutated to Val in the Norwegian family. Aligned sequences are Mus musculus (derived from EST databases), D. melanogaster (CG3362 gene product), Caenorhabditis elegans (hypothetical protein F25B5.3), and Arabidopsis thaliana (AAC67350.1).
Amino acid sequence homology of human P5′N-1 to similar proteins.
Dash indicates gaps in the aligned sequences, and x indicates amino acids identical to those in the equivalent position of the wild-type human protein. Residue Asp98 (boxed) is mutated to Val in the Norwegian family. Aligned sequences are Mus musculus (derived from EST databases), D. melanogaster (CG3362 gene product), Caenorhabditis elegans (hypothetical protein F25B5.3), and Arabidopsis thaliana (AAC67350.1).
The hematologic phenotype of P5′N-1 deficiency is well defined—moderate hemolytic anemia, jaundice, splenomegaly, and marked basophilic stippling. Blood transfusions are rarely necessary, and splenectomy has generally given little benefit,1,3,4,25though experience is limited. The enzyme deficiency has been linked to learning difficulties of variable severity in 7 patients, including 3 Peruvian siblings.26,27 The significance of this association is unclear but can be further explored now that the molecular basis of this disease has been elucidated. There is also evidence that P5′N-1 deficiency can interact with hemoglobin E to produce marked hemolytic anemia,28 raising the possibility that heterozygosity for P5′N-1 deficiency is one of the unidentified factors that contribute to the marked variability seen in HbE/β thalassemia and other forms of thalassemia.29 This possibility can now be investigated by DNA analysis. Previous studies were made difficult by the acquired deficiency of P5′N-1 associated with β thalassemia trait.19,30 It is also possible that genetic analysis will identify atypical forms of P5′N-1 deficiency that may not produce the classical phenotype but could explain some of the many cases of inherited hemolysis with no identifiable cause.5 Database searches show expression of P5′N-1 cDNA in various tumors (lung, ovary, colon, bladder), fetal tissues (lung, heart, spleen, liver), adult testis, and brain. This suggests that P5′N-1 has a fundamental role in pyrimidine metabolism and raises questions about why the phenotype is largely confined to the erythron.
We thank Professor Sir David Weatherall, Professor John Clegg, and Dr J. T. Reilly for advice and Sue Butler for establishing some of the cell lines. We also thank Professor Sverre O. Lie for helping to prepare the manuscript.
Supported in part by CNR Target Project on Biotechnology and by EC grant BMH4-CT98-3079 (A.M.M.).
Martin Seip died during the preparation of this manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David C. Rees, Department of Haematology, Royal Hallamshire Hospital, Glossop Rd, Sheffield, S10 2JF, United Kingdom; e-mail: david.rees@csuh.nhs.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal