Abstract
The absence of immunoglobulin (Ig) expression in B-cell–derived Hodgkin and Reed-Sternberg (HRS) cells of classical Hodgkin disease (cHD) was initially suggested to be caused by crippling mutations in the Ig promoter or coding region. More recent investigations have, however, challenged this concept. This study addressed the role of mutations in the Ig promoter region in HRS cells. Nine cases of cHD and 3 B-cell–derived HD lines were analyzed for mutations in the TATA box and octamer motif of the Ig promoter. Mutations in the octamer motif were found in only 1 of the 9 cases and in 1 of the 3 HD cell lines (L1236). Furthermore, in all cases either a complete lack or strong reduction in the expression of the Oct2 transcription factor and the BOB.1/OBF.1 coactivator were found. The relevance of the rare promoter mutations was investigated by assaying the activity of Ig promoter reporter constructs transfected into the HD cell line L1236, which harbors a mutated octamer motif. These Ig reporter constructs were completely inactive in L1236 cells; however, their activity could be reconstituted by the cotransfection of a BOB.1/OBF.1 expression vector. The additional transfection with an Oct2 expression vector did not further enhance the Ig promoter activity. The conclusions drawn from these results are that crippling mutations in the Ig promoter and coding region are not the sole cause for the lack of Ig expression in HRS cells and that defects in the transcription machinery such as absence of BOB.1/OBF.1 are more important for this phenomenon.
Introduction
It has recently been demonstrated that classical Hodgkin disease (cHD) is, in approximately 98% of cases, a B-cell lymphoma,1 and in about 2% of cases a T-cell lymphoma.2 In light of these findings it is still unaccountable why B-cell–type Hodgkin and Reed-Sternberg (HRS) cells do not show any (or show only a very low level of) transcription of the immunoglobulin (Ig) light (L)1,3,4-6 and heavy (H) chain genes,1 although they contain rearranged IgL and IgH genes1,7There are 4 possible explanations for this phenomenon: (1) crippling mutations within the V region gene render the coding sequence nonfunctional,8 (2) crippling mutations in the octamer motif of the Ig promoter or other regulatory sequences inactivate the Ig transcription,9 (3) defects in Ig transcription factors prevent Ig gene transcription,1 or (4) any combination of these. In a recent study1 we demonstrated that crippling mutations in the Ig coding region are absent in the majority of cases, making it unlikely that crippling mutations within the V region gene are generally responsible for the lack of Ig gene transcription in HRS cells. We also demonstrated that the 2 HD-derived cell lines, KM-H2 and L428, have largely lost their Ig gene transcription ability, supporting the view that the Ig transcription machinery might indeed be defective in HRS cells. Support for a role of alterations in the Ig octamer-binding region came from work by Jox and coworkers9 in which a mutation in the octamer region of the HD-derived cell line L1236 was reported, raising the possibility that mutations in the Ig gene regulatory element might contribute to the blockage of Ig transcription in HRS cells.10 11
To clarify the role of alterations in the Ig regulatory sequences conclusively, we investigated the TATA box and the octamer motif of the Ig promoters in single HRS cells isolated from 9 HD cases with functional (n = 6) and nonfunctional (n = 3) Ig genes and with absence of Ig expression. Furthermore, in addition to the Ig promoter region, we studied the intronic Eμ enhancer in the 3 HD cell lines with rearranged Ig genes. Moreover, these cell lines were also analyzed for expression of the Ig transcription factors Oct1, Oct2, and BOB.1/OBF.1 as well as for mutations in the coding regions of these factors. Finally, the activity of Ig promoter–driven reporter constructs or synthetic octamer–dependent reporters transfected into L1236 cells with or without cotransfection of BOB.1/OBF.1 and/or Oct2 was determined. The results obtained from these experiments show that mutations in the Ig regulatory motifs are rare and do not significantly contribute to the absence of Ig expression. Instead, the absence of BOB.1/OBF.1 appears to be the most decisive cause for the lack of Ig expression in classical HD.
Materials and methods
Cell culture and transfection
All cell lines (Namalwa, Raji, L1236, KM-H2, L428) were grown in RPMI 1640 medium (Life Technologies, Karlsruhe, Germany) supplemented with 10% fetal bovine serum (FCS), antibiotics,l-glutamate, and 50 μM β-mercaptoethanol at 37°C and 5% CO2. For electroporation 5 × 106 cells were pelleted and resuspended in a final volume of 300 μL RPMI medium supplemented with 10% FCS. Fifteen micrograms of the respective luciferase reporter plasmid12 and 100 ng pRL-TK (Promega, Mannheim, Germany) were added to the cells. The mixture was transferred to a cuvette with a gap of 0.4 cm and electroporated at 230 V and 950 μF with a Biorad Gene Pulser (Biorad, München, Germany). Cells were immediately transferred to a Petri dish containing 10 mL RPMI/10% FCS and incubated at 37°C in 5% CO2 for 20 hours. The cells were harvested and the lysates were analyzed with the dual-luciferase reporter assay system (Promega). All luciferase values were calculated against the renilla-luciferase values to correct for different transfection efficiencies. The details of the various constructs are described elsewhere.12
Single cell isolation
Single HRS cells were isolated from frozen tissue sections as previously described.1 In brief, HRS cells were identified by immunostaining for CD30 (Ber-H2) and morphology. Selected cells were released from the surrounding tissue by means of a closed glass capillary fixed to a hydraulic micromanipulation device, pushed into an open glass capillary, and transferred with a minimal volume of overlaying buffer to a polymerase chain reaction (PCR) tube. For negative control, small aliquots of buffer covering the tissue specimen during the cell isolation procedure were drawn and subjected to single-copy PCR.
PCR
Amplification of the Ig promoter.
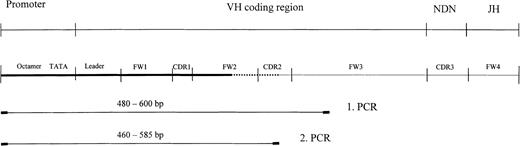
The Ig promoter region comprising heptamer and octamer motifs, TATA box, and pyrimidin region as well as parts of the coding region (leader, FW1, and CDR1) were amplified from single cells and from HD-derived cell lines by seminested (PCR; Figure1). The upper strand primers were located between 160 to 230 bp (depending on the VH segment involved in the Ig rearrangement) upstream of the leader1 segment13,14 The lower strand primers were placed in highly mutated CDR2 and FW3 sections of the already sequenced VH rearrangements.1 This ensured the nearly exclusive amplification of the promoter of the rearranged Ig gene. PCR was performed with 40 cycles of amplification for both rounds of PCR. However, each case required the selection of individual primers and thus different conditions for primer annealing and MgCl2 concentration. The initial 5 cycles of the first PCR consisted of 96°C for 15 seconds, 60°C to 64°C for 150 seconds, and 72°C for 40 seconds. In the remaining 35 cycles, the cycling conditions were kept constant except for the annealing temperature (56°-59°C) and time (60 seconds). The reamplification was carried out with an aliquot (1%) of the first PCR under the same conditions but with higher annealing temperatures throughout the PCR (60°-65°C) The buffer conditions were kept constant throughout both rounds of PCR consisting of 1.5 to 2.5 mM MgCl2 (depending on the case-specific primers), 150 ng forward primer, 100 ng reverse primer, and 0.8 mM each dNTP in Taq buffer supplied with 2 U Taq polymerase (Applied Biosystems, Weiterstadt, Germany).
Amplification strategy.
Schematic representation of the amplification strategy used for amplification of the Ig promoter region in 9 cases of cHD and 3 HD-derived cell lines. Thick line, newly amplified proportion; dotted line, overlapping region; thin line, published Ig sequence proportion.
Amplification strategy.
Schematic representation of the amplification strategy used for amplification of the Ig promoter region in 9 cases of cHD and 3 HD-derived cell lines. Thick line, newly amplified proportion; dotted line, overlapping region; thin line, published Ig sequence proportion.
Amplification of the intronic Eμ enhancer.
The amplification of an approximately 500-bp proportion of the intronic Eμ enhancer was carried out with DNA extracted from 3 HD-derived cell lines using a seminested PCR approach. Briefly, the first round of amplification was performed with the upstream primer ENHup 5′-ACA GCA GGT GGC AGG AAG-3′ (100 ng) and the downstream primer ENHlow 5′-AAA ATG AAG GCC ACT CTA TCA G-3′ (150 ng). In the second round of amplification the ENHup was replaced by the upstream primer ENHRup (5′-CAC CGC GAG AGT CTA TTT TAG G-3′; 100 ng). The cycling conditions consisted of 5 initial cycles at 96°C for 15 seconds, 60°C for 120 seconds, and 72°C for 60 seconds and 35 further cycles with an annealing at 56°C for 60 seconds. The cycling conditions were kept constant in the second round of amplification with the exception that the primer annealing was performed at 60°C throughout the PCR. The buffer conditions were identical throughout both rounds of PCR consisting of 2 U Taq polymerase (Perkin-Elmer), 2.5 mM MgCl2, and 0.8 mM each dNTP.
Amplification of transcribed Ig rearrangements.
Reverse transcriptase PCR (RT-PCR) for the amplification of the Ig transcripts was performed on HD-derived cell lines as well as on control B-cell and T-cell lines. For this purpose, total RNA (1 μg) was transcribed into single-stranded complementary DNA (cDNA) with random hexamer oligonucleotides and reverse transcriptase. For the first round of amplification a set of 6 family-specific framework (FW) I primers was applied in combination with a JH-specific consensus primer (LJH). Aliquots (6 μL) of the first PCR were analyzed on 6% polyacrylamide gels for the presence of Ig-specific PCR products. For the detection of very small amounts of Ig transcripts, a reamplification was performed with 1 μL of the first PCR as a template using a set of family-specific FWII primers in conjunction with a second JH-specific primer (VLJH). The conditions for the first and second PCR were as described elsewhere.1
The PCR products of Ig promoter, intronic Eμ enhancer, and rearrangedIg genes were separated on 0.8% agarose gels, isolated, and sequenced in both directions (BigDye sequencing system, Applied Biosystems) using the primers of the reamplification separately.
RT-PCR for the detection of Oct1, Oct2, and BOB.1/OBF.1
The RT-PCR for the detection of Oct1, Oct2 and BOB.1/OBF.1 transcripts was performed with RNA from various cell lines. In brief, 1 μg total RNA was subjected to 30 cycles (Oct2) or 40 cycles (Oct1 and BOB.1/OBF.1) of PCR after reverse transcription. The primers used for amplification of Oct1, Oct2, and BOB.1/OBF.1 RNA were located within the coding region (Oct1 divided in to 3 parts: 5′-segment-up 5′-CAG CAG CAG CAG ACT CAA G-3′, 5′-segment-low 5′-GTC TTG GCA AAC TGC TCA AG-3′; middle segment-up 5′-GCC CAG TGA CCT TGA GGA G-3′, middle segment-low 5′-ACG TGA CTG CTG AGG AAG C-3′; 3′-segment-up 5′-CAC AGC AAC CGT GAT TTC C-3′, 3′-segment-low 5′-GCT TCT GGC AGC CCA GC-3′; Oct2-up 5′-CCT GCT CAG TTC CTG CTA CC-3′ and Oct2-low 5′-GAT GCT GGT CCT CTT CTT GC-3′; BOB.1-up 5′-CAT CCT GTC ACA GGC CAT G-3′ and BOB.1-low 5′-AGG ACT CAG GTG GGA GCC AC-3′). The conditions for PCR consisted of 30 seconds at 95°C, 40 seconds at 60°C (Oct1), 63°C (Oct2), or 56°C (BOB.1) and 40 seconds at 72°C. All reactions were performed with 2 U Taq polymerase (Perkin-Elmer) and Taqbuffer supplemented with 1.5 mM MgCl2 and 0.8 mM each dNTP and 100 ng of each primer. The PCR products were cloned by using the TOPO TA Cloning Kit (Invitrogen, San Diego, CA) and sequenced in both directions.
Northern blot analysis
Conditions for Northern blots using 10 μg total RNA and probes specific for BOB.1/OBF.1, Oct1, Oct2, and GAPDH were as described.12
Western blot analysis and electromobility shift assays
For the detection of Oct2 and BOB.1/OBF.1 proteins in various cell lines, 100 μg whole cell extracts were separated on a 12% sodium dodecyl sulfate-polyacrylamide gel, proteins were transferred to polyvinylidene difluoride (PVDF) membrane and incubated with polyclonal rabbit anti-BOB.1/OBF.1 antibodies,12 Oct1 and Oct2 antibodies (Santa Cruz Technologies, Santa Cruz, CA). After incubation with appropriate secondary antibodies blots were developed using the enhanced chemiluminescence detection system (Amersham, Uppsala, Sweden). The DNA-binding activity of Oct2 was determined as previously described12 using whole cell extracts of L1236, L428, and KM-H2 as well as Raji and Namalwa (controls).
Results
Immunoglobulin transcripts in HD-derived cell lines
The expression of Ig genes in HD-derived cell lines was analyzed by RT-PCR. No PCR products were detectable after the first round of amplification in all T-cell–derived as well as in all B-cell–derived HD cell lines, whereas all control B-cell lines investigated were clearly positive. After reamplification, the control B-cell lines gave huge amounts of PCR products in contrast to the B-cell–derived HD cell lines L1236, L428, and KM-H2, which displayed only faint Ig-specific PCR products. All T-cell–derived HD cell lines and T-cell lines were completely negative. This indicates that B-cell–derived HD cell lines contain only traces of Ig messenger RNA (mRNA).
Mutations in IgH promoter and coding regions of cultivated and primary Reed-Sternberg cells
Cell lines.
To elucidate the reasons for the near complete absence of Ig transcripts in B-cell–derived HD cell lines, we studied their Ig promoter region comprising TATA box and octamer motif as well as their intronic Eμ enhancer site. All HD-derived cell lines were devoid of mutations with the exception of the already described mutation in the octamer binding site of the cell line L12369 (Table1) and a few mutations outside the transcription factor recognition sites, which most likely represent irrelevant interindividual polymorphisms (not shown).
Mutations in the immunoglobulin promoter, intronic Eμ enhancer, and complete coding region of 3 Hodgkin disease-derived cell lines
| Cell lines . | Somatic mutations . | Coding capacity . | Family . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Promoter . | Leader . | VH region . | Eμ enhancer . | ||||||
| Octamer motif . | TATA box . | Leader and intron . | FW* . | CDR† . | ATTTGCGT 5′octamer . | ATTTTCAT 3′octamer . | |||
| L1236 | 1 | 0 | 4 | 33 | 18 | 0 | 0 | + | VH-1 |
| KM-H2 | 0 | 0 | 18 | 30 | 13 | 0 | 0 | + | VH-3 |
| L428 | 0 | 0 | 6 | 29 | 10 | 0 | 0 | + | VH-5 |
| Cell lines . | Somatic mutations . | Coding capacity . | Family . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Promoter . | Leader . | VH region . | Eμ enhancer . | ||||||
| Octamer motif . | TATA box . | Leader and intron . | FW* . | CDR† . | ATTTGCGT 5′octamer . | ATTTTCAT 3′octamer . | |||
| L1236 | 1 | 0 | 4 | 33 | 18 | 0 | 0 | + | VH-1 |
| KM-H2 | 0 | 0 | 18 | 30 | 13 | 0 | 0 | + | VH-3 |
| L428 | 0 | 0 | 6 | 29 | 10 | 0 | 0 | + | VH-5 |
Number of mutations in all framework regions.
Number of mutations in CDR1 and CDR2.
Primary HRS cells.
To determine the frequency of mutations in the IgH promoter region in primary tissues, 335 single HRS cells were isolated by micromanipulation from immunostained (CD30) frozen sections of 9 cases of nodular sclerosing HD whose rearranged VH region sequences were known from a previous study.1 The VH promoter regions containing the octamer motif and the TATA box were successfully amplified from 65 HRS cells. The amplification of the correct Ig promoter segment was confirmed by a sequence overlap of the amplified proportion with the known VH sequences in these cases. Mutations in the octamer region were found only in 1 of the 9 cases (Table2), whereas the TATA box was found to be devoid of mutations in all instances. In addition, single base substitutions were present outside the transcription factor binding sites, most likely representing interindividual modifications (not shown).
Mutations in the immunoglobulin promoter and complete coding region of Reed-Sternberg cells isolated from 9 cases of classical Hodgkin disease
| Case . | Number of somatic mutations . | Coding capacity . | Family . | ||||
|---|---|---|---|---|---|---|---|
| Promoter . | Leader . | VH-region . | |||||
| Octamer motif . | TATA box . | Leader and intron . | FW* . | CDR† . | |||
| 1 | 0 | 0 | 1 | 15 | 9 | − | VH-1 |
| 2 | 0 | 0 | 1 | 26 | 8 | + | VH-2 |
| 3 | 0 | 0 | 17 | 41 | 21 | − | VH-3 |
| 4 | 0 | 0 | 5 | 21 | 22 | + (−)‡ | VH-3 |
| 5 | 0 | 0 | 24 | 34 | 13 | + | VH-3 |
| 6 | 0 | 0 | 0 | 5 | 7 | + | VH-3 |
| 7 | 2 | 0 | 13 | 38 | 18 | + | VH-3 |
| 8 | 0 | 0 | 2 | 10 | 8 | − | VH-3 |
| 9 | 0 | 0 | 9 | 36 | 17 | + | VH-4 |
| Case . | Number of somatic mutations . | Coding capacity . | Family . | ||||
|---|---|---|---|---|---|---|---|
| Promoter . | Leader . | VH-region . | |||||
| Octamer motif . | TATA box . | Leader and intron . | FW* . | CDR† . | |||
| 1 | 0 | 0 | 1 | 15 | 9 | − | VH-1 |
| 2 | 0 | 0 | 1 | 26 | 8 | + | VH-2 |
| 3 | 0 | 0 | 17 | 41 | 21 | − | VH-3 |
| 4 | 0 | 0 | 5 | 21 | 22 | + (−)‡ | VH-3 |
| 5 | 0 | 0 | 24 | 34 | 13 | + | VH-3 |
| 6 | 0 | 0 | 0 | 5 | 7 | + | VH-3 |
| 7 | 2 | 0 | 13 | 38 | 18 | + | VH-3 |
| 8 | 0 | 0 | 2 | 10 | 8 | − | VH-3 |
| 9 | 0 | 0 | 9 | 36 | 17 | + | VH-4 |
Number of mutations in all framework regions.
Number of mutations in CDR1 and CDR2.
Two HRS cells with an insertion of 22 base pairs resulting in a stop codon.
VH mutations.
The largest proportion of the rearranged IgH coding regions (FW2 to JH) of the 9 classical HD cases used for this study have already been described in previous publications1 showing functional Ig rearrangements in 6 cases and crippling Ig mutations in 3 cases. In the present study, we extended the analysis of the Ig coding region from leader 1 to FW2. No further crippling mutations were found in this proportion of the Ig coding region of all cases, although there was a high load of mutations in most instances (Table 2).
Analysis of Ig gene promoter activity in L1236 cells
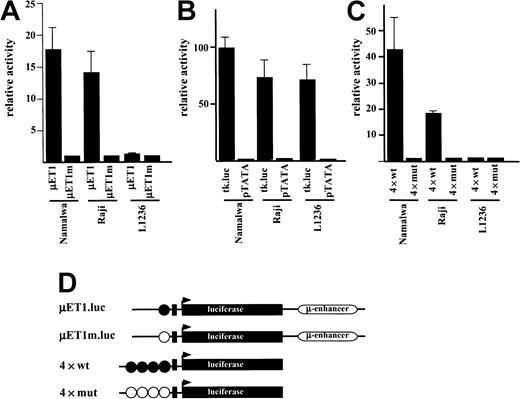
The lack of Ig gene transcription in L1236 cells has been attributed to the mutation in the octamer motif. Therefore, we wondered whether a wild-type Ig promoter would be active in L1236 cells. Reporter constructs driven by either wild-type or mutated Ig promoters were transfected into L1236 cells (Figure2D). As a control, the B-cell lines expressing Ig on the RNA and protein level were investigated in parallel. Following transfection into the control B-cell lines, the wild-type construct was 12 to 18 times more active than the mutated promoter (Figure 2A). Strikingly, in the L1236 cells the wild-type promoter was as inactive as the mutated one (Figure 2A). To clarify the cause of this promoter inactivity, we transfected synthetic luciferase Ig promoter constructs driven by multimerized octamer motifs. Again, the unmutated promoter construct showed a high activity in the control B-cell lines but virtually no activity in the L1236 cells (Figure 2C). This absence of Ig promoter activity was, however, not due to a general transcription deficiency as shown by transfection of L1236 cells with a herpes simplex virus thymidine kinase promoter. This construct revealed the same level of activity in the L1236 cells as in the control B-cell line (Figure 2B).
Transcriptional activity of Ig reporter constructs.
(A) Transfection of Namalwa and Raji (positive control) and L1236 (HD cell line) with the wild-type promoter-driven luciferase reporters (μET1) and the mutant promoter-driven reporters (μET1m). (B) The same cell lines as in panel A were transfected for control with luciferase reporters containing the intact tk-promoter (tk.luc, −109 to +52) or a truncated version of this promoter (pTATA, −38 to +52). (C) Transfection of Namalwa, Raji, and L1236 cell lines with wild-type (4 × wt) or mutant (4 × mut) octamer-dependent Ig reporter constructs. (D) Schematic representation of the reporter constructs used. Wild-type octamer motifs in the Ig promoter or synthetic promoter are indicated as filled circles, mutant octamer motifs are shown as open circles. Details about the reporter constructs can be found in Laumen and colleagues12 (for μET1.luc and μET1m.luc) or Pfisterer and coworkers22 (for 4 × wt and 4 × mut). All transfections were independently repeated minimally 3 times and in all cases a tk-driven renilla-luciferase reporter was cotransfected to correct for differences in transfection efficiencies. Relative activity is shown and the cotransfections with empty expression vectors were arbitrarily set to 1.
Transcriptional activity of Ig reporter constructs.
(A) Transfection of Namalwa and Raji (positive control) and L1236 (HD cell line) with the wild-type promoter-driven luciferase reporters (μET1) and the mutant promoter-driven reporters (μET1m). (B) The same cell lines as in panel A were transfected for control with luciferase reporters containing the intact tk-promoter (tk.luc, −109 to +52) or a truncated version of this promoter (pTATA, −38 to +52). (C) Transfection of Namalwa, Raji, and L1236 cell lines with wild-type (4 × wt) or mutant (4 × mut) octamer-dependent Ig reporter constructs. (D) Schematic representation of the reporter constructs used. Wild-type octamer motifs in the Ig promoter or synthetic promoter are indicated as filled circles, mutant octamer motifs are shown as open circles. Details about the reporter constructs can be found in Laumen and colleagues12 (for μET1.luc and μET1m.luc) or Pfisterer and coworkers22 (for 4 × wt and 4 × mut). All transfections were independently repeated minimally 3 times and in all cases a tk-driven renilla-luciferase reporter was cotransfected to correct for differences in transfection efficiencies. Relative activity is shown and the cotransfections with empty expression vectors were arbitrarily set to 1.
Analysis of the transcription factors Oct1 and Oct2 and their coactivator BOB.1/OBF.1
A potential explanation for the lack of Ig promoter activity could be the absence of the Oct transcription factors or BOB.1/OBF.1 or both. We therefore investigated their expression at the RNA (Northern blotting and RT-PCR) and protein level (immunohistochemistry, Western blotting, and electromobility shift assays [EMSAs] and super shift assays).
Transcripts specific for Oct2 were completely absent from the HD-derived cell lines L1236 and L428 and only low levels were detectable in the cell line KM-H2. BOB.1/OBF.1 transcripts were not present in KM-H2 and L428 and were expressed at very low levels in L1236 cell line cells. In contrast, the methods applied revealed significant amounts of mRNA for Oct2 and BOB.1/OBF.1 in all 3 sIg+ B-cell lines investigated for control. Oct1 mRNA was present in all B-cell and HD-derived cell lines (Table3).
Expression of Oct1, Oct2 and BOB.1/OBF.1 in Hodgkin disease–derived cell lines and control cell lines
| Cell line . | Oct1 . | Oct2 . | BOB.1/OBF.1 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RNA . | Mutations . | Protein . | RNA . | Mutations . | Protein . | RNA . | Mutations . | Protein . | |
| L1236 | + | 0 | + | − | / | − | +3-152 | 0 | − |
| KM-H2 | + | 3 | + | + | nd | − | − | / | − |
| L428 | + | 23-151 | + | − | / | − | − | / | − |
| RAJI B-cell line | + | 0 | + | +3-150 | nd | + | +3-150 | nd | + |
| Cell line . | Oct1 . | Oct2 . | BOB.1/OBF.1 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RNA . | Mutations . | Protein . | RNA . | Mutations . | Protein . | RNA . | Mutations . | Protein . | |
| L1236 | + | 0 | + | − | / | − | +3-152 | 0 | − |
| KM-H2 | + | 3 | + | + | nd | − | − | / | − |
| L428 | + | 23-151 | + | − | / | − | − | / | − |
| RAJI B-cell line | + | 0 | + | +3-150 | nd | + | +3-150 | nd | + |
Mutations are the number of replacement mutations in the coding region.
− indicates negative; nd, not done; /, no analysis possible due to absence of expressed RNA.
Very strong signal intensity.
One in-frame deletion of a triplet (amino acid).
Only trace amount of RNA.
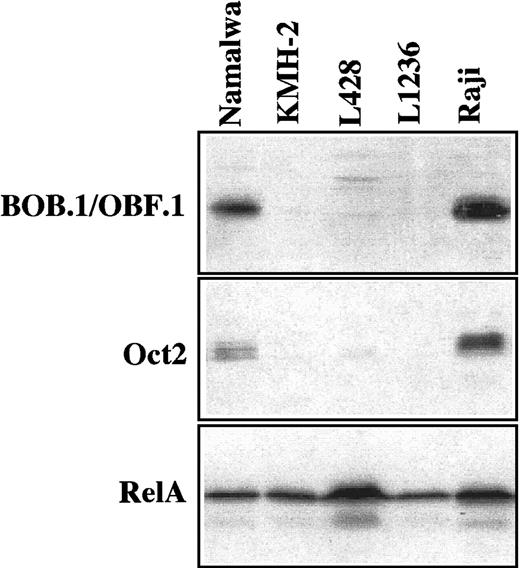
The analysis of the protein expression revealed absence of BOB.1/OBF.1 and Oct2 in all 3 HD-derived cell lines, whereas an expression of these factors was easily detectable in all control B-cell lines (Figure3 and Table 3). The absence of Oct2 protein was confirmed by EMSA. The 3 HD-derived cell lines did not exhibit any Oct2 DNA-binding activity in contrast to the control B-cell lines (Figure 4). Oct1 protein was present in large amounts in all cell lines (Figure 4).
Western blot analysis for BOB.1/OBF.1 and Oct2.
Expression of BOB.1/OBF.1 and Oct2 was analyzed with 50 μg whole cell protein extracts; RelA was investigated in parallel for control. There was no expression of Oct2 and BOB.1/OBF.1 in the HD-derived cell lines KMH-2, L428, and L1236, whereas Raji and Namalwa (B-cell control cell lines) were clearly positive. The presence of RelA demonstrates equal amounts of intact protein in all cell lines.
Western blot analysis for BOB.1/OBF.1 and Oct2.
Expression of BOB.1/OBF.1 and Oct2 was analyzed with 50 μg whole cell protein extracts; RelA was investigated in parallel for control. There was no expression of Oct2 and BOB.1/OBF.1 in the HD-derived cell lines KMH-2, L428, and L1236, whereas Raji and Namalwa (B-cell control cell lines) were clearly positive. The presence of RelA demonstrates equal amounts of intact protein in all cell lines.
Electromobility shift and supershift analysis (Oct2).
The presence of Oct2 with DNA binding activity was investigated in the HD-derived cell lines KM-H2, L428, and L1236 and B-cell control cell lines Raji and Nalmalwa. No Oct2 DNA binding activity was observed in the HD-derived cell lines, whereas both control cell lines showed high DNA binding activity (left panel). Super shift analysis with Oct2-specifc antibodies (right panel) demonstrated the specificity of our technique. Note: Oct1 was present in all cell lines investigated.
Electromobility shift and supershift analysis (Oct2).
The presence of Oct2 with DNA binding activity was investigated in the HD-derived cell lines KM-H2, L428, and L1236 and B-cell control cell lines Raji and Nalmalwa. No Oct2 DNA binding activity was observed in the HD-derived cell lines, whereas both control cell lines showed high DNA binding activity (left panel). Super shift analysis with Oct2-specifc antibodies (right panel) demonstrated the specificity of our technique. Note: Oct1 was present in all cell lines investigated.
Cotransfection with BOB.1/OBF.1 and Oct2
To elucidate whether the down-regulation of BOB.1/OBF.1 and Oct2 is causally involved in silencing Ig expression in classical HD, we cotransfected both transcription factors into L1236 cell line cells containing wild-type or mutated Ig promoter constructs. Transfection of BOB.1/OBF.1 but not of Oct2 reestablished the activity of the wild-type promoter. Cotransfection of both BOB.1/OBF.1 and Oct2 did not further enhance the promoter activity. The transfection of BOB.1/OBF.1 or Oct2 (or both) had no effect on the mutated Ig promoter construct demonstrating the specificity of our transfection assay (Figure5).
Transient cotransfections of HD-derived L1236 cells.
L1236 cells were cotransfected with Ig promoter luciferase reporters (wild-type [left panel] and mutated [right panel]) and expression vectors for BOB.1/OBF.1 or Oct2 as indicated. Ig promoter activity could be reestablished by transfection with BOB.1/OBF.1 expression vectors but not with Oct2. No activity was observed by transfection with the mutated Ig reporter construct.
Transient cotransfections of HD-derived L1236 cells.
L1236 cells were cotransfected with Ig promoter luciferase reporters (wild-type [left panel] and mutated [right panel]) and expression vectors for BOB.1/OBF.1 or Oct2 as indicated. Ig promoter activity could be reestablished by transfection with BOB.1/OBF.1 expression vectors but not with Oct2. No activity was observed by transfection with the mutated Ig reporter construct.
Discussion
We report on studies aimed at clarifying whether mutations in noncoding elements of the IgH gene with special reference to the Ig promoter octamer motif are involved in silencing Iggene transcription in HRS cells. For this purpose we amplified, by PCR, the TATA box and the octamer motif of the Ig promoter and the Eμ enhancer octamers in 3 bona fide HD cell lines, specifically, L1236, KM-H2, and L428. With the exception of the already described octamer mutation in L1236 cells,9 no mutations were found in the octamer motifs, the TATA boxes, and intronic Eμ enhancers of these cell lines.
To determine how generally valid these cell line findings are for classical HD, we extended our investigations to primary HRS cells. For this purpose, we isolated 335 HRS cells by micromanipulation from 9 classical HD cases and analyzed their Ig promoter and neighboring coding sequences. In a previous study we had already analyzed the largest proportion of the Ig coding region (FW2 to JH) of these 9 cases and had found a preserved coding capacity in 6 cases.1 The completion of the analysis of coding regions by amplification with promoter-specific primers in conjunction with HRS cell-specific CDR2 primers revealed no additional crippling mutations in any instance despite the presence of many somatic mutations. The Ig promoter region comprising the TATA box and the octamer motif was unaffected by Ig expression preventing mutations with the exception of one case (case 7) where the octamer motif was modified by 2 base substitutions. This clearly indicates that mutations in the Ig promoter are rare events in primary HRS cells; therefore, they cannot be the general cause for the absence of Ig expression in classical HD.
To confirm or dismiss this assumption, we investigated whether a transfected wild-type Ig promoter construct would be active in the L1236 cells containing a mutated endogenous Ig promoter. Strikingly, the transfected wild-type Ig promoter was as inactive in L1236 cells as the mutated promoter transfected for control. In contrast, in control B-cell lines the wild-type promoter showed a 12- to 18-fold higher activity than the mutated promoter. To determine whether this inactivity of the wild-type promoter construct is due to defects in the octamer-dependent transcription, we transfected an Ig promoter construct driven by multimerized octamer motifs. Even with this construct no promoter activity was seen in the L1236 cells, demonstrating that these cells are incapable of activating octamer-dependent transcription. These results, in conjunction with previous data, clearly demonstrate that a defect in the Ig transcription machinery appears to be the real reason for the lack of Ig expression in HRS cells and not mutations in regulatory or coding Ig sequences
Because the transcription factors Oct1 and Oct2 and their coactivator BOB.1/OBF.1 are critical for octamer-dependent transcription in B cells,15-17 the question arose whether these factors are missing in classical HD. To test this possibility we investigated the expression of Oct1, Oct2, and BOB.1/OBF.1 at the RNA and protein level in all 3 bona fide HD-derived cell lines (ie, L1236, L428, and KM-H2). Transcripts of Oct2 and BOB.1/OBF.1 proved to be absent (Northern blot analysis) or present at very low levels (RT-PCR). At the protein level neither BOB.1/OBF.1 nor Oct2 was detectable by all methods applied. In contrast, Oct1 was expressed at the RNA and protein level in all HD and control B-cell lines. These findings prompt the conclusion that the missing expression of Oct2 and/or BOB.1/OBF.1 is involved in the down-regulation of Ig expression in classical HD.
To further strengthen the validity of this conclusion we cotransfected BOB.1/OBF.1 or Oct2 together with wild-type or mutated Ig reporter constructs into L1236 cells. The transfection of BOB.1/OBF.1 alone completely reconstituted the activity of the wild-type Ig promoter, whereas the transfection of Oct2 had no effect. These findings are in keeping with the observation that Oct1 can compensate the absence of Oct2.18 This conclusion indicates that the lack of BOB.1/OBF.1 is responsible for the missing Ig gene transcription in HRS cells. The importance of BOB.1/OBF.1 was also demonstrated by the observation that both synthetic octamer-dependent promoters as well as a naturally occurring promoter are inactive in B cells that lack the coactivator BOB.1/OBF.1. This inactivity of the Ig promoter can be overcome by induction of BOB.1/OBF.1 expression, demonstrating that BOB.1/OBF.1 is a nonredundant protein in B cells, which is unconditionally required for octamer-dependent transcriptional activity.12
To further strengthen the validity of this conclusion, we cotransfected BOB.1/OBF.1 or Oct2 together with wild-type or mutated Ig reporter constructs into L1236 cells. The transfection of BOB.1/OBF.1 alone completely reconstituted the activity of the wild-type Ig promoter, whereas the transfection of Oct2 had no or no additional effect. These findings are in keeping with the observation that Oct1 can compensate for the absence of Oct2 but appears to be in conflict with the phenotype of BOB.1/OBF.1-deficient mice because they show an almost normal development of B cells with expression of normal levels of IgM.19 However, later stages of B-cell differentiation are affected by the BOB.1/OBF.1 deficiency causing an absence of germinal center formation and a 10-fold decrease in serum isotype levels. Because this reduction does not appear to be due to a defect in class switch recombination,19-21 it might reflect reduction in the transcription rates at the IgH locus. That this really is the case could recently be verified by the introduction of octamer-dependent promoter constructs into BOB.1/OBF.1-deficient cells. These constructs were—like in HRS cells—totally inactive in BOB.1/OBF.1-deficient cells, whereas they showed normal activity in wild-type B cells. This raises the possibility that mutations of theBOB.1/OBF.1 gene might be responsible for the absence of BOB.1/OBF.1 protein in HRS cells. Our sequence analysis of BOB.1/OBF.1 mRNA detectable in trace amounts in the L1236 cell line revealed no alterations. However, further studies are needed to clarify whether alterations of the BOB.1/OBF.1 gene or defects in the transcription factors regulating the expression of BOB.1/OBF.1 are causal to the absence of BOB.1/OBF.1 protein in the HRS cells. Such studies are in progress.
In summary, we have clearly demonstrated that absent Ig expression is not associated with the presence of crippling mutations in the coding region or Ig promoter. Instead, our data indicate that the coactivator BOB.1/OBF.1 is alone or predominantly responsible for the inactivity of the Ig promoter in HRS cells of cHD.
We thank H. Protz, A. Foerster, and H. Lammert for their excellent technical assistance and L. Udvarhelyi for his help with the preparation of the manuscript.
Supported by grants from the Deutsche Forschungsgemeinschaft and Deutsche Krebshilfe.
J.T. and H.L. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Harald Stein, Institute of Pathology, Benjamin Franklin University Hospital, Free University Berlin, Hindenburgdamm 30, 12200 Berlin, Germany; e-mail: stein@ukbf.fu-berlin.de.


![Fig. 5. Transient cotransfections of HD-derived L1236 cells. / L1236 cells were cotransfected with Ig promoter luciferase reporters (wild-type [left panel] and mutated [right panel]) and expression vectors for BOB.1/OBF.1 or Oct2 as indicated. Ig promoter activity could be reestablished by transfection with BOB.1/OBF.1 expression vectors but not with Oct2. No activity was observed by transfection with the mutated Ig reporter construct.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/10/10.1182_blood.v97.10.3191/6/m_h81011034005.jpeg?Expires=1769087458&Signature=woxxaKfq8SLOI15jAVewbDT-LvAUJSg6Rr7t1VKz~uup885PL3sMukh1wZZ5pUlV-ViGnWnTwAY5zjDlRMQ7wS8Qmle3jVCn1MwbkuVn2x2l1hRf-NOt6YMNDWATXgXC5NMfIOA8jlTno~knZ3mboxIRFqBuV6n5wQobQ9GAJD2S9QshZywfIFl1MjAv1SbrlAFpkJ4AzVp0aiGJMBp1XK7gZUm6eVEEj6L4CTFweHk8LZG5UHbQB0A8Ktvgu9SsyJUuVwymLqxiKN5hFsawRiiMQrXf5tL5V0DTn9~YLKiSdaRER45ogk7G~bh~zkAfWxP~jXCM6nYht9~dAWggMw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal