Abstract
Cell signaling by coagulation factor Xa (Xa) contributes to pro-inflammatory responses in vivo. This study characterizes the signaling mechanism of Xa in a HeLa cell line that expresses protease-activated receptor 1 (PAR-1) but not PAR-2, -3, or -4. Xa induced NF-κB in HeLa cells efficiently but with delayed kinetics compared to thrombin. This delay caused no difference in gene expression patterns, as determined by high-density microarray analysis. Both proteases prominently induced the angiogenesis-promoting geneCyr61 and connective tissue growth factor. Inhibition of PAR-1 cleavage abolished MAP kinase phosphorylation and gene induction by Xa, demonstrating that Xa signals through PAR-1 and not through a novel member of the PAR family. Activation of cell surface prothrombin with the snake venom enzyme Ecarin also produced PAR-1–dependent signaling. However, though the response to Ecarin was completely blocked by the thrombin inhibitor hirudin, the response to Xa was not. This suggests that the Xa response is not mediated by locally generated thrombin. The concentration dependence of Xa for PAR-1 activation is consistent with previously characterized Xa-mediated PAR-2 signaling, suggesting that local concentration of Xa on the cell surface, rather than sequence-specific recognition of the PAR scissile bond, determines receptor cleavage. This study demonstrates that PAR-1 cleavage by Xa can elicit the same cellular response as thrombin, but mechanistic differences in receptor recognition may be crucial for specific roles for Xa in signaling during spatial or temporal separation from thrombin generation.
Introduction
In addition to maintaining normal hemostasis, coagulation proteases play important roles in cell signaling. A family of homologous G-protein–coupled receptors that are activated by proteolytic cleavage rather than ligand binding have been identified in recent years.1-5 Among the 4 currently known protease-activated receptors (PARs), 3 are cleaved by thrombin, whereas PAR-2 can be activated by trypsin2 and mast cell tryptase.6 Only the recently identified thrombin receptor PAR-4 is also activated by trypsin,4,5 indicating protease selectivity in this receptor family. Studies have now clearly shown that coagulation proteases upstream of thrombin can also elicit cellular signals through proteolytic mechanisms. When allosterically induced for full function on binding to its cellular receptor tissue factor (TF), factor VIIa (VIIa) can induce Ca++oscillations,7 activate the MAP kinase pathway,8 and induce gene expression.9-11Coagulation factor Xa (Xa) has been shown to be mitogenic for smooth muscle cells12-14 and to elicit inflammatory responses in endothelial cells, including nitric oxide–mediated hypotension15,16 and cytokine induction.15,17Although it is controversial whether a cell surface receptor for Xa, termed EPR-1,18 is involved in the Xa-dependent gene induction,9,15,17,19 all recent studies concur that blocking the proteolytic function of Xa with specific inhibitors abolishes Xa signaling. Consistent with a requirement for proteolytic function, a recent study20 indicated that the activation of murine fibroblasts by Xa and VIIa can be mediated by PAR-2, though other studies21 22 concluded that PAR-2 is not the receptor activated by VIIa in different cell types.
Therapeutic intervention in animal models of gram-negative septicemia indicates that the upstream coagulation proteases contribute significantly more than thrombin action to the inflammatory responses that lead to the lethality of septic shock. Inhibition of TF-initiated coagulation by anti-TF antibodies,23 tissue factor pathway inhibitor,24 or active site-blocked VIIa25all reduce lethality in a particular primate model, whereas blocking prothrombin activation by the infusion of active site-blocked Xa was ineffective in improving survival.26 These data emphasize the importance of understanding the mechanism of signaling by upstream coagulation serine proteases. Numerous inflammatory genes are regulated by the nuclear factor kappa B (NF-κB) family of transcription factors, and we identified a HeLa cell line that induced NF-κB specifically in response to thrombin and Xa. Here we demonstrate that thrombin and Xa both activate HeLa cells through PAR-1 and that Xa responses are not indirectly mediated by thrombin generated from cell-associated prothrombin. Xa may thus substitute for thrombin in proteolytic signaling under conditions in which the activation of coagulation does not proceed to thrombin generation.
Materials and methods
Proteins and inhibitors
Thrombin was generated from prothrombin by activation with Ecarin (Sigma, St Louis, MO) and purified as previously described.27 Recombinant VIIa was produced in Chinese hamster ovary cells and purified by sequential affinity and ion exchange chromatography.28 Plasma-derived Xa was either purchased from Hematologic Technologies (Essex Junctions, VT) or generated from purified factor X by activation with Russell viper venom, and then it was purified on benzamidine agarose. Sequencing-grade trypsin was purchased from Boehringer Mannheim (Indianapolis, IN). Activated protein C (aPC) and factor Va were obtained from Hematologic Technologies. Agonist peptides corresponding to the tethered ligands of PAR were synthesized as carboxyl amides and purified by reverse-phase high-performance liquid chromatography, and the peptide composition was confirmed by mass spectrometry. Monoclonal antibodies ATAP229 and WEDE15,30 which bind to intact and cleaved PAR-1, and SPAN11,31 which binds only to intact PAR-1, have previously been described. The hookworm-derived Xa inhibitor nematode anticoagulant protein 5 (NAP5)32 and tick anticoagulant peptide (TAP)33 were kindly provided by Dr George Vlasuk (Corvas International, San Diego, CA). Hirudin was obtained from Calbiochem (La Jolla, CA).
Cell culture
HeLa cells were grown in Dulbecco modified Eagle medium supplemented with 10 mM HEPES (pH 7.5), 2 mM L-glutamine, nonessential amino acids, penicillin (100 IU/mL), streptomycin (100 μg/mL), and 10% fetal bovine serum. Cells were passaged by detachment with 1 mM EDTA in phosphate-buffered saline. Human umbilical vein endothelial cells (CRL 1730) and DAMI cells were obtained from the American Type Culture Collection (Rockville, MD) and were grown as described previously.34 35
RNA isolation and Northern blot analysis
Total RNA was prepared from 1 to 2 × 106 cells by using the Trizol reagent (Life Technologies, Gaithersburg, MD). Samples of RNA (10 μg) were separated on 1% agarose–formaldehyde gel and transferred to nylon membrane (GeneScreen; NEN, Boston, MA), according to standard procedures.36 The blots were hybridized to DNA probes corresponding to nucleotides 345-1622 for PAR-1,11-1451 for PAR-2,2 603-1003 for PAR-3,3 1-787 for PAR-4,5 589-1093 for Cyr61,37 and 1055-1467 for CTGF.38 The DNA probe for IκB mRNA was prepared from a 1.1-kb EcoRI fragment of the mouse IκB cDNA.39 The probes were labeled by random priming using32P-dCTP (ICN, Costa Mesa, CA), the DECAprime II DNA labeling kit (Ambion, Austin, TX), and Sephadex G-50 mini-spin columns (Worthington, Lakewood, NJ) for probe purification. Hybridization was performed in QuikHyb hybridization solution (Stratagene, La Jolla, CA) for 1 hour at 65°C, and the blots were washed to final stringencies of 0.2 × SSC, 0.1% sodium dodecyl sulfate (SDS), at 65°C. The probe for glyceraldehyde-3–phosphate dehydrogenase (GAPDH) mRNA was prepared from synthetic oligonucleotides as described previously.40
Nuclear extract preparation and EMSA
HeLa cells were grown to subconfluence, incubated for 48 hours in growth medium without serum, and stimulated at 37°C as described. Nuclear extracts were prepared from approximately 106 cells and used for electrophoretic mobility shift assays (EMSA) as described previously.41 SP1- and NFκB-specific probes for EMSA were obtained from Promega (Madison, WI).
Microarray analysis
Subconfluent HeLa cells in T150 flasks were serum-deprived for 48 hours, as described above. Quiescent cells were either not stimulated or stimulated by adding 50 nM thrombin or 100 nM hirudin and then 50 nM Xa. After 90 minutes at 37°C, total RNA was isolated from the control and protease-treated cells as described above, and poly(A) RNA was purified using 2 passes over Oligotex mRNA columns (Qiagen, Valencia, CA) according to the manufacturer's protocols. The mRNA samples were sent for high-density gene expression profiling on the UniGEM V microarray (Incyte Genomics, St Louis, MO) using the fluorogenic dyes Cy3 and Cy5 for control and protease-treated conditions, respectively.
Analysis of MAP kinase phosphorylation
Cells were grown and serum-deprived in 12 well plates as described above; after stimulation, proteins were extracted in 300 μL reducing SDS sample buffer. Aliquots (5 μL) were analyzed by SDS–polyacrylamide gel electrophoresis (PAGE), and this was followed by transfer to nylon membrane for Western blotting. Phosphorylated MAP kinases p44/42 were reacted with a monoclonal antibody (New England Biolabs, Beverly, MA), and bound primary antibody was detected with horseradish peroxidase–conjugated secondary antibody (Amersham Pharmacia, Piscataway, NJ) and enhanced chemiluminescence. Signal intensities on the autoradiographs were quantified by laser densitometry using ImageQuant software (Molecular Dynamics, Piscataway, NJ).
Analysis of PAR-1 cleavage by flow cytometry
HeLa cells were grown, serum-deprived, and stimulated at 37°C in T25 flasks in the presence of 20 nM thrombin or 50 nM Xa/100 nM hirudin for 30 seconds or 5 minutes. The protease was inactivated by adding 100 nM hirudin or 1 μM NAP5, respectively, and the cells were detached with Enzyme-Free Cell Dissociation Buffer (Life Technologies) and kept on ice for all subsequent steps. Cells were pelleted in serum-free HeLa growth medium, washed, and resuspended in staining buffer (phosphate-buffered saline, 0.2% bovine serum albumin, pH 7.2). Cells were stained with a 1:300 dilution of ascites of monoclonal antibody SPAN11 for 30 minutes; this was followed by washing and a 30-minute incubation with a 1:50 dilution of FITC-labeled goat F(ab')2 antimouse IgG (Southern Biotechnology Associates, Birmingham, AL). After another washing step, cells were analyzed on a FACScan flow cytometer (Becton Dickinson, Mountain View, CA).
Results
Xa and thrombin induce NF-κB binding activities in HeLa cells
Because thrombin is known to activate NF-κB,42 we focused on this central pathway as a possible mechanism by which Xa can induce inflammatory responses. To identify Xa-responsive cell lines, serum-starved cells were exposed to 50 nM Xa or thrombin for 45 minutes; this was followed by nuclear extract preparation for EMSA with an NF-κB consensus oligonucleotide probe. Although other cell lines also showed some response to Xa stimulation, HeLa cells displayed a similar induction of nuclear NF-κB binding activity by thrombin and Xa (Figure 1A). Because HeLa cells could be easily passaged without protease (trypsin) treatment for cell detachment, we focused on this cell line as a model system to study the mechanism of Xa-dependent signaling. Induction of NF-κB was specific for thrombin and Xa because the serine proteases VIIa, activated protein C (aPC), factor IXa, and trypsin did not induce nuclear translocation of NF-κB (Figure 1A). HeLa cells that were not serum-starved and cells that were serum-starved under our experimental conditions expressed similar levels of TF, as determined by a one-stage clotting assay, but this excluded that the failure to respond to VIIa resulted from the absence of the essential cofactor TF. Thus, previously described protease cell signaling by VIIa7,43likely involves a receptor that is distinct from the Xa receptor on HeLa cells. These data demonstrate that HeLa cells are selectively responsive to thrombin and Xa, indicating that a putative Xa-responsive receptor is not activated by a broad range of homologous serine proteases. More specifically, PAR-2, which has been previously implicated as an Xa receptor,20 is presumably not the Xa receptor on HeLa cells because PAR-2 can be activated by trypsin2 and the TF-VIIa complex.20
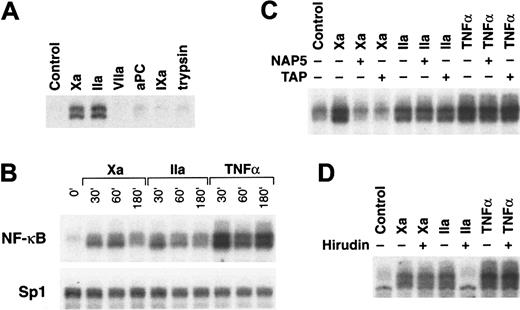
Induction of NF-κB DNA binding activity by Xa and thrombin.
(A) Protease selectivity. Cells were incubated for 45 minutes with 10 nM of the indicated serine proteases Xa, thrombin (IIa), VIIa, activated protein C (aPC), factor IXa, or trypsin for analysis by EMSA (6% gel) using an oligonucleotide containing an NF-κB site. (B) Time dependence. Cells were incubated with Xa (50 nM), thrombin (50 nM), or TNF-α (20 ng/mL) for the indicated times, followed by nuclear extract preparation for EMSA using oligonucleotide probes specific for NF-κB (top panel) or, as a control for equal loading, for Sp1 (lower panel). (C) Effect of specific inhibitors of Xa. HeLa cells were incubated with 10 nM Xa, 10 nM thrombin, or 20 ng/mL TNF-α in the presence or absence of either 5 μM TAP33 or 1 μM NAP5.32 Nuclear extracts prepared after 45 minutes of treatment were analyzed by EMSA. (D) Effect of the thrombin inhibitor hirudin. HeLa cells were stimulated with 10 nM Xa, 10 nM thrombin, or 20 ng/mL TNF-α in the presence or absence of 100 nM hirudin for 45 minutes, followed by nuclear extract preparation for EMSA.
Induction of NF-κB DNA binding activity by Xa and thrombin.
(A) Protease selectivity. Cells were incubated for 45 minutes with 10 nM of the indicated serine proteases Xa, thrombin (IIa), VIIa, activated protein C (aPC), factor IXa, or trypsin for analysis by EMSA (6% gel) using an oligonucleotide containing an NF-κB site. (B) Time dependence. Cells were incubated with Xa (50 nM), thrombin (50 nM), or TNF-α (20 ng/mL) for the indicated times, followed by nuclear extract preparation for EMSA using oligonucleotide probes specific for NF-κB (top panel) or, as a control for equal loading, for Sp1 (lower panel). (C) Effect of specific inhibitors of Xa. HeLa cells were incubated with 10 nM Xa, 10 nM thrombin, or 20 ng/mL TNF-α in the presence or absence of either 5 μM TAP33 or 1 μM NAP5.32 Nuclear extracts prepared after 45 minutes of treatment were analyzed by EMSA. (D) Effect of the thrombin inhibitor hirudin. HeLa cells were stimulated with 10 nM Xa, 10 nM thrombin, or 20 ng/mL TNF-α in the presence or absence of 100 nM hirudin for 45 minutes, followed by nuclear extract preparation for EMSA.
NF-κB binding activity was induced by Xa at 30 minutes and was maximal at 60 minutes, returning toward control levels at 180 minutes (Figure 1B). Thrombin-dependent induction of NF-κB decreased already at 60 minutes toward baseline levels that were approached after 180 minutes of stimulation (Figure 1B). Tumor necrosis factor (TNF)-α–mediated induction showed the typical biphasic induction profile previously described for this cell type.44 TNF-α was consistently a more potent inducer of NF-κB binding activity. Nuclear binding activity for an oligonucleotide containing an Sp1 site did not change under these experimental conditions (Figure 1B). To discriminate between signaling events through high-affinity binding of Xa versus proteolysis mediated by Xa, specific protease inhibitors were used. The hookworm-derived inhibitor NAP532 and TAP33 abolish Xa catalytic function by noncovalently occupying the active site cleft. Both inhibitors blocked Xa-mediated NF-κB induction but did not influence thrombin or TNF-α signaling (Figure 1C). Xa that was mutated at the catalytic triad serine to generate a catalytically inactive mutant and Xa that was covalently active site-modified with the small tripeptide inhibitor, Glu-Gly-Arg-chloromethylketone, did not activate the cells, which provided further evidence for a catalytic mechanism (results not shown). The thrombin-specific inhibitor hirudin only blocked thrombin- but not Xa-mediated NF-κB induction, suggesting that Xa can activate cells directly rather than indirectly through the generation of thrombin (Figure 1D). Northern blot analysis for the inhibitor of NF-κB, IκBα, demonstrated induction by both Xa and thrombin (results not shown). Thus, protease-induced nuclear translocation of NF-κB in HeLa cells is sufficient for gene induction.
HeLa cells express only PAR-1
Xa induced the nuclear translocation of NF-κB with similar efficiency as the prototypical signaling of thrombin that can act through the previously identified protease activated receptors PAR-1, -3, and -4. Expression of the known PAR transcripts in HeLa cells was evaluated by Northern blotting (Figure2A). Total RNA was prepared from serum-starved and nonstarved HeLa cells and, as controls, from non–serum-starved human umbilical vein endothelial cells and from the megakaryocytic DAMI cell line.45 Only PAR-1 mRNA was detected in HeLa cells, whereas endothelial cells also expressed significant amounts of PAR-2 mRNA, consistent with previous results.46 PAR-3 and PAR-4 mRNA were expressed in DAMI cells but were not found at detectable levels in the HeLa cells. Like serum-starved HeLa cells, non–serum-starved HeLa cells expressed only PAR-1, excluding the possibility that PAR -2, -3, or -4 remained expressed on the protein level after serum starvation because of slow protein degradation. The expression of mRNA for PAR-1 is consistent with the responsiveness of this cell type to thrombin. PAR -1, -2, and -4 can be activated using small synthetic peptides that correspond to the tethered ligand sequence of the specific PAR. To analyze the cell surface expression of these PARs at the functional level, HeLa cells were stimulated with agonist peptides (Figure 2B). HeLa cells were responsive to the PAR-1 agonist peptides SFLLRN1 and TFLLRNPNDK.47 Reduced induction by agonist peptides, in comparison with that by proteases, might have resulted from suboptimal peptide concentrations used to assure receptor specificity. In contrast, the PAR-2–specific peptide SLIGRL2 and the PAR-4–specific peptide GYPGKF4 48 did not induce NFκB translocation even at high concentrations, corroborating mRNA data at the functional level.
Expression of protease-activated receptors in HeLa cells.
(A) Northern blotting. Total RNA (10 μg) from the indicated cell lines was probed using specific probes for human PAR-1, -2, -3, and -4 and for GAPDH as a loading control. Only PAR-1 mRNA was detected in non–serum-starved and serum-starved HeLa cells. Human umbilical vein endothelial cells expressed PAR-1 and –2, and the megakaryocytic DAMI cell line expressed PAR-3 and -4. (B) Stimulation with tethered ligand peptides for PAR-1, -2, and -4. Serum-starved HeLa cells were incubated for 45 minutes in the presence of 20 nM Xa or thrombin (IIa) as controls or the PAR-1 peptides SFLLRN (5 μM) or TFLLRNPNDK (5 μM), the PAR-2 peptide SLIGRL (5 and 100 μM), or the PAR-4 peptide GYPGKV (5 and 200 μM), followed by nuclear extract preparation for EMSA using NF-κB–specific probes.
Expression of protease-activated receptors in HeLa cells.
(A) Northern blotting. Total RNA (10 μg) from the indicated cell lines was probed using specific probes for human PAR-1, -2, -3, and -4 and for GAPDH as a loading control. Only PAR-1 mRNA was detected in non–serum-starved and serum-starved HeLa cells. Human umbilical vein endothelial cells expressed PAR-1 and –2, and the megakaryocytic DAMI cell line expressed PAR-3 and -4. (B) Stimulation with tethered ligand peptides for PAR-1, -2, and -4. Serum-starved HeLa cells were incubated for 45 minutes in the presence of 20 nM Xa or thrombin (IIa) as controls or the PAR-1 peptides SFLLRN (5 μM) or TFLLRNPNDK (5 μM), the PAR-2 peptide SLIGRL (5 and 100 μM), or the PAR-4 peptide GYPGKV (5 and 200 μM), followed by nuclear extract preparation for EMSA using NF-κB–specific probes.
Microarray analysis identifies Cyr61 andCTGF as Xa- and thrombin-inducible genes
Taken together, these data suggest that Xa acts either through PAR-1 or a novel member of the PAR family to induce NF-κB in HeLa cells. To define further more downstream cellular consequences of thrombin versus Xa signaling in HeLa cells, we analyzed the effects of thrombin and Xa on gene expression levels by high-density microarray analysis. RNA was isolated from serum-starved cells after 90-minute stimulation with either 50 nM thrombin or Xa. Hirudin (100 nM) was included during stimulation with Xa to inhibit thrombin that might have been generated from cell-associated prothrombin. mRNA samples from stimulated and unstimulated control cells were labeled with the fluorogenic dyes Cy5 and Cy3, respectively, and simultaneously hybridized to the UniGEM V chip (Incyte Genomics). The UniGEM V array contains a panel of approximately 7000 probes for different human genes. The most prominent induction on the panel was observed for the transcript of the angiogenesis-promoting genes Cyr61 and connective tissue growth factor (CTGF). Expression of Cyr61 and CTGF compared to the unstimulated cells was 12.6-fold and 5.2-fold higher for thrombin and 5.3-fold and 2.7-fold higher for Xa/hirudin. Cyr61 and CTGF are both members of the CCN family of extracellular matrix-associated proteins that regulate cellular processes such as adhesion, migration, proliferation, and survival and that have been implicated in angiogenesis, wound healing, tumor biology, and fibrotic diseases.49 50 Additional genes were induced to a much lesser extent by both thrombin and Xa/hirudin—eg, apolipoprotein D, retinoid X receptor gamma, apoptosis inhibitor 1, macrophage scavenger receptor 1—and several expressed sequence tags for novel genes. No genes with significantly reduced expression levels in response to Xa and thrombin were identified, and overall no significant divergence in gene expression patterns in response to the 2 coagulation proteases was seen. Northern blotting confirmed the up-regulation of Cyr61 and CTGFmRNA by thrombin as well as Xa in the presence of hirudin. Both genes were highly induced after 15 minutes by thrombin and more slowly with peak levels after 1 hour by Xa (Figure3A). Dose titrations demonstrate that Xa as low as 10 nM could induce Cyr61 and CTGF mRNA (Figure 3B). Despite the differences in the kinetics of gene induction, these data demonstrate that Xa and thrombin induce a similar set of genes in HeLa cells.
Induction of
Cyr61 and CTGF mRNA by Xa/hirudin. (A) Time dependence. Total RNA from serum-starved HeLa cells was purified after the indicated periods of stimulation in the presence of 50 nM thrombin or 50 nM Xa/100 nM hirudin. Transcripts forCyr61, CTGF, and GAPDH (G3) as a loading control were detected by Northern blotting. (B) Dose dependence. Cells were preincubated with 100 nM hirudin for 2 minutes, followed by the addition of the indicated concentrations of Xa for 90 minutes. Transcripts for Cyr61, CTGF, andG3 were detected by Northern blotting.
Induction of
Cyr61 and CTGF mRNA by Xa/hirudin. (A) Time dependence. Total RNA from serum-starved HeLa cells was purified after the indicated periods of stimulation in the presence of 50 nM thrombin or 50 nM Xa/100 nM hirudin. Transcripts forCyr61, CTGF, and GAPDH (G3) as a loading control were detected by Northern blotting. (B) Dose dependence. Cells were preincubated with 100 nM hirudin for 2 minutes, followed by the addition of the indicated concentrations of Xa for 90 minutes. Transcripts for Cyr61, CTGF, andG3 were detected by Northern blotting.
Xa activates cells through PAR-1 but independent of thrombin generation
The cleavage of PAR-1 can be blocked with specific monoclonal antibodies,51 allowing us to analyze directly whether PAR-1 was a candidate receptor that mediated the Xa response of HeLa cells. In a first set of experiments, activation of the MAP kinase pathway in serum-starved HeLa cells was analyzed by Western blotting for the phosphorylated forms of p44/42. MAP kinase phosphorylation by 20 nM thrombin for 10 minutes was blocked to background levels in the presence of hirudin, whereas preincubation of the cells with cleavage-blocking anti–PAR-1 antibodies resulted in 98% ± 2% (n = 4) inhibition. Thus, hirudin can effectively block cell activation by exogenously added thrombin, and PAR-1 is the only thrombin receptor in these cells leading to MAP kinase phosphorylation. Xa also induced MAP kinase phosphorylation on HeLa cells. There was a slight reduction in MAP kinase phosphorylation on preincubation of the cells with hirudin, but Xa responses were completely abolished by anti–PAR-1 antibodies (Figure 4). The slight reduction of MAP kinase phosphorylation on hirudin inhibition indicated that some locally generated thrombin might have potentiated the Xa response. To test whether prothrombin, at concentrations that could contribute to cell signaling events, was associated with cells under our experimental conditions, we used the snake venom prothrombin activator Ecarin52 53 to convert prothrombin to thrombin in our assay system. Ecarin stimulated MAP kinase phosphorylation in the HeLa cells. This response was completely blocked by hirudin and by anti–PAR-1 antibody (Figure 4), providing evidence that the stimulation resulted from the conversion of prothrombin to thrombin, which, in turn, activated PAR-1. Notably, thrombin generated in situ from cell-associated prothrombin was efficiently blocked by hirudin. The failure of hirudin to inhibit the Xa response thus argues against thrombin as an intermediate in Xa signaling through PAR-1.
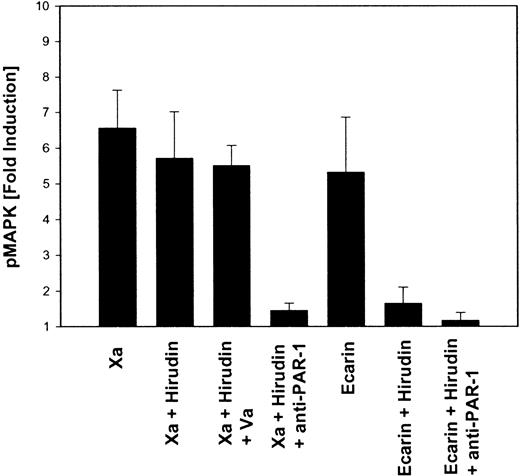
MAP kinase phosphorylation in response to Xa is mediated by PAR-1 but does not involve intermediate thrombin generation.
HeLa cells were serum-starved, and, where indicated, the cells were incubated with 100 nM hirudin, 2 nM factor Va, or monoclonal antibodies to PAR-1 (10 μg/mL ATAP2, 25 μg/mL WEDE15) for 20 minutes before the addition of 50 nM Xa or 0.5 U/mL Ecarin for 10 minutes. Cells were lysed in SDS-sample buffer; this was followed by SDS-PAGE and Western blotting with ECL detection of phosphorylated MAP kinases p44/42. Autoradiographs were analyzed by laser densitometry, and the fold induction of phosphorylated MAP kinase was calculated (mean ± SEM; n = 3-5).
MAP kinase phosphorylation in response to Xa is mediated by PAR-1 but does not involve intermediate thrombin generation.
HeLa cells were serum-starved, and, where indicated, the cells were incubated with 100 nM hirudin, 2 nM factor Va, or monoclonal antibodies to PAR-1 (10 μg/mL ATAP2, 25 μg/mL WEDE15) for 20 minutes before the addition of 50 nM Xa or 0.5 U/mL Ecarin for 10 minutes. Cells were lysed in SDS-sample buffer; this was followed by SDS-PAGE and Western blotting with ECL detection of phosphorylated MAP kinases p44/42. Autoradiographs were analyzed by laser densitometry, and the fold induction of phosphorylated MAP kinase was calculated (mean ± SEM; n = 3-5).
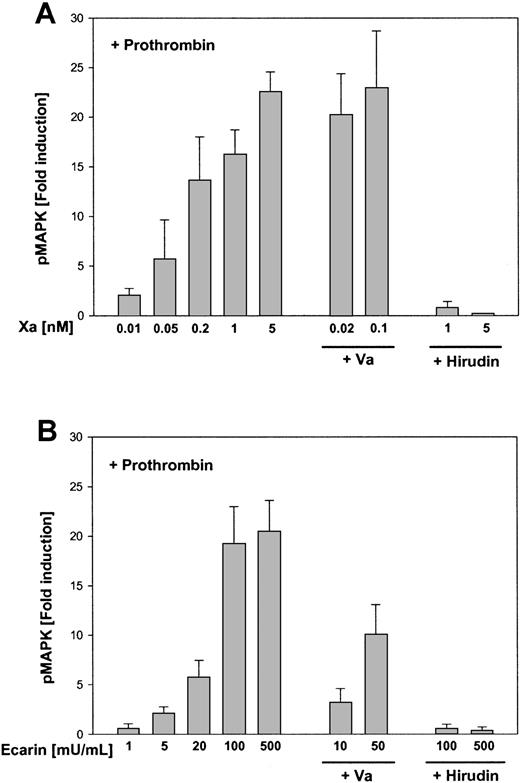
However, it was possible that thrombin generated by cell surface Xa activates cells more effectively than Ecarin-generated thrombin. PAR-1 activation by Xa in the presence of hirudin required Xa at concentrations of 10 nM or higher. We reasoned that indirect signaling through prothrombin activation could occur at much lower concentrations of Xa. Cells were supplemented with a defined amount of exogenous prothrombin (10 nM) and were stimulated with increasing concentrations of either Xa (Figure 5A) or Ecarin (Figure 5B). Half-maximal MAP kinase phosphorylation was obtained with 0.1 nM Xa or 40 mU/mL Ecarin. Even when cells were maximally stimulated by thrombin generated in situ by 5 nM Xa or 500 mU/mL Ecarin, hirudin was a potent and complete inhibitor of these signaling events. These data exclude that the observed activation of PAR-1 by higher concentrations of Xa in the presence of hirudin is indirect through thrombin generation that escaped inhibition by hirudin. Another difference between thrombin-dependent and thrombin-independent signaling of Xa is evident from the effect of factor Va (Va). As expected from the role of this cofactor in Xa-dependent prothrombin conversion, the addition of Va shifted the dose-response curve of Xa in indirect, thrombin-mediated signaling (Figure 5). However, the addition of Va did not affect MAP kinase phosphorylation by higher concentrations of Xa in the presence of hirudin (Figure 4), suggesting that Va does not function as a cofactor in the thrombin-independent activation of PAR-1 by Xa.
MAP kinase phosphorylation in response to Xa and Ecarin in the presence of a defined amount of prothrombin.
Serum-starved HeLa cells were preincubated for 20 minutes with prothrombin (10 nM) and activated for 10 minutes with increasing concentrations of Xa (A) or Ecarin (B). Where indicated, the cells were also preincubated for 20 minutes with factor Va (2 nM) or hirudin (100 nM) before the addition of the proteases. Cells were lysed in SDS-sample buffer followed by SDS-PAGE and Western blotting and ECL to detect phosphorylated MAP kinases p44/42. Autoradiographs were analyzed by laser densitometry, and the fold induction of phosphorylated MAP kinase was calculated (mean ± SEM; n = 3).
MAP kinase phosphorylation in response to Xa and Ecarin in the presence of a defined amount of prothrombin.
Serum-starved HeLa cells were preincubated for 20 minutes with prothrombin (10 nM) and activated for 10 minutes with increasing concentrations of Xa (A) or Ecarin (B). Where indicated, the cells were also preincubated for 20 minutes with factor Va (2 nM) or hirudin (100 nM) before the addition of the proteases. Cells were lysed in SDS-sample buffer followed by SDS-PAGE and Western blotting and ECL to detect phosphorylated MAP kinases p44/42. Autoradiographs were analyzed by laser densitometry, and the fold induction of phosphorylated MAP kinase was calculated (mean ± SEM; n = 3).
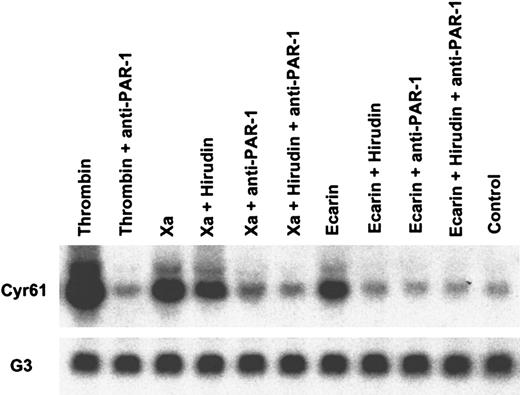
Figure 6 shows inhibition experiments analogous to Figure 4 at the level of gene induction. Activation of cell-associated prothrombin by Ecarin induced Cyr61expression, which was completely inhibited by hirudin. Hirudin did not block Cyr61 expression induced by Xa, but gene induction was prevented by preincubation of the cells with anti–PAR-1 antibodies, consistent with the results obtained by measuring MAP kinase phosphorylation. Taken together, these data provide conclusive evidence that gene induction by Xa in HeLa cells is independent of thrombin but dependent on PAR-1. Furthermore, the nuclear translocation of NF-κB was blocked by anti–PAR-1 (data not shown). Cleavage of PAR-1 by Xa was also directly confirmed by analyzing epitope loss of the activation-sensitive antibody SPAN11.31 Thrombin treatment of HeLa cells resulted in detectable epitope loss within 30 seconds, but no epitope loss was observed with Xa at early time points. After 5 minutes of thrombin treatment, the mean fluorescence of SPAN11 staining, as determined by flow cytometry, was reduced by 80% ± 5% (mean ± SD; n = 3). Activation of HeLa cells by Xa for 5 minutes resulted in a 16% ± 2% reduction in SPAN11 staining, demonstrating that only a limited amount of the cell surface PAR-1 is cleaved by Xa. These experiments confirm that PAR-1 is cleaved in an Xa-dependent manner and identify the rate and extent of receptor cleavage as probable cause for the kinetic differences that distinguish Xa from thrombin-mediated PAR-1 activation.
Cyr61 induction by Xa is independent of thrombin but mediated by PAR-1 activation.
Serum-starved HeLa cells were stimulated for 90 minutes with 20 nM thrombin, 50 nM Xa, or 0.5 U/mL Ecarin. Where indicated, the cells were incubated with monoclonal antibodies to PAR-1 (10 μg/mL ATAP2, 25 μg/mL WEDE15) or 100 nM hirudin for 20 minutes before the addition of the proteases. Total RNA was purified, and transcripts forCyr61 and GAPDH (G3) as a loading control were detected by Northern blotting.
Cyr61 induction by Xa is independent of thrombin but mediated by PAR-1 activation.
Serum-starved HeLa cells were stimulated for 90 minutes with 20 nM thrombin, 50 nM Xa, or 0.5 U/mL Ecarin. Where indicated, the cells were incubated with monoclonal antibodies to PAR-1 (10 μg/mL ATAP2, 25 μg/mL WEDE15) or 100 nM hirudin for 20 minutes before the addition of the proteases. Total RNA was purified, and transcripts forCyr61 and GAPDH (G3) as a loading control were detected by Northern blotting.
Discussion
The presented data demonstrate that Xa induces NF-κB binding activity, gene expression, and MAP kinase phosphorylation in HeLa cells. High-density microarray analysis indicated that a limited and similar set of genes was induced by both Xa and thrombin. HeLa cells used in this study expressed only PAR-1 of the 4 known protease-activated receptors, and inhibition studies with specific antibodies demonstrated that Xa responses were mediated through PAR-1. Independent lines of evidence further excluded that thrombin is an intermediate in Xa-dependent PAR-1 activation. First, the thrombin inhibitor hirudin did not block the Xa responses but completely abolished signaling that originated from the in situ conversion of prothrombin by the snake venom activator Ecarin (Sigma). Second, low concentrations of Xa are shown to produce a Va-dependent signal through prothrombin conversion that was completely blocked by hirudin. In contrast, higher concentrations of Xa produced a Va-independent signal that was not inhibited by hirudin. Thus, Xa can activate PAR-1 on HeLa cells independent of thrombin formation, and, in this cell type, there is no evidence for an unidentified, novel member of the PAR family that can mediate Xa responses.
The activation of PAR-1 by Xa differs from thrombin stimulation in 2 important aspects—the kinetics of cleavage and the effective concentration of enzyme required to cleave PAR-1. Activation by thrombin shows faster kinetics with earlier induction of NF-κB binding activity and of downstream protease-responsive genes. This efficient cleavage likely results from the mode of recognition of PAR-1 by thrombin. Thrombin receptors PAR-1 and PAR-3 both have hirudin-like sequences that show excellent charge complementarity with the basic exosite I of thrombin. This allows for a substrate-like recognition of PAR-1 by thrombin, followed by rapid cleavage of essentially all the receptors expressed on the cell surface. Because Xa is acidic in the substrate binding exosite I,54 binding of Xa to the hirudin-like region of PAR-1 is unlikely. Therefore, Xa cannot interact with PAR-1 in the same substrate-like manner as thrombin, nor can affinity for the receptor be created by this mechanism. As a consequence, only a small percentage of PAR-1 appears to be susceptible for cleavage by Xa, as determined by epitope loss of the cleavage-sensitive antibody SPAN11. However, Xa, unlike thrombin, has cell membrane–binding properties through its amino-terminal Gla-domain. Gene induction required 10 to 100 nM Xa, a concentration range consistent with cell membrane-binding affinity of the Gla-domain. The presented concentration range for PAR-1 activation is also similar to the dose range determined for Xa-mediated PAR-2 activation in fibroblasts.20 Thus, it appears that the major determinant for Xa signaling is the cell surface localization of Xa in proximity to the receptor rather than sequence specific recognition of the cleavage site in the respective PAR. Although the delayed kinetics of PAR-1 cleavage by Xa can reasonably be explained by rate limitation arising from the requirement for cell surface binding before the proteolytic activation of the PAR, more complex scenarios are difficult to exclude. For example, Xa might activate a membrane-associated serine protease that in turn cleaves and activates PAR-1. However, Xa has been found to activate PAR-1 to some extent in a number of heterologous expression systems,3 20 and the assumption of a direct cleavage of PAR-1 by Xa is therefore reasonable.
Platelets express PAR-1 along with PAR-4,48 but Xa fails to activate platelets in the presence of hirudin. Platelets are unique in their rapid change in procoagulant membrane properties after the generation of trace amounts of thrombin that lead to PAR-1 cleavage. Before this event, membrane-binding sites for Xa may be of insufficient affinity or availability to localize Xa in proximity to PAR-1. More important, membrane binding of Xa to platelets is mediated by Va,55 whereas tumor cells apparently have membrane-binding sites for factor X that are distinct from the procoagulant membrane environment that supports prothrombinase assembly.56 Va neither enhanced nor inhibited thrombin-independent Xa signaling in our HeLa cell model. However, this lack of inhibition in the presence of Va-independent membrane binding of Xa does not exclude that Va may inhibit Xa-mediated PAR-1 activation when the only cell surface binding site for Xa is Va. Indeed, Xa in the prothrombinase may be inhibited from activating PAR-1, and the unresponsiveness of platelets may be a consequence of these conformational effects of Va on the active site of Xa.
Whether Xa signaling plays an important physiological role in vivo can be questioned in light of the fairly high concentrations of Xa required to elicit cellular responses. One must consider whether the in vitro analysis of cell signaling by Xa faithfully mimics concentration dependence during the activation of coagulation in vivo. Typically, the zymogen factor X assembles with the cell surface to become activated by the TF-VIIa complex. At the plasma concentration, the zymogen factor X likely saturates cell surface binding sites, eliminating a requirement for de novo binding of the protease. Zymogen conversion is the rate-limiting step for cell signaling under these conditions, and the cell surface half-life of newly generated Xa may be of sufficient duration to stimulate cells. Together with diffusion- or flow-dependent clearance of locally generated thrombin, a more stably membrane-localized protease, such as Xa, may provide sustained PAR activation in local activation of coagulation by the TF pathway. Furthermore, the conversion of approximately 5% to 10% of the plasma zymogen factor X is sufficient for gene induction. Massive activation of coagulation, such as during disseminated intravascular coagulopathy in gram-negative septic shock, could locally generate these relevant concentrations of Xa.
Activation of coagulation within the vasculature typically proceeds to the generation of thrombin that will be the primary activator of PAR-1. However, in specific vascular compartments, potent anticoagulant mechanisms, such as high local concentrations of thrombomodulin, can antagonize thrombin-mediated PAR-1 activation, and Xa may serve to activate PAR-1 in these microenvironments. During anticoagulant therapy with potent thrombin57 or prothrombinase inhibitors,26 one can envision that the excessive generation of Xa can propagate PAR-1 activation normally achieved by thrombin. Xa signaling may be particularly important in temporal or spatial separation from thrombin generation, when cells exit into extravascular spaces that do not support downstream activation of the coagulation cascade. Monocytes/macrophages are known to express PAR-158 and the αMβ2 integrin (Mac1) that is a receptor for the zymogen factor X.59 Thus, factor X can stay associated with cells that migrate out of the bloodstream. Subsequent extracellular activation of X to Xa, now spatially separated from prothrombin, may elicit specific signaling in these migratory cells.
Notwithstanding the experimental evidence that PAR-2, expressed in endothelial cells60 and vascular smooth muscle cells,61 is efficiently activated by Xa,20,62endogenously expressed PAR-1 is here clearly shown to be a target for activation by Xa in a tumor cell line. The Xa-dependent activation of PAR-1 suggests that Xa may function as a more generalized proteolytic signal by bypassing the restricted expression of members of the PAR family in certain vascular (eg, monocytes/macrophages) and extravascular (eg, tumor cells) cell types that frequently stain positively for Xa.63 The apparent membrane-binding requirement for Xa signaling strongly suggests primarily autocrine functions of this protease, representing the most significant difference from thrombin that has a central role in cell–cell communication because of its free diffusion away from cell surfaces.
We thank Pablito Tejada and Aaron Donner for excellent technical assistance and Dr George Vlasuk (Corvas International, San Diego, CA) for the generous gift of the Xa inhibitors TAP and NAP5.
Supported by National Institutes of Health grants HL16411 (W.R.) and HL40387 (L.F.B.) and by an RPR/ISTH Thrombosis Research Fellowship Award (M.R.). W.R. is an Established Investigator of the American Heart Association.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Wolfram Ruf, Department of Immunology, C204, The Scripps Research Institute, 10550 North Torrey Pines Rd, La Jolla, CA 92037; e-mail: ruf@scripps.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal