Abstract
The properties of 9 δ-aminolevulinate dehydratase (ALAD) mutants from patients with ALAD porphyria (ADP) were examined by bacterial expression of their complementary DNAs and by enzymologic and immunologic assays. ALADs were expressed as glutathione-S-transferase (GST) fusion proteins inEscherichia coli and purified by glutathione-affinity column chromatography. The GST-ALAD fusion proteins were recognized by anti-ALAD antibodies and were enzymatically active as ALAD. The enzymatic activities of 3 ALAD mutants, K59N, A274T, and V153M, were 69.9%, 19.3%, and 41.0% of that of the wild-type ALAD, respectively, whereas 6 mutants, G133R, K59N/G133R, F12L, R240W, V275M, and delTC, showed little activity (< 8%). These variations generally reflect the phenotype of ALAD in vivo in patients with ADP and indicate that GST-ALAD fusion protein is indeed useful for predicting of the phenotype of ALAD mutants. The location of F12L mutation in the enzyme's molecular structure indicates that its disturbance of the quaternary contact of the ALAD dimer appears to have a significant influence on the enzymatic activity. Mouse monoclonal antibodies to human ALAD were developed that specifically recognized a carboxy terminal portion of ALAD, or other regions in the enzyme. This study represents the first complete analysis of 9 mutants of ALAD identified in ADP and indicates the highly heterogeneous nature of mutations in this disorder.
Introduction
Delta-aminolevulinate dehydratase (ALAD) deficiency porphyria (ADP) is an autosomal recessive disorder caused by a homozygous ALAD deficiency. Patients with this condition have clinical symptoms of acute hepatic porphyria such as abdominal pain, vomiting, pain in the arms and legs, and neuropathy. ALAD is the second enzyme in the heme biosynthetic pathway. It catalyzes the Knorr-type condensation of 2 molecules of aminolevulinate acid (ALA) to form a monopyrrole, porphobilinogen (PBG).1 This enzyme activity is present in large excess compared to other enzymes in the heme biosynthetic pathway, and its partial deficiency does not usually result in any clinical consequences. Seven patients with ADP have been reported to date,2-6 though only 4 of them have been confirmed by immunochemical or molecular analysis.2-4 Of these 4 cases, the first 2 were German patients reported by Doss and coworkers,7 the third was a Swedish baby boy studied by Thunell and colleagues,8 and the fourth was an elderly Belgian man reported by Hassoun and associates.9 Recently, an asymptomatic Swedish baby girl has been identified to have a markedly decreased ALAD activity.10 Thus far, a total of 9 point mutations of the alad gene have been identified in these 5 subjects, which resulted in different amino acid changes. However, the enzymatic activity of ALAD expressed by mutant ALAD was investigated only for 4 mutants, A274T and R240W, in German patient B,3 and V153M and delTC in German patient H11(single-letter amino acid codes). This previous study with German patient B demonstrated that ALAD activity contributed by the R240W allele was less than 10% of that of the wild type, whereas the A274T allele produced an unstable protein.3 Thus the compound heterozygosity of an inactive protein and an unstable protein accounted for an almost complete lack of ALAD activity (< 2% of normal) in the proband. German patient H had also compound heterozygous mutations, V153M with a decreased ALAD activity,12 whereas delTC was a frame-shift mutation resulting in no ALAD protein.13The first Swedish child with ADP studied by Plewinska and coworkers4 was also compound heterozygous for ALAD deficiency (G133R and V275M).
Although homozygous or compound heterozygous deficiency of ALAD is extremely rare, enzyme activities below 50% of normal are not exceptional in the population, as found in 16 of 880 individuals examined in a Swedish study.8 If these individuals are heterozygous carriers of ALAD deficiency, the prevalence of this condition is about 2% in a normal population. Clinical implications of these findings are obvious in that these individuals with low ALAD activity may be more vulnerable than normal subjects to toxic effects of chemicals or compounds such as lead,14,15trichloroethylene,16 styrene oxide,17 or bromobenzene,18 which inhibit ALAD activity.19 It is thus important to identify heterozygous carriers of ALAD deficiency and to define the nature of their enzymatic defect.
With this view in mind, we examined the enzymologic and immunochemical properties of 9 mutant ALADs so far identified in patients with ADP. We used pGEX plasmid, which can be used for inducible, intracellular expression of genes as fusion proteins with the Schistosoma japonicum glutathione S-transferase (GST) gene encoding a 26-kd protein. Fusion proteins can be purified in a single step directly from bacterial lysates by using glutathione-affinity chromatography.20 The fusion proteins have also been successfully used in enzyme assays without removing GST.21-24 In this study, we purified 9 different ALAD mutants as GST-fusion proteins that had been expressed in pGEX-3X plasmid, and used them to characterize their phenotype. Characterization of the mutant proteins was performed by enzymatic assays of ALAD activity, immunoblot analysis of ALAD-GST protein using both polyclonal and monoclonal antibodies (MAbs) against antihuman ALAD, and examination of thermostability.
Patients, materials, and methods
Case reports
German patient 1.7
This is one of the first 2 patients with ADP reported by Doss and coworkers.7 The patient was referred to as patient B in the original report.7 The patient developed sudden abdominal cramps, with nausea, constipation, and vomiting when he was 15 years old. Because of the “strong abdominal pain,” he underwent an appendectomy. A distal motor polyneuropathy, hypertonus of the muscle, tachycardia, and loss of appetite also developed with paralysis of extensors, during acute phases. Urinary ALA excretion, but not PBG, was markedly elevated (Table 1). Erythrocyte ALAD activities of the patient, the father, the mother, one brother, and a sister were less than 1%, 43%, 25%, 38%, and 46% of normal, respectively (Table 1). Thus, these subjects can be considered heterozygous for ALAD deficiency, whereas the patient was homozygous for the enzymatic defect. Another brother had an enzymatic activity within the normal range, representing a normal family member (Table 1). Similar ALAD deficiency was also confirmed in lymphocytes isolated from these subjects.25 Molecular analysis of thealad gene defect demonstrated that the patient was compound heterozygous for A274 and R240W mutations.3
Biochemical findings and mutations of ALAD in 5 subjects with inherited ALAD deficiency
| Subject . | . | German 1*7 44 . | German 2†7 44 . | Belgian9 28 . | Swedish 18 . | Swedish 211 . |
|---|---|---|---|---|---|---|
| Sex | Male | Male | Male | Male | Female | |
| Age of onset | 15 | 15 | 63 | At birth | At birth | |
| Disease | Moderate | Moderate | Mild | Severe | None | |
| Mutations of ALAD | Allele 1 | A2743 | V153M12 | G133R/K59N29 | G133R4 | F12L11 |
| Allele 2 | R240W3 | delTC13 | None29 | V275M4 | None11 | |
| Biochemical assay | % of normal | % of normal | % of normal | % of normal | % of normal | |
| Plasma | ALA | — | — | 1155 | — | — |
| PBG | — | — | Normal | — | — | |
| Coproporphyrin | 300 | 167 | — | — | — | |
| Protoporphyrin | 127 | 327 | — | — | — | |
| Urine | ALA | 2449 | 2373 | 794 | 8150 | 163 |
| PBG | 238 | 200 | Normal | 447 | Normal | |
| Uroporphyrin | 361 | 224 | 888 | — | — | |
| Coproporphyrin | 5380 | 4698 | 2987 | — | 200 | |
| Total porphyrin | — | — | — | 7247 | — | |
| Stool | Coproporphyrin | Normal | Normal | 285 | 284 | Normal |
| Protoporphyrin | Normal | Normal | 300 | Normal | Normal | |
| Erythrocytes | Protoporphyrin | 3192 | 3164 | 408 | 663 | 300 |
| ALAD activity | 0 | 1 | 1 | 1 | 12 | |
| PBGD activity | Normal | Normal | Normal | Normal | Normal |
| Subject . | . | German 1*7 44 . | German 2†7 44 . | Belgian9 28 . | Swedish 18 . | Swedish 211 . |
|---|---|---|---|---|---|---|
| Sex | Male | Male | Male | Male | Female | |
| Age of onset | 15 | 15 | 63 | At birth | At birth | |
| Disease | Moderate | Moderate | Mild | Severe | None | |
| Mutations of ALAD | Allele 1 | A2743 | V153M12 | G133R/K59N29 | G133R4 | F12L11 |
| Allele 2 | R240W3 | delTC13 | None29 | V275M4 | None11 | |
| Biochemical assay | % of normal | % of normal | % of normal | % of normal | % of normal | |
| Plasma | ALA | — | — | 1155 | — | — |
| PBG | — | — | Normal | — | — | |
| Coproporphyrin | 300 | 167 | — | — | — | |
| Protoporphyrin | 127 | 327 | — | — | — | |
| Urine | ALA | 2449 | 2373 | 794 | 8150 | 163 |
| PBG | 238 | 200 | Normal | 447 | Normal | |
| Uroporphyrin | 361 | 224 | 888 | — | — | |
| Coproporphyrin | 5380 | 4698 | 2987 | — | 200 | |
| Total porphyrin | — | — | — | 7247 | — | |
| Stool | Coproporphyrin | Normal | Normal | 285 | 284 | Normal |
| Protoporphyrin | Normal | Normal | 300 | Normal | Normal | |
| Erythrocytes | Protoporphyrin | 3192 | 3164 | 408 | 663 | 300 |
| ALAD activity | 0 | 1 | 1 | 1 | 12 | |
| PBGD activity | Normal | Normal | Normal | Normal | Normal |
German patient 2.7
This patient was referred to as patient H in the original report.7 The patient, a 15-year-old boy, also suddenly developed vomiting, pains in arms and legs with sensory disturbances, followed by loss of appetite. He was admitted to a neurologic clinic because of paralysis, which included respiratory muscles. Urinary ALA and porphyrin excretion was elevated (Table 1). Molecular analysis of the alad gene defect demonstrated that the patient was compound heterozygous for V153M and delTC mutations.12 13
Both patients showed a significant elevation of urinary porphyrin excretion, that is, coproporphyrin III was about 50-fold higher than the upper limit of normal (Table 1). There was also an elevated concentration of erythrocyte protoporphyrin in both patients (∼30-fold). Immunoreactive ALAD in red cell lysates from the 2 patients showed that both patients had cross-reactive immunologic material (CRIM+) that amounted to 20% and 33% of the control level.2 This finding suggested that the molecular defect of ALAD in these patients may be due to the expression of a nonfunctional enzyme subunit, which is contributed by at least one of the 2 mutant alleles.26 Both patients had moderate ADP and acute crises were successfully treated with glucose and heme arginate infusion. Both patients remained in clinical remission for a long period of time, and they are currently still alive 20 years after the onset of the disease.27
Belgian patient.9
The patient was a 63-year-old man. At the age of 63, he developed a gradually worsening loss of strength in both arms.28 The patient showed markedly elevated plasma ALA concentration (110 μg/dL; normal < 10); plasma PBG was undetectable. Urinary ALA excretion (85 mg/dL; normal < 5) was also abnormally elevated, without an increase in PBG concentration. Erythrocyte ALAD activity of the patient was less than 1% of normal controls,9 whereas lymphocyte ALAD activity was about 20% of normal controls25 (Table 1). Other causes of ALAD deficiency, such as lead poisoning, tyrosinemia, alcoholism, smoking, cirrhosis, renal insufficiency, and diabetes mellitus were excluded.9 Both oral glucose and intravenous infusion of glucose were effective in ameliorating biochemical and clinical abnormalities related to porphyria. Family studies showed that, except for a brother who had normal levels of enzyme activity, a sister, a daughter, and a granddaughter of the patient had about 50% ALAD activity compared with normal controls,28 indicating that these subjects are heterozygous carriers of ALAD deficiency. Molecular analysis revealed that the patient was heterozygous for G133R and K59N mutations in one ALAD allele.29
Swedish symptomatic child (Swedish 1).8
In contrast to the 3 previous patients, severe symptoms were already present in this patient at birth. This is highly unusual for patients with acute hepatic porphyria, who commonly develop clinical disease at puberty or after.30 At the age of 3, he had recurrent attacks of pain, vomiting, and hyponatremia and symptoms of polyneuropathy involving motor functions including respiratory muscles. The patient excreted large amounts of ALA and coproporphyrin in the urine, but only moderately elevated levels of PBG (∼5-fold of the normal upper limit; Table 1). Erythrocyte protoporphyrin was also moderately increased (6-fold of the normal control). Erythrocyte ALAD activity was less than 5% of normal in the patient, and between 26% and 51% in both parents, both grandfathers, and a sibling. Attempts to treat the porphyria syndrome of the patient by glucose, hematin, or blood were unsuccessful.4 At the age of 6, the patient underwent liver transplantation.31 Although the postoperative period was complicated by episodes of severe paralysis and respiratory failure requiring respirator support, the patient's condition gradually improved, and the patient was free from pain.4 He needed assisted ventilation intermittently, but his condition permitted him to attend school at times. Two years and 9 months after the transplantation, at the age of 9 years, the boy died. The immediate cause of death was pneumonia, respiratory insufficiency, and finally pneumothorax. At autopsy his diaphragm and intercostal muscles looked unusually thin. Histopathologic investigation showed demyelinization of peripheral nerves and neurogenous muscular atrophy within several muscle groups. The transplanted liver was macroscopically and microscopically normal (S. Thunell, L. Holmberg, and J. Lundgren, personal oral communications, July 1998). Molecular analysis of the alad gene defect indicated that the patient was compound heterozygous for G133R and V153M mutations.4
Swedish asymptomatic girl (Swedish 2).10
This asymptomatic newborn child had a marked reduction in erythrocyte ALAD activity (∼12% of normal). The child is in good health and has normal growth and normal neuromuscular and intellectual development. There are signs of deranged heme biosynthesis because urinary ALA and coproporphyrin are slightly increased, as is erythrocyte protoporphyrin concentration. Urinary PBG is normal. Hereditary tyrosinemia and lead poisoning have been excluded by specific assays. Molecular analysis of the alad gene defect demonstrated that the child is heterozygous for F12L mutation.11
Plasmidconstruction
Mutant ALAD complementary DNAs (cDNAs) (K59N, G133R, K59N/G133R, F12L, V153M, delTC), which have been identified in patients with ADP,12,13 32 were cloned by reverse transcription-polymerase chain reaction (RT-PCR). Mutant R240W, A274T, and V275M ALAD cDNA were prepared using QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using the wild-type human ALAD cDNA as a template according to the manufacturer's protocol. Three sets of oligomers harboring the point mutation of interest (indicated by a bold letter) were used as a mutagenic primer: For R240W: 5′-CCGAGCTGTGGACTGGGATGTACGGG-3′ (sense), and 5′-CCCGTACATCCCAGTCCACAGCTCGG-3′ (antisense), and A274T: 5′-CCCTGACCTCCCTCTCACCGTGTACCACG-3′ (sense), and 5′-CGTGGTACACGGTGAGAGGGAGGTCAGGG-3′ (antisense), and V275M: 5′-CCTCCCTCTCGCCATGTACCACGTCTCTGG-3′ (sense), and 5′-CCAGAGACGTGGTACATGGCGAGAGGGAGG -3′ (antisense). These mutant cDNAs were inserted into pGEM-T Easy, or pGEM-T plasmids (Promega, Madison, WI), and were reconstructed into BamHI site-attached plasmids as follows. The wild-type ALAD and F12L mutant ALAD containing BamHI site at the 5′ end were synthesized by PCR, using each ALAD cDNA as a template. An oligomer, 5′-GGATCCATATGCAGCCCCAGTCCGTTCT-3′, was synthesized and used as a sense primer, whereas an oligomer, 5′-GGAAACAGCTATGACCATG-3′ (M13 reverse, Promega) was used as an antisense primer. The reaction mixture (100 μL) contained 100 pmol of each primer, 0.25 μg human ALAD cDNA as a template, 2.5 U Taq DNA polymerase (Roche, Nutley, NJ), dNTPs (200 μmol each), and 10 μL 10 × PCR buffer (Roche). PCR was carried out using a Thermal Cycler (Perkin Elmer Cetus, Norwalk, CT). The samples were denatured at 94°C for 2 minutes, followed by 15 cycles of amplification that included heating at 94°C for 1 minute, 60°C for 1 minute, and 72°C for 90 seconds. PCR products were purified using QIAEX II Agarose Gel Extraction System (Qiagen, Chatsworth, Canada), and were inserted into pGEM-T Easy vector. K59N, G133R, K59N/G133R, V153M, R240W, A274T, and delTC mutant cDNA plasmids were digested with restriction enzymes, and DNA fragments harboring these mutation(s) or deletion were inserted into aBamHI site-attached wild-type ALAD plasmid, by digestion with the identical restriction enzyme(s). After confirmation of both wild-type and mutant ALAD sequences by nucleotide sequencing, the plasmids were digested with BamHI and EcoRI, then cDNA fragments were ligated to pGEX-3X plasmids (Pharmacia, Uppsala, Sweden). Their sequences were again confirmed by sequencing, or by restriction analysis (data not shown).
Expression and purification of the wild-type and mutant ALAD as GST-fusion proteins
The GST-fusion proteins were expressed and purified by a minor modification of the method as previously described.20 33The pGEX-3X plasmids coding the wild-type and mutant aladgenes were transformed into Escherichia coli BL21 (DE3)PlysS (Novagen, Madison, WI). Transformed cells were grown in 400 mL Luria-Bertani (LB) medium containing 50 μg/mL ampicillin and 170 μg/mL chloramphenicol at 37°C for 3 hours. Then, isopropyl-1-thio-α-d-galactoside (IPTG, Sigma Chemical, St Louis, MO) was added to a final concentration of 0.05 mM to induce fusion protein expression. Induced cells were grown at 25°C for an additional 7 hours and centrifuged at 4°C. The collected cells were resuspended in 16 mL buffer A, which consisted of 50 mM HEPES (pH 7.5), 1 mM EDTA, and 5 mM dithiothreitol (DTT) containing 0.1% Triton X-100, 10 μg/mL antipain (Sigma), 10 μg/mL leupeptin (Sigma), and 10 μg/mL pepstatin A (Sigma). Then, the samples were frozen at −80°C overnight. After thawing the samples on ice, DNase, Rnase, and MgCl2 were added to a final concentration of 50 μg/mL, 100 μg/mL, and 5 mM, and centrifuged. The supernatant fractions were applied to glutathione Sepharose 4B affinity column (Pharmacia), and then washed 3 times with washing buffer (50 mM Hepes [pH 7.5], 1 mM ZnSO4, and 5 mM DTT, 10 μg/mL antipain, 10 μg/mL leupeptin, and 10 μg/mL pepstatin A). The fusion proteins were eluted with an elution buffer (50 mM Hepes [pH 7.5], 1 mM ZnSO4, 5 mM DTT, and 10 mM glutathione, adjusted to pH 7.5) and stored at −80°C until used.
Antibodies against human ALAD
A rabbit antihuman ALAD polyclonal antibody had been prepared by immunizing 3 rabbits with a homogeneously purified ALAD preparation from normal human erythrocytes, and had been purified to its IgG, as described previously,34 and used as a polyclonal antibody. MAbs against human ALAD were prepared using male BALB/c mice. The spleen cells were fused to mouse myeloma cells after 2 or 3 injections of purified human ALAD34 to animals. Hybridomas were cloned and selected by enzyme-linked immunosorbent assay (ELISA) using purified human ALAD. As a result, 3 MAbs, that is, MAb4, MAb330, and MAb350, were selected for positive binding to the wild-type ALAD.
Immunoblotanalysis of fusion proteins
The purified fusion proteins were heated at 100°C for 3 minutes in sodium dodecyl sulfate (SDS) gel-loading buffer (50 mM Tris-Cl [pH 6.8], 100 mM DTT, 2% SDS, 0.1% bromophenol blue, and 10% glycerol). The samples were loaded on to a 10% polyacrylamide gel containing 0.01% SDS (SDS-PAGE), and electrophoresed according to the method described previously.35 Samples were then transferred to a sheet of nitrocellulose membrane and incubated overnight at 4°C with 5% skim milk in Tris-buffered saline-0.1% Tween 20. The membrane was incubated with the primary antibody for 1 hour at room temperature. Goat antimouse IgG (Santa Cruz Biotech, Santa Cruz, CA), or goat antirabbit IgG (Santa Cruz Biotech), conjugated with horseradish peroxidase was used as a secondary antibody. Protein bands to which antibodies were bound were visualized with the enhanced chemoluminescence (ECL) immunoblotting system (Amersham International, Buckinghamshire, United Kingdom), which was performed according to manufacturer's protocol.
ALAD activity assay
The ALAD activity of purified GST-fusion proteins was determined colorimetrically as described previously, and expressed as units per milligram protein.1 Protein concentration was determined by using the Biorad Protein Assay System (Biorad, Hercules, CA). ALAD activity was expressed as the percent of that of the wild-type ALAD.
Results
Expression and purification of fusion proteins
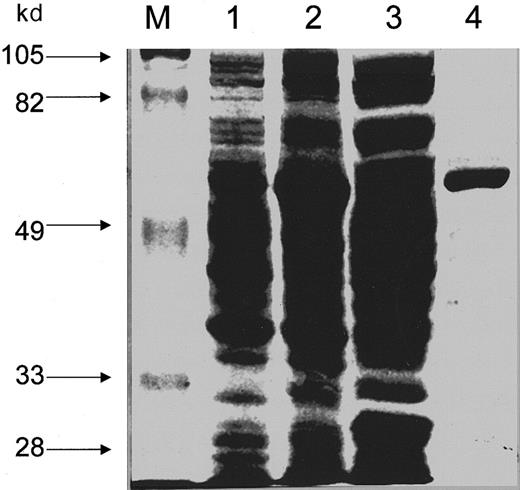
The wild-type and mutant ALADs were ligated to pGEX-3X plasmid and transformed into E coli BL21 (DE3)PlysS. These ALADs were expressed as GST-fusion proteins and were purified using glutathione-affinity column chromatography. Fusion protein synthesis inE coli was induced by IPTG, and then soluble proteins were collected. The estimated molecular size of all GST-fusion ALADs is 62 kd, except for that of delTC which is 58 kd, based on the assumption of the sizes of 26 kd, 36 kd,36 and 32 kd, for GST, ALAD, and mutant delTC, respectively. The size for the mutant delTC was estimated on the basis of the total length of the coding region of its cDNA. A single major protein band corresponding to a monomer of GST-ALAD fusion protein was observed for the purified fusion protein preparation by staining with Coomassie brilliant blue R (Figure1). This finding suggests that the GST-ALAD fusion protein has been highly purified by glutathione-affinity chromatography.
SDS-PAGE analysis of the wild-type ALAD-GST fusion protein.
Expression and purification of GST-ALAD fusion protein and SDS-PAGE were carried out as described in “Materials and methods.” One percent (v/v) of the total volume was loaded to each lane. Proteins were stained with Coomassie brilliant blue. Lane M: molecular weight protein standards; lane 1: transformed E coli PLysS cells without induction; lane 2: transformed E coli PLysS cells after induction with IPTG; lane 3: supernatant from the lysate of induced cells; lane 4: GST-fusion protein purified by glutathione-affinity chromatography.
SDS-PAGE analysis of the wild-type ALAD-GST fusion protein.
Expression and purification of GST-ALAD fusion protein and SDS-PAGE were carried out as described in “Materials and methods.” One percent (v/v) of the total volume was loaded to each lane. Proteins were stained with Coomassie brilliant blue. Lane M: molecular weight protein standards; lane 1: transformed E coli PLysS cells without induction; lane 2: transformed E coli PLysS cells after induction with IPTG; lane 3: supernatant from the lysate of induced cells; lane 4: GST-fusion protein purified by glutathione-affinity chromatography.
Immunoblotanalysis
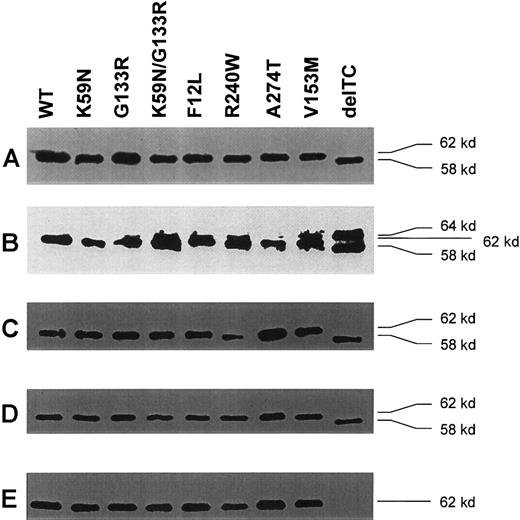
All GST-ALAD fusion proteins, including the wild-type and mutant ALADs, were detected as a single band at the predicted size by immunoblot analysis using mouse MAb against GST (Figure2A). The protein expressed by delTC was smaller than by other ALAD cDNAs by about 4 kd, as expected by its premature stop codon (Figure 2A). When rabbit antihuman ALAD antibody was used, there was a major band at the same size that was detected by anti-GST MAb for all fusion proteins, but there was additionally a minor band with a higher molecular size (∼64 kd), for most samples (Figure 2B). The extra band detected by a polyclonal antibody to human ALAD was at the same size for all samples, but not detectable by any MAbs against human ALAD, suggesting that the extra band was derived from E coli.
Immunoblot analysis of GST-ALAD fusion proteins.
(A) Immunoblot analysis with MAb anti-GST. The molecular size of the wild type and mutants were 62 kd, except for delTC, which was 58 kd, indicating that ALAD expressed by delTC is shorter than the wild-type ALAD by about 4 kd. (B) Immunoblot analysis with rabbit IgG against antihuman ALAD. Essentially similar results are seen as in panel A, except for the presence of an additional band at about 64 kd, including delTC. This finding suggests that the additional band is likely due to a nonspecific binding of a bacterial protein with IgG. (C) Immunoblot analysis with mouse MAb4. Results are essentially similar to row A. (D) Immunoblot analysis with mouse MAb350. Results are essentially similar to row A. (E) Immunoblot analysis with mouse MAb330. In contrast to polyclonal IgG, MAb4 and MAb350, MAb330 completely failed to recognize delTC.
Immunoblot analysis of GST-ALAD fusion proteins.
(A) Immunoblot analysis with MAb anti-GST. The molecular size of the wild type and mutants were 62 kd, except for delTC, which was 58 kd, indicating that ALAD expressed by delTC is shorter than the wild-type ALAD by about 4 kd. (B) Immunoblot analysis with rabbit IgG against antihuman ALAD. Essentially similar results are seen as in panel A, except for the presence of an additional band at about 64 kd, including delTC. This finding suggests that the additional band is likely due to a nonspecific binding of a bacterial protein with IgG. (C) Immunoblot analysis with mouse MAb4. Results are essentially similar to row A. (D) Immunoblot analysis with mouse MAb350. Results are essentially similar to row A. (E) Immunoblot analysis with mouse MAb330. In contrast to polyclonal IgG, MAb4 and MAb350, MAb330 completely failed to recognize delTC.
The MAbs against human ALAD were prepared as IgG using male BALB/c mice. The spleen cells were fused with mouse myeloma cells after 2 or 3 times injection of purified human ALAD to animals. Hybridomas were cloned and evaluated by ELISA for their binding to purified human ALAD. As a result, 3 MAbs, that is, MAb4, MAb330, and MAb350, were selected for positive binding to the wild-type ALAD. The MAbs against human ALAD were diluted to a 1000-fold in buffered saline for Western blotting. Both MAb4 and MAb350 detected the same size protein as did anti-GST MAb (Figure 2C,D), but there was no extra band detected by the polyclonal antibody (Figure 2B). MAb330 did detect the same protein for all samples, except for delTC (Figure 2E). Although the wild-type and mutants except delTC were recognized by the 3 MAbs, delTC failed to cross-react with MAb330. The delTC mutation, namely,818T819C deletion,13 encoded completely abnormal amino acids after 272P, and resulted in a new stop codon at 293I, as opposed to the wild-type ALAD, which consists of 330 amino acids. Accordingly, the molecular size of delTC ALAD should be 32 kd, instead of 36 kd. Thus, the C-terminal sequence of delTC ALAD should be completely different from that of the wild-type ALAD. These findings suggest that, although MAb4 and MAb350 recognize epitopes encoded by amino acid sequences N-terminal to 272P, MAb330 recognizes an epitope encoded by sequences C-terminal to this site.
ALAD activity of fusion proteins
The ALAD activity of mutant ALADs was determined using GST-ALAD fusion proteins.1 The activity of each mutant ALAD was expressed as the ratio to that of the wild-type ALAD (Figure3). The wild-type ALAD is fully active as a GST-fusion protein. Among mutant-GST fusion proteins, K59N showed the highest activity, that is, 69.9% of the wild type, followed by V153M and A274T, which showed 41.0% and 19.3% activity of the wild type, respectively. In contrast, the activity of other mutants were all under 10% of the wild type, specifically, 8.1%, 8.1%, 0.5%, 3.5%, and 1.2%, for G133R, K59N/G133R, F12L, R240W, V275M, and delTC, respectively. These results demonstrate that mutants G133R, F12L, R240W, V275M, and delTC had little enzymatic activity, whereas mutants K59N, A274T, and V153M had a partial but significant ALAD activity, as a GST-ALAD fusion protein. It is also clear that the activity of K59N/G133R was determined by the low activity of G133R in the same allele.
ALAD activity of mutant ALAD.
ALAD activity was measured in the form of GST-fusion protein as described in “Materials and methods.” Enzyme activity was expressed as the percent of that of the wild type. ALAD activity of the wild type was 1.73 nmol PBG formed/mg protein, h, 37°C. Data are the mean ± 1 SD of triplicate assays.
ALAD activity of mutant ALAD.
ALAD activity was measured in the form of GST-fusion protein as described in “Materials and methods.” Enzyme activity was expressed as the percent of that of the wild type. ALAD activity of the wild type was 1.73 nmol PBG formed/mg protein, h, 37°C. Data are the mean ± 1 SD of triplicate assays.
To examine thermostability of ALAD mutants, ALAD activity of GST-fusion proteins that had a significant enzyme activity were determined after preincubation for 15 minutes at 37°C, 40°C, 50°C, 60°C, or 70°C. Enzyme activity was expressed as the ratio to that which was measured at 37°C without preincubation. The wild-type enzyme, K59N, A274T, and V153M proteins showed no decline in ALAD activity up to 50°C, whereas the activity was reduced to about 60% to 75% by preincubation at 60°C or 70°C. These findings indicate that K59N, A274T, and V153M proteins had similar thermostability to that of the wild-type ALAD.
Discussion
In this study, we have prepared 9 mutant ALAD cDNAs so far identified in patients with ADP, 6 mutants by site-directed mutagenesis, and 3 mutants by subcloning from patients' lymphoid cells, expressed them as a GST-fusion protein in E coli, and purified the fusion protein by glutathione-affinity column chromatography. We then studied immunochemical and enzymologic properties of the purified proteins by using a polyclonal antibody and MAbs raised against the normal human ALAD, and by examining enzyme activity and thermostability. ALAD-GST fusion proteins were detectable by immunoblot analysis by using rabbit IgG and mouse MAbs against human ALAD, and a mouse MAb against E coli GST.
It is of special interest to note that ALAD activity of the expressed proteins was measurable as a GST-fusion protein. In comparison to ALAD activity expressed by the wild-type ALAD cDNA, enzyme activities of mutants were 69.9%, 8.1%, 8.1%, 0.5%, 3.5%, 19.3%, 7.1%, 41.0%, and 1.2% for K59N, G133R, K59N/G133R, F12L, R240W, A274T, V275M, V153M, and delTC, respectively. Results with A274T and R240W ALAD were consistent with our previous findings on the expression of these proteins in Chinese hamster ovary (CHO) cells3; that is, mutant R240W ALAD showed little enzymatic activity, whereas mutant A274T ALAD showed partial activity. Results with V153M and delTC, which had been identified in German patient B, also confirmed our findings on the phenotype of these proteins expressed in CHO cells.11
The effect of heat treatment for 15 minutes prior to enzyme assay revealed that 3 mutants with significant ALAD activity had similar thermostability. These findings suggest that these mutant proteins have no overt structural abnormality, despite the fact that they have some reduced catalytic activity. Human ALAD has a Mr of 280 000 and 36 000 kd for the homooctamer and for the subunit, respectively. It is thought that mammalian ALAD is active only as the homooctamer, though the prokaryotic enzyme appears to be active as a monomer.37According to the x-ray crystallographic analysis of yeast ALAD, the dominant feature of the monomer is the closed (α/β)8 or TIM barrel with the active site located at the C-terminal ends of the β-strands, and the dimer is composed by the extensive association of 2 monomers about an intervening 2-fold axis and their arm regions wrap around the TIM barrel domain of the neighboring subunit.38Then 4 dimers form an octamer by interacting with each other by identical manner by the arm regions that are on the surface of each dimer. It is not known whether an ALAD-GST fusion protein forms a homooctamer as does the wild-type ALAD, or functions as a catalytically active ALAD-GST monomer. However, if ALAD activity is associated with a monomeric ALAD-GST protein, F12L mutant should have active ALAD activity, because its disturbance in the molecular structure is at the dimeric interaction domain, rather than at the catalytic site of the enzyme. Because F12L-GST protein had little enzymatic activity, it appears that, despite its substantially larger size (62 kd) than the subunit of ALAD itself (36 kd), the ALAD-GST fusion protein perhaps forms a homooctamer that may then carry out the Knorr-type condensation for the formation of PBG from 2 molecules of ALA.
Many GST-fusion proteins are known to be immunoreactive as well as enzymologically active.21-23 It is important to note that ALAD deficiency in vivo in patients with ADP is largely consistent with the phenotype of GST-ALAD fusion proteins, suggesting that the bacterial expression of mutant proteins is useful for studies of human mutant ALADs. The advantage of the GST expression system is that the expressed protein can be readily purified by a single-step glutathione-affinity chromatography. Because ALAD is exceptionally sensitive to the toxic effects of certain chemicals such as lead, trichloroethylene, styrene oxide, and succinylacetone, the enzyme holds a unique place in toxicology as the sensitive target of environmental insults.17,19 39-41 It is thus important to examine the enzymologic and toxicologic characteristics of mutant ALAD phenotypes, and purified GST-fusion ALAD offers a useful model for these studies.
The structural analysis of the yeast ALAD has permitted insights into the basis for the malfunction of 5 mutant ALA enzymes.38For example, the K59N mutation is thought not to prevent the folding or the functioning of the enzyme. This is consistent with our finding that demonstrated that K59N mutant had the highest ALAD activity (∼70% of the wild type) among all mutants examined. It is also consistent with the fact that this mutant is highly abundant in the normal population (∼10% of the population).42,43 In this context, K59N should more appropriately be termed as a variant, rather than a mutant of ALAD. Four other mutants identified in ADP, G133R, R240W, A274T, and V275M, were thought to have significant structural effects on the enzyme molecule.38 For example, glycine 133, which is adjacent to the 3 cysteine residues (cysteine 132 in human ALAD, equivalent to 143 in yeast) that form the zinc binding site at the active center, can be disrupted by substitution of an arginine for glycine 133, which may alter the zinc binding and catalytic activity of the enzyme. Consistent with this prediction, our findings indicate that ALAD activity expressed by G133R was 8.1% of that of the wild-type ALAD. F12L, a new mutant ALAD identified in an asymptomatic Swedish child with heterozygous ALAD deficiency, had ALAD activity less than 1% of the wild type. This mutation occurred in the α1 helix of the N-terminal arm, which is involved in the extensive quaternary interactions among the subunit, thus it is unlikely that this mutant protein can fold properly to form even a dimer.
Amino acids encoded by mutant delTC are completely different from the wild type at amino acids after 272P due to T816C817 deletion. The 2-base substitution occurred at the site next to A274T mutation characterized by us previously, which was known to accompany little enzyme activity.3 This mutation also directly influences α8 helix, which makes quaternary contacts located from V278 to A291.38 Consistent with this prediction, the enzymatic activity of mutant delTC was only 1.2% of the wild type.
In contrast, V153M mutation is located in the α4 helix, one of the secondary structure elements that form the main TIM barrel fold of the enzyme. Because this mutation is not in the close vicinity of the metal binding site, and may not destabilize the buried core of the barrel, it is expected that such a mutant may show some enzyme activity. The enzyme activity expressed by mutant V153M was in fact 41.0% of the wild type.
In the study of German patient H who showed little ALAD activity in erythrocytes, we identified mutants V153M and delTC.11 The activity of mutant V153M ALAD and delTC were, however, 41% and 1.2% of the wild-type ALAD. Why the combination of V153M and delTC resulted in little enzyme activity in vivo in the patient is unclear, but it appears that delTC produces a dominant negative effect on the expression of ALAD in vivo in the patient.
The Belgian patient with a late-onset ADP is particularly noteworthy. In this patient, 2 mutations of the alad gene, K59N and G133R, were detected in the same allele; however, the other allele was entirely normal. The protein encoded by G133R alone had little activity (∼8% compared with that of the wild type), and the mutant containing both K59N/G133R had a similar activity as G133R alone. This patient was unique in that he developed the disease only at age 63, which was remarkably late.7-9 This patient also developed ADP in conjunction with polycythemia.28 Polycythemia is a clonal disorder and progeny of a single cell can gain ascendancy in the bone marrow, which presumably carried the mutant alad allele, resulting in clinical ADP in the patient.29
The study using antihuman ALAD mouse MAb in immunoblot analysis of ALAD deficiency in ADP has proven extremely useful in characterizing distinct immunologic epitopes of mutant ALAD identified in patients. Namely, although MAb4 and MAb350 recognized all ALAD mutants similarly as did a rabbit polyclonal IgG against human ALAD, both in immunoblot analysis in this study and in ELISA (data not shown), MAb330 failed to recognize protein encoded by delTC in both assays. Based on the mutant amino acid sequences, and the recognition by MAbs, it can be concluded that MAb330 recognizes an epitope that is encoded by amino acid sequences after 272P, whereas MAb4 and MAb350 recognize epitopes encoded by amino acids prior to this residue. These findings indicate the usefulness of a panel of MAbs in the structural analysis of mutant ALAD proteins. The findings in this study also indicate that ALAD mutations identified in ADP are highly heterogeneous, and the mechanisms of developing ADP are also variable.
We are grateful to Dr George Drummond for his reading this manuscript and for his helpful comments.
Supported in part by a General Clinical Research Center grant (M01-RR00102) from the National Center for Research Resources at the National Institutes of Health; U.S. Public Health Service grant DK32890 (to S.S.); Chugai Fund for Molecular Hematology (to S.S.); Yamanouchi Molecular Medicine Fund (to S.S.); the German Research Association grant GR 1363/2-2 (to M.O.D.); and Karolinska Institute Research Grant (to P.H.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Shigeru Sassa, Head, Laboratory for Biochemical Hematology, Rockefeller University, New York, NY 10021; e-mail:sassa@rockvax.rockefeller.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal