Abstract
The course of clonal evolution of 2 related clones in the blood of a patient with Waldenstrom macroglobulinemia (WM) indicates the functional importance for the expression of the B-cell receptor for the survival of these malignant cells. Protein and nucleotide sequencing of the paraproteins' variable regions revealed 2 predominant Vλ and 2 VH sequences, each set comprised in the ratio 1:1.5. The 2 VH sequences and 2 Vλ sequences shared the same VDJ and VJ junctional sequences, respectively, indicating that 2 malignant clones had evolved from a common ancestor. This is the first report on intraclonal heterogeneity in WM. Comparison of the Vλ and VH sequences with the closest matching known germline genes showed that they contained approximately 10 somatic mutations each. The distribution and type of mutations demonstrate that mutations have continued to accumulate in the malignant clones and that selection has been operating to preserve immunoglobulin structure.
Introduction
Waldenstrom macroglobulinemia (WM) is a malignancy of pleomorphic lymphoid cells accompanied by high concentrations of monoclonal serum immunoglobulin M (IgM). Although in WM the malignant cells are found primarily in bone marrow, 20% to 70% of blood mononuclear cells are malignant B cells.1 2
Analyses of the V regions revealed that malignant cells in WM often express somatically mutated IgM.3 Distribution of replacement (R) and silent (S) mutations suggests that the IgM in WM is derived from cells at a late stage of differentiation that have undergone antigenic selection in germinal centers but failed to coordinate somatic mutation and class switching.4
Evidence has accumulated indicating malignant B cells display a variety of physiologic activities and requirements possessed by normal cells at similar stages of differentiation. Some of the well-documented examples are the extensive clonal evolution of follicular lymphomas5-7 and Burkitt lymphomas.8 9
Although by classical definition WM is monoclonal, the presence of 2 unrelated clones in the same patient has been described.10 11 Here we report a case of clonal evolution in WM in which 2 related clones were identified in the blood of a patient. This is the first report of intraclonal heterogeneity in WM. Our results indicate that mutations continued to accumulate after the transformation event, and mutations in the malignant clones were selected by requirements for the preservation of the B-cell receptor (BCR) structure rather than for antigen binding.
Study design
Patient history
A 62-year-old woman was evaluated at the Mayo Clinic in 1985. She was found to have a monoclonal IgMλ protein measuring 13.3 g/L (1.33 g/dL). The patient was monitored until 1993 when her hemoglobin had fallen to 97 g/L (9.7 g/dL). The monoclonal IgM was 32.6 g/L (3.26 g/dL), and 3 g/L (0.3 g/dL) of free λ chain was detected in the urine. A bone marrow biopsy demonstrated 90% replacement of hematopoietic elements with small lymphocytes in a diffuse and interstitial pattern, consistent with WM. The patient was placed on chlorambucil for 5 years through 1998. After a period of observation through November 1999, the monoclonal IgM rose to 38 g/L (3.8 g/dL), the platelet count fell to 94 000, and chlorambucil was resumed. The patient's blood was collected for analysis just before resumption of chlorambucil treatments.
Characterization of the IgMλ sH-IgM.22
IgM antibody was isolated from patient plasma, and variable fragments (Fv) were produced as described.12 Fv fragments were sequenced in an automated sequencer (PE Biosystems Procise 492 HT, Foster City, CA), using standard procedures. Complementary DNA (cDNA) was generated by reverse transcriptase–polymerase chain reaction (RT-PCR) of peripheral blood messenger RNA (mRNA), using oligonucleotide primers based on the protein sequence. The cDNA was cloned and sequenced, using standard procedures.
Results and discussion
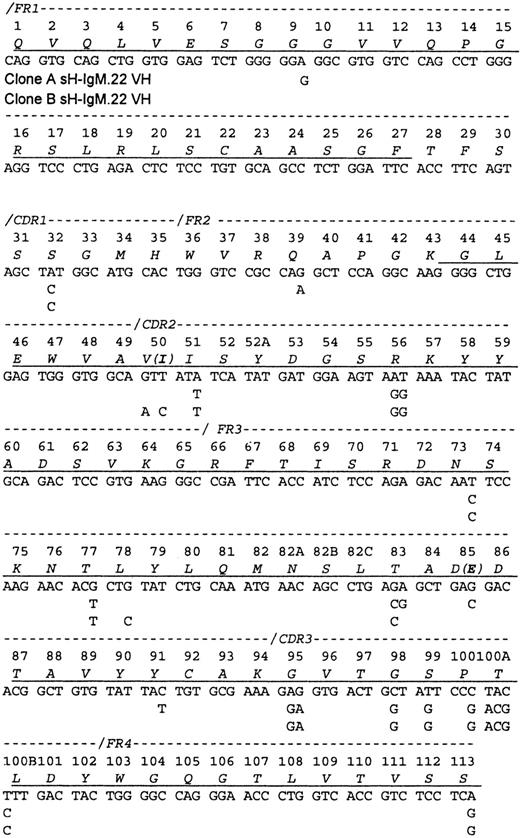
A partial protein sequence of the variable heavy chain (VH) was obtained, including the N-terminal 27 amino acids and the segment spanning amino acids 43 through 113 (Figure1). Sequencing of the PCR-amplified cDNA (2 cDNA preparations and 6 PCR reactions) revealed a mixture of related VH sequences. Therefore, PCR products were cloned, and the plasmids isolated from 42 bacterial colonies were sequenced. Two VH sequences (A and B) that differ by 2 R and 6 S mutations but with identical CDR3 regions that corresponded to the protein sequence were discovered. Only peptides characteristic of VH IgM-A were identified by amino acid sequence analyses. A total of 242 bacterial colonies containing cDNA bearing the sH-IgM.22 VDJ junctional sequence were identified. With the use of hybridization, 60% were identified as type A and 40% as type B.
Sequence of the sH-IgM.22 VH.
The sequence is aligned according to the numbering system of human VH sequences in Kabat et al.26 The sH-IgM 22 VH is a member of VH subgroup III. Underlined amino acids have been confirmed by protein sequencing. Amino acid sequence corresponds to sH-IgM.22 nucleotide sequence. sH-IgM.22 VH type A and type B sequences are represented only with nucleotides that differ from IGHV3-30/3-30-05*01, IGHJ4*02, and IGHD2-21*02 germline sequences.27 Two amino acid replacements in the protein sequence of SH-IgM.22 VH type B are printed in parentheses.
Sequence of the sH-IgM.22 VH.
The sequence is aligned according to the numbering system of human VH sequences in Kabat et al.26 The sH-IgM 22 VH is a member of VH subgroup III. Underlined amino acids have been confirmed by protein sequencing. Amino acid sequence corresponds to sH-IgM.22 nucleotide sequence. sH-IgM.22 VH type A and type B sequences are represented only with nucleotides that differ from IGHV3-30/3-30-05*01, IGHJ4*02, and IGHD2-21*02 germline sequences.27 Two amino acid replacements in the protein sequence of SH-IgM.22 VH type B are printed in parentheses.
The sequences of both sH-IgM.22 VH most closely matched the IGHV3-30/3-30-5*01 germline sequence (96% homology). Clone A shares 3 nucleotides with IGHV3-30/3-30-5*01 that are mutated in clone B, and clone B has 5 nucleotides in common with IGHV3-30/3-30-5*01 that are not present in clone A. This suggests that clones A and B evolved independently from a common ancestral clone that had 17 mutations in VH, compared to the germline genes, including D and J segments. Except for the 8 mutations that differentiate sequences A and B, we do not know which of these other mutations were acquired after the malignant transformation of the ancestral B cell.
In comparison with IGHV3-30/3-30-5*01, clone A acquired 5 (1 R + 4 S) mutations located in the frameworks (FRWs), and clone B has mutations distributed as follows: FRWs (1 S) and complementary determining regions (CDRs) (1 R and 1 S). Random mutations yield an R:S ratio of 2.9. In regions, such as the FRW of antibodies that need to be conserved, only some R mutations are tolerated, dropping this ratio to about 1.5. Ratios higher than 2.9 can be achieved only through positive selection of R mutations. Most such mutations are expected to be in the CDRs.13 However, studies suggest that the frequency of mutations in CDRs is twice that found in FRWs. Additional bias of this process is focusing of R mutations to the CDRs.14 15 Consequently, the R:S ratio of 2.9 in the CDRs should not be used as the sole basis to determine whether antigen-driven selection is occurring. However, accumulation of S mutations in the FRWs can be attributed to selective pressure toward preservation of immunoglobulin structure. The presence of 6 S (individual expected frequency of 0.255) and 2 R (individual expected frequency of 0.745) mutations would be expected to occur randomly in less than 1 per 100 cases (P = .009). This strong bias toward the accumulation of S mutations since the time when clones A and B diverged, indicates selective pressure for the maintenance of the BCR structure.
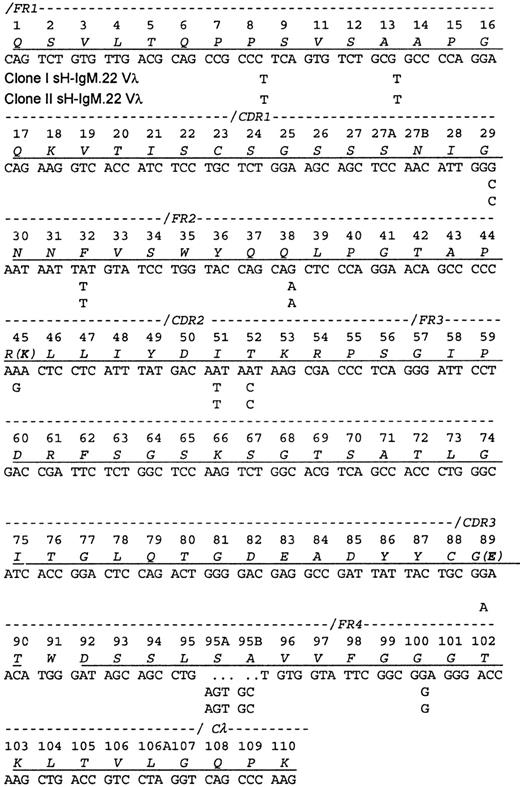
A cDNA library contained 2 related Vλ sequences in approximately equal frequency (7 of 18 clones of one and 11 of 18 clones of the other), which is consistent with the ratio of the related VH sequences. The protein sequence obtained contained peptides from each of the encoded Vλ, indicating that both clones are synthesizing and secreting IgM. Each differed from the protein sequence by a single amino acid. Because the isolation procedure only recovered intact IgM molecules, the identification of both light chains implies also that both heavy chains were present in the serum.
The Vλ sequences from sH-IgM22 most closely matched the IGLV1-51*01 germline sequence (97% homology). The 2 genes differ from their common ancestor by a single nucleotide change (Figure2). Because only the 2 mutations differentiating the 2 clones could be placed after the transformation event, no conclusions could be drawn regarding the selective forces on the light chain.
Sequence of the sH-IgM.22 Vλ.
The sequence is aligned according to the numbering system of human Vλ sequences in Kabat et al.26 Vλ of sH-IgM.22 is a member of the lambda subgroup I. The underlined amino acids have been confirmed by protein sequencing. Amino acid sequence corresponds to sH-IgM.22 nucleotide sequence. sH-IgM.22 Vλ type I and II sequences are represented only with nucleotides that differ from IGLV1-51*01 and IGLJ3*01 germline sequences.27 Two amino acid replacements in the protein sequence of SH-IgM.22 Vλ type II are printed in parentheses. Reference for germline sequences on both figures are IMGT.27
Sequence of the sH-IgM.22 Vλ.
The sequence is aligned according to the numbering system of human Vλ sequences in Kabat et al.26 Vλ of sH-IgM.22 is a member of the lambda subgroup I. The underlined amino acids have been confirmed by protein sequencing. Amino acid sequence corresponds to sH-IgM.22 nucleotide sequence. sH-IgM.22 Vλ type I and II sequences are represented only with nucleotides that differ from IGLV1-51*01 and IGLJ3*01 germline sequences.27 Two amino acid replacements in the protein sequence of SH-IgM.22 Vλ type II are printed in parentheses. Reference for germline sequences on both figures are IMGT.27
It has been established that in vivo BCR expression is needed for B-cell development and maintenance. Progression through the pre–B-cell stage of differentiation depends on signals from the BCR or its precursor. This phase of B-cell development is antigen independent, which means that successful assembly and expression of the BCR itself delivers a survival signal. A mature B cell, even in the absence of antigen stimulation, must retain continuous BCR expression to avoid apoptotic death.16-20
The idea that many B-cell lymphomas need expression of the BCR and potentially antigen stimulation to maintain viability is also supported by numerous observations.21-25 This requirement does not apply to all types of B-cell malignancies (eg, multiple myelomas). Our data suggest a preservation of BCR structure in WM, which implies that the presence of BCR is necessary to generate a survival signal in these malignant cells.
Supported in part by grant NS24180 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Larry R. Pease, Guggenheim 3, Dept of Immunology, Mayo Clinic/Foundation, 200 First St SW, Rochester, MN 55905.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal