Abstract
Lutheran blood group glycoproteins (Lu gps) are receptors for the extracellular matrix protein, laminin. Studies suggest that Lu gps may contribute to vaso-occlusion in sickle cell disease and it has recently been shown that sickle cells adhere to laminin isoforms containing the α5 chain (laminin 10/11). Laminin α5 is present in the subendothelium and is also a constituent of bone marrow sinusoids, suggesting a role for the Lu/laminin interaction in erythropoiesis. The objectives of the current study were to define more precisely the molecular interactions of the extracellular and intracellular regions of human Lu and to clone and characterize a mouse homologue. To this end, complementary DNA and genomic clones for the mouse homologue were sequenced and the mouse Lu gene mapped to a region on chromosome 7 with conserved synteny with human 19q13.2. Mouse and human Lu gps are highly conserved (72% identity) at the amino acid sequence level and both mouse and human Lu gps specifically bind laminin 10/11 with high affinity. Furthermore, the first 3, N-terminal, immunoglobulin superfamily domains of human Lu are critical for this interaction. The results indicated that the cytoplasmic domain of BRIC 221-labeled human Lu gp is linked with the spectrin-based skeleton, affording the speculation that this interaction may be critical for signal transduction. These results further support a role for Lu gps in sickle cell disease and indicate the utility of mouse models to explore the function of Lu gp-laminin 10/11 interaction in normal erythropoiesis and in sickle cell disease.
Introduction
The Lutheran blood group is composed of a complex set of antigens expressed on 2 integral membrane glycoprotein isoforms of 85 and 78 kd.1,2 The complementary DNA (cDNA) encoding the 85-kd isoform has been cloned,3 and the predicted structure is that of a type 1 membrane protein. There are 5 disulfide-bonded, extracellular, immunoglobulin superfamily (IgSF) domains, a single hydrophobic membrane span, and a cytoplasmic tail of 59 residues.3 The composition of the extracellular IgSF domains puts Lutheran blood group glycoproteins (Lu gps) in the subset of adhesion molecules that includes the human tumor marker MUC18/MCAM4 and the chicken neural adhesion molecule Gicerin.4-6Chicken gicerin binds neurite outgrowth factor, a variant of the extracellular matrix (ECM) protein laminin7,8 and, interestingly, recent studies suggest that Lu gp also functions as a laminin receptor.9-11
The Lu gp cytoplasmic tail contains an SH3 binding motif and 5 potential phosphorylation sites, consistent with receptor signaling function. Of note, differences in the structure of the cytoplasmic tail distinguish the 2 isoforms. The 78-kd isoform (also termed B-CAM12 or Lu[v13]13) is generated by alternative splicing of intron 13 and differs from the larger form by having a truncated cytoplasmic tail lacking the SH3 binding motif as well as the potential phosphorylation sites. A recent study in epithelial cells14 suggests that the cytoplasmic tail may, in addition, contain basolateral membrane sorting signals. Analysis of transfected MDCK cells showed that a cytoplasmic di-leucine motif in the 85-kd isoform regulated its sorting to the basolateral membrane while the smaller isoform demonstrated a nonpolarized expression.14
Normal and sickle red cells, as well as stably transfected cells expressing either the 85- or 78-kd isoform, bind laminin.9-11,15-17 The degree of binding correlates with the level of Lu gp expression and sickle red cells bind more laminin than normal red cells.9-11 It therefore seems reasonable to postulate that this newly characterized laminin receptor may function not only during normal erythropoiesis in cell-ECM interactions but it may also contribute to the pathophysiology of sickle cell disease by mediating adhesion of sickle cells to vascular endothelial cells.
Many vital questions regarding the molecular characterization and the biologic function of the Lu-laminin interaction remain. In this paper we show that the Lu extracellular region binds specifically and with high affinity to human laminin isoforms that contain the α5 chain (laminins 10/11) and that laminin binding is mediated by the N-terminal 3 IgSF domains of human Lu. We also show that Lu, labeled with fluorescent antibody BRIC 221, is linked to the membrane skeleton in intact erythrocytes and we speculate that the interaction of the Lu cytoplasmic tail with the spectrin-based skeleton may be important for laminin-induced signal transduction. Because we believe that the development of Lu knockout mice is a key requirement for obtaining a detailed understanding of the role of Lu in both normal erythropoiesis and in sickle cell disease, we have cloned, sequenced, and localized the mouse gene encoding the murine homologue to human Lu gp. Importantly, using recombinant murine Lu gp extracellular IgSF domains, we document that the laminin binding property of murine and human Lu gp molecules is conserved.
Materials and methods
Sequencing of cDNA
Mouse EST clones that were closely related to human Lu gp were obtained from IMAGE Consortium (info@image.llnl.gov). Sequencing primers were sense and antisense vector primers, as well as 2 sense mouse cDNA primers (5′-AGATTGCCACCTGCAGC and 5′-GCAACTTGTCAAGAAGC) and 2 antisense mouse cDNA primers (5′-AAAGGCCCCCTCCCAG and 5′-TGTTAGACGCTTCACAG). DNA sequencing was performed using dye-labeled terminator chemistry on Applied Biosystems 373 or 373A automated DNA sequencers (PerkinElmer, Foster City, CA and PerkinElmer, Warrington, United Kingdom, respectively).18
Genomic cloning
The mouse Lu gene was characterized by analysis of bacterial artificial chromosome (BAC) clones obtained by polymerase chain reaction (PCR) screening of mouse BAC library CitbCJ7 derived from embryonic stem (ES) cell lines of the 129/SvJ strain (Research Genetics, Huntsville, AL). Briefly, the putative organization of the murine gene was deduced by aligning sequence of the human gene with sequences of murine EST clones. One pair of primers was designed based on sequence of the 3′ untranslated region (forward primer: 5′-CAATCTCTGGATCTGGAACTTTGG-3′; reverse primer: 5′-AGCCCCCACCTCCCTCTTC-3′). Of 4 BAC clones that were positive using this primer set, one was found to contain the entire Lugene, as determined by sequencing of PCR products and by subcloning of BAC DNA. Subcloning of BAC DNA in a bluescript vector enabled sequencing of the entire murine Lu gene.
Chromosomal localization
The Jackson Laboratory BSS interspecific back-cross ([C57BL/6JEi X SPRET/Ei]F1 X SPRET/Ei) panel was used for mapping.19 A HhaI polymorphism (C57BL/6JEi, 95 and 125 base pairs [bp] fragments; SPRET/Ei, 220 bp) within a 220-bp PCR fragment amplified with the Lu specific forward and reverse primers 5′-GCAGTGGAGGCTTTGGAGAT-3′ and 5′-CTCCCTCTTTCCCTCCCCA-3′, respectively, was used to follow segregation of alleles in the 94 back-cross progeny from the BSS panel on ethidium-stained 2% agarose gels.
The mouse Lu gene was also mapped by fluorescence in situ hybridization (FISH) analysis, using BAC DNA labeled by random priming with digoxigenin and stained with Texas red on mouse metaphase chromosome from C57 black mouse.20 Chromosome-specific probes were random labeled with biotin and stained with fluorescein isothiocyanate (FITC).21
Transfections
Stable, transfected K562 cells expressing human Lu gp and domain deletion mutants were previously described.22 A control, stable transfectant expressing LW glycoprotein was prepared as outlined.22 Prior to use in cell adhesion assays, cell surface expression of Lu gp and mutants was confirmed by flow cytometric analysis using monoclonal antibody BRIC 224 against the N-terminal domain.22 Mean fluorescence intensity (MFI) values obtained and percent positive cells were respectively: control LW transfectant-MFI 4, 0%; full length Lu (FL)-MFI 557, 98%; domain 5 deleted (Del 5)-MFI 401, 98%; Del 4-5–MFI 219, 92%; Del 3-5–MFI 560, 98%; and Del 2-5–MFI 176, 94%.
Chimeric fusion proteins comprising the extracellular domains of full-length human or mouse Lu gp or of human Lu domain deletion mutants and the hinge region and Fc domains of human IgG (Lu-Fc) were expressed as follows. Regions of human Lu cDNA encoding the extracellular domains (GenBank accession X83425, nucleotides 20-1660 or the corresponding regions in domain deleted cDNAs) were amplified by PCR using a sense primer with a 5′ HindIII restriction site (TGGGATCCAGATAGGCCACGCGCAGCTCC) and an antisense primer with an in-frame 5′ consensus splice sequence and BamHI site (ACGGATCCACTTACCTGTCTCTGGCGAGCCTTGGACCA). The PCR templates were full-length Lu cDNA or domain deletion Lu cDNAs in pBluescript prepared as previously described.22 PCR products were subcloned into pIg vector as described23 and verified by sequence analysis. The region of mouse Lu cDNA encoding the predicted extracellular domains was amplified by PCR using a sense primer with a 5′ HindIII restriction site (AATATAAGCTTAACATGGAACCCCCTGACGCC) and an antisense primer with an in-frame, 5′ SalI site (CTAGTCTAGACTGGGCAGTCTGAGGGGCCACA). The PCR product was subcloned into pIg+ vector (R&D Systems, Oxford, United Kingdom) and verified by sequence analysis. Recombinant proteins were overexpressed by transient transfection of COS-7 cells as described23 and either directly captured on anti-Fc on the cuvette of the optical biosensor or purified on protein A sepharose. Full-length Lu-Fc and domain deletion mutants were tested in the biosensor for binding of monoclonal antibodies against the first N-terminal IgSF domain (BRIC 108 and BRIC 224) or the fourth IgSF domain (BRIC 221) of human Lu gp22prior to use in laminin binding experiments. Clones encoding the control Fc fusion proteins MUC18-Fc and NCAM-Fc were gifts from Dr David Simmons (SmithKline Beecham, Harlow, United Kingdom). Control LW-Fc and CD47-Fc were prepared as above.
Laminin preparations
Human placental laminin, affinity purified using antibody against laminin α5 chain (human laminin 10/11), was obtained from Calbiochem (Nottingham, United Kingdom) or from Gibco (Paisley, United Kingdom); crude, human placental laminin (human merosin) from Gibco or from Sigma (Poole, United Kingdom); and mouse EHS laminin 1 from Sigma. The protein concentration of each preparation was confirmed using the “Nano-orange” dye binding technique (Molecular Probes, Leiden, The Netherlands). Molar concentrations of each laminin species were calculated using published apparent molecular weights.24 25 Control ECM protein, human thrombospondin were purchased from Calbiochem.
Cell adhesion assay
Laminin or bovine serum albumin (BSA, Sigma) in 0.1 M sodium bicarbonate pH 9.6 was coated on microplates (Immulon-4, Dynex Technologies, Billingshurst, United Kingdom) at 4°C for 18 hours. Various amounts of laminin (0-5 × 10−12 mole) were added to each well and allowed to bind passively. K562 transfectants were fluorescently labeled using 2′, 7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester (BCECF, Sigma). Ten microliters of 1 mg/mL BCECF was added to 107 cells suspended in 1 mL Iscove modified Eagle medium containing 5% human AB serum (IMEM-AB), incubated at 37°C for 15 minutes, then washed in IMEM-AB. Then, 104 cells, suspended in IMEM-AB, were added to each well and allowed to adhere at 37°C for 1 hour before unbound cells were removed by washing with IMEM-AB. Fluorescence was measured before and after washing and the percentage bound cells quantitated.
Optical biosensor assay
Binding of laminin to Lu-Fc was examined in an IAsys optical biosensor (Affinity Sensors, Cambridge, United Kingdom). Affinity purified goat antihuman Fc (Sigma) was covalently coupled to a carboxy methyl dextran reaction cuvette (Affinity Sensors) according to the manufacturer's instructions. Fc fusion proteins in phosphate-buffered saline (PBS) pH 7.4, 0.05% Tween 20 (PBSt) were captured on this matrix and tested for interaction with laminin. To characterize the kinetic rate and equilibrium constants for the interaction of Lu-Fc with human laminin 10/11 soluble, recombinant, full-length Lu-Fc was added to PBSt buffer (final concentration 10 μg/mL) and captured on the anti-Fc surface until the biosensor recorded a response of 100 arc seconds; then the buffer was changed. Dissociation of Lu-Fc from anti-Fc was negligible. Soluble, human laminin 10/11 was applied (final concentration 24 nM) and association recorded for 4 minutes before the laminin was exchanged for PBSt and dissociation of the complex recorded. In all subsequent experiments Lu-Fc or control recombinant Fc fusion proteins were applied to the anti-Fc matrix to give an instrument response of 100 arc seconds before washing with PBSt and examination of laminin interaction. Association and dissociation rates were estimated by curve fitting using biosensor software.
Fluorescence-imaged microdeformation
The redistribution of membrane components in response to mechanical deformation was analyzed by fluorescence-imaged microdeformation (FIMD).26-28 The membrane component of interest was fluorescently labeled in situ and then individual cells were aspirated into a glass micropipette (diameter 1.0-1.2 μm). Aspiration created an in-plane deformation of the membrane skeleton, which condensed to enter the micropipette and subsequently dilated down the micropipette. Aspiration length scaled by the micropipette radius (L/Rp) was predetermined by the surface-to-volume ratio of the cell (osmotically controlled) and by the large compressibility modulus of the lipid bilayer. The relative density of a fluorescently labeled component at different locations along the aspiration length was quantified as the fluorescence intensity at that location normalized by the fluorescence intensity of the labeled molecules in the spherical portion of the aspirated cell.
Results
Cloning and characterization of mouse Lu
By scanning the GenBank database with the human Lu protein sequence we identified 5 mouse EST clones (accession numbers AA034658,AA008742, W48082, W78289, and AA038169), which were more closely related to the human Lu protein than to any other member of the human IgSF. The clones were obtained from IMAGE Consortium and found to contain overlapping DNA sequences, suggesting that they were derived from the same cDNA. We characterized the cDNA insert sizes of these 5 ESTs and determined that the cDNA insert in IMAGE clone 466937 (GenBank accession number AA034658) was ∼2.4 kb. Because the human Lu cDNA protein-coding region comprises ∼1.89 kb, we anticipated that this ∼2.4-kb cDNA insert would provide us with the complete protein coding region. The clone was sequenced and found to contain a cDNA insert of 2.34 kb with a single open reading frame of 1869 bp encoding the putative mouse Lu gp, the 3′ nontranslated region and poly-A tail. The cDNA and translated amino acid sequences have been submitted to GenBank (accession number AF221507).
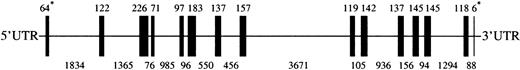
To clone the murine Lu gene we screened a BAC murine genomic library (CitbCJ7) derived from ES cell lines of the 129/SvJ mouse strain. Using primers in the 3′ untranslated region, we isolated a BAC clone 97J17, which was shown by PCR and hybridization with cDNA probes to contain the entire Lu gene (GenBank accession numberAF246667). Sequencing of the BAC DNA showed that the murine Lugene was 14.1 kb and contained 15 exons, ranging in size from 6 to 226 bp (Figure 1). All of the intron/exon boundary sequences exhibited the characteristic 5′ splice donor GT and the 3′ splice acceptor AG motifs (Table1). The exonic sequences derived from our genomic sequence were completely homologous with the sequence of the Lu cDNA derived from the EST clone with the exception of 6 nucleotides (nt 4830, 5035, 5103, 6317,10114, and 12046). Three of these 6 nucleotide differences would result in amino acid changes. Specifically, nt 5035 in exon 6 was G in the genomic sequence and A in the cDNA sequence, generating codons for aspartic acid and asparagine, respectively. Nucleotide 6317 in exon 8 was G in the genomic sequence and C in the cDNA sequence, encoding glycine and arginine respectively. Finally, nt 10114 in exon 9 was C in the genomic sequence and T in the cDNA sequence, coding for alanine and valine, respectively. The size and organization of the murine gene was very similar to that of its human counterpart, which also contains 15 exons distributed over 13 kb.22 29 Mouse Lu, like human LU, does not have Kozak consensus sequence around the initiating ATG. In addition, both mouse and human genes have stop codons at the beginning of intron 13 (CCgtaagtg and CCgtgagtg, respectively). In the case of the human gene this structure results in alternative splicing of intron 13 that generates 2 different stop codons, producing the 85-kd and 78-kd isoforms of human Lu gp. Further study will be necessary to determine whether similar splicing occurs in the mouse.
Mouse
Lu gene. A cartoon depicting the 13575 bpLu gene. Vertical bars represent the 15 exons ofLu. The sizes of exons are shown above the cartoon and sizes of introns below. *The sizes of the 1st and 15th exons indicate the number of bases in coding sequence only. UTR indicates untranslated region.
Mouse
Lu gene. A cartoon depicting the 13575 bpLu gene. Vertical bars represent the 15 exons ofLu. The sizes of exons are shown above the cartoon and sizes of introns below. *The sizes of the 1st and 15th exons indicate the number of bases in coding sequence only. UTR indicates untranslated region.
Organization of the mouse Lu gene
| Exon . | Nucleotide number* . | IgSF domain . | Splice acceptor site . | Splice donor site . |
|---|---|---|---|---|
| 1 | 1-64 | LS | CTCAG/gtatgacttggggct | |
| 2 | 1899-2020 | D1a | Tgcactgctctgcag/GTGCC | TCCTC/gtgagtgctctgtgc |
| 3 | 3386-3611 | D1b | Ccaacacacctacag/GTTGA | GTTTG/gtaagtgtccccaaa |
| 4 | 3688-3758 | D2a | acatctttccaacag/CAACA | AGGAG/gtaatatggactgga |
| 5 | 4744-4840 | D2b | ccctgtcttccccag/ATTGC | CCAAA/gtgagccatcgggag |
| 6 | 4937-5119 | D2c | atgcctcccttacag/AGGGC | GCACT/gtgagtccccatgac |
| 7 | 5670-5806 | D3a | cctctcctcccacag/ATCCC | AGCAG/gtaactcactctgtg |
| 8 | 6263-6419 | D3b | catcctcccgctcag/GGTAC | GGCCT/gtgagagctttggga |
| 9 | 10 090-10 208 | D4a | ttttccccactgcag/ACCTG | CCAAG/gtgactggaggaaag |
| 10 | 10 314-10 455 | D4b | ctcttccttccctag/GATTC | CCAAG/gtttggatggttggg |
| 11 | 11 392-11 528 | D5a | ccctgtcacccacag/GAGCA | ATACT/gtaaggggctttcct |
| 12 | 11 685-11 829 | D5b | tccaagcctccacag/CCAGC | CTCTG/gtgggtacccaagaa |
| 13 | 11 924-12 068 | TM | tttttctcattacag/TGGCC | GCGCC/gtaagtgcttgcaag |
| 14 | 13 363-13 480 | Cyto | actctcctcccacag/GCCAG | ATGAG/gtgagtgatggtctg |
| 15 | 13 569-13 574 | Cyto | ccgttctgtccccag/TGTTG |
| Exon . | Nucleotide number* . | IgSF domain . | Splice acceptor site . | Splice donor site . |
|---|---|---|---|---|
| 1 | 1-64 | LS | CTCAG/gtatgacttggggct | |
| 2 | 1899-2020 | D1a | Tgcactgctctgcag/GTGCC | TCCTC/gtgagtgctctgtgc |
| 3 | 3386-3611 | D1b | Ccaacacacctacag/GTTGA | GTTTG/gtaagtgtccccaaa |
| 4 | 3688-3758 | D2a | acatctttccaacag/CAACA | AGGAG/gtaatatggactgga |
| 5 | 4744-4840 | D2b | ccctgtcttccccag/ATTGC | CCAAA/gtgagccatcgggag |
| 6 | 4937-5119 | D2c | atgcctcccttacag/AGGGC | GCACT/gtgagtccccatgac |
| 7 | 5670-5806 | D3a | cctctcctcccacag/ATCCC | AGCAG/gtaactcactctgtg |
| 8 | 6263-6419 | D3b | catcctcccgctcag/GGTAC | GGCCT/gtgagagctttggga |
| 9 | 10 090-10 208 | D4a | ttttccccactgcag/ACCTG | CCAAG/gtgactggaggaaag |
| 10 | 10 314-10 455 | D4b | ctcttccttccctag/GATTC | CCAAG/gtttggatggttggg |
| 11 | 11 392-11 528 | D5a | ccctgtcacccacag/GAGCA | ATACT/gtaaggggctttcct |
| 12 | 11 685-11 829 | D5b | tccaagcctccacag/CCAGC | CTCTG/gtgggtacccaagaa |
| 13 | 11 924-12 068 | TM | tttttctcattacag/TGGCC | GCGCC/gtaagtgcttgcaag |
| 14 | 13 363-13 480 | Cyto | actctcctcccacag/GCCAG | ATGAG/gtgagtgatggtctg |
| 15 | 13 569-13 574 | Cyto | ccgttctgtccccag/TGTTG |
+1 taken as the first nucleotide of the initiation codon in the cDNA sequence.
LS indicates leader sequence; D, IgSF domain; TM, membrane-spanning domain; Cyto, cytoplasmic domain.
Chromosomal localization of mouse Lu
The chromosomal localization of the mouse Lu gene was mapped using The Jackson Laboratory BSS interspecific back-cross panel.19Lu was nonrecombinant with Atp6c2 and D7Mit56, placing Lu ∼4 cM distal to the centromere on mouse chromosome 7 (Figure 2). This region of mouse chromosome 7 shows conserved synteny with human 19q13.2,30 where human LUlocalizes.3 13 The typing data have been placed in the Mouse Genome Database (accession number J:60990) and can be accessed through the World Wide Web (http://www.jax.org). BAC DNA containing theLu gene localized to the proximal end of chromosome 7 by FISH analysis (data not shown), confirming the linkage data.
Lu localizes to region of mouse chromosome 7 that shows conserved synteny with human 19q13.2.
Chromosomal localization ofLu on the Jackson Laboratory BSS interspecific back-cross panel. The map is depicted with the centromere toward the top. The percent recombination (R) between adjacent loci is given to the left of the chromosome bar, with the SE for each R. Loci mapping to the same position are listed in alphabetical order. Missing typings were inferred from surrounding data where assignment was unambiguous. References for mapping the other loci are publicly available from The Jackson Laboratory Mapping Resource through http://www.jax.org /resources/documents/cmdata.
Lu localizes to region of mouse chromosome 7 that shows conserved synteny with human 19q13.2.
Chromosomal localization ofLu on the Jackson Laboratory BSS interspecific back-cross panel. The map is depicted with the centromere toward the top. The percent recombination (R) between adjacent loci is given to the left of the chromosome bar, with the SE for each R. Loci mapping to the same position are listed in alphabetical order. Missing typings were inferred from surrounding data where assignment was unambiguous. References for mapping the other loci are publicly available from The Jackson Laboratory Mapping Resource through http://www.jax.org /resources/documents/cmdata.
Amino acid sequence comparisons of mouse and human Lu
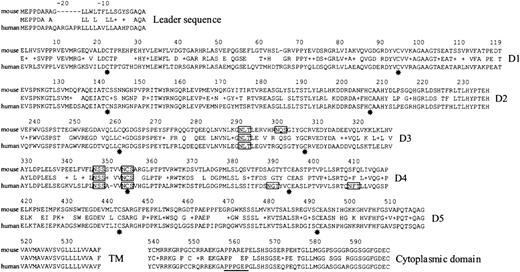
Characterization of mouse Lu cDNA allowed us to align the deduced amino acid sequences of murine and human Lu gp3 using the “BLAST 2 sequences” program (NCBI Entrez, Figure3). The level of amino acid identity between the putative translated products was 72%. The mouse Lu cDNA we have sequenced encodes the 85-kd Lu glycoprotein isoform. Analysis of the extracellular region of the murine homologue of Lu revealed several interesting features. It appears likely that the disulfide-bonded IgSF domains will be similarly folded in the murine and human polypeptides because the critical cysteine residues, 22, 93, 140, 205, 260, 305, 353, 393, 441, and 492, as well as other key residues within each of the predicted IgSF domains, are conserved. In addition, 3 of the 4 potential glycosylation sites in the mouse third and fourth extracellular domains are conserved. Within the cytoplasmic tail the proline-rich SH3 binding motif P557PPXXP present in human is partially conserved in mouse (P557PAXXP) and each of the 5 serine residues is conserved, suggesting that ligand-induced receptor signaling may be similar in mouse and human cells.
Alignment of mouse and human Lutheran glycoprotein sequences.
Identical amino acid residues and conservative substitutions (+) between the mouse and human sequences are indicated. Residues are numbered from the known N-terminal residue in human Lu gp. The 2 cysteine residues (*) within each of the 5 predicted IgSF domains are conserved. Potential N-glycosylation sites are boxed—3 of the 4 sites in the mouse sequence are conserved. The proline-rich, putative SH3-binding site within the cytoplasmic domain of human Lu gp is underlined—this motif is partly conserved in the mouse sequence. D1-D5 indicates 5 extracellular, IgSF domains; TM, transmembrane region. The cDNA and translated amino acid sequences have been submitted to GenBank (accession number AF221507).
Alignment of mouse and human Lutheran glycoprotein sequences.
Identical amino acid residues and conservative substitutions (+) between the mouse and human sequences are indicated. Residues are numbered from the known N-terminal residue in human Lu gp. The 2 cysteine residues (*) within each of the 5 predicted IgSF domains are conserved. Potential N-glycosylation sites are boxed—3 of the 4 sites in the mouse sequence are conserved. The proline-rich, putative SH3-binding site within the cytoplasmic domain of human Lu gp is underlined—this motif is partly conserved in the mouse sequence. D1-D5 indicates 5 extracellular, IgSF domains; TM, transmembrane region. The cDNA and translated amino acid sequences have been submitted to GenBank (accession number AF221507).
Human and mouse Lu-Fc fusion proteins specifically bind human laminin 10/11 with high affinity
Although there is now very good evidence that Lu gp serves as a laminin receptor, the particular laminin proteins that function as ligands in this interaction have not yet been characterized. Laminins are a large family of proteins, each composed of 3 polypeptide chains (α, β, and γ).25 There are at least 5 distinct α chains and similar numbers of β and γ chains. Earlier studies on laminin adhesion have used laminins prepared from mouse Engelbreth Holm Schwarm (EHS) tumor cell cultures or from human placenta. Mouse EHS-laminin is the prototype laminin 1 (α1β1γ1). In contrast, crude human placental laminin preparations (merosin) contain mainly laminin 2 (α2β1γ1), laminin 4 (α2β2γ1), and a comparatively small proportion of laminins 10 (α5β1γ1) and 11 (α5β2γ1).24 Recently it has been discovered that laminins 10 and 11 can be specifically purified from human placental laminin,24 using a monoclonal antibody that recognizes the laminin α5 chain.31 Thus a number of distinct laminin isoforms are now available for adhesion assays. In the present study we explored the possibility that a specific form of laminin is a high-affinity ligand of Lu gp.
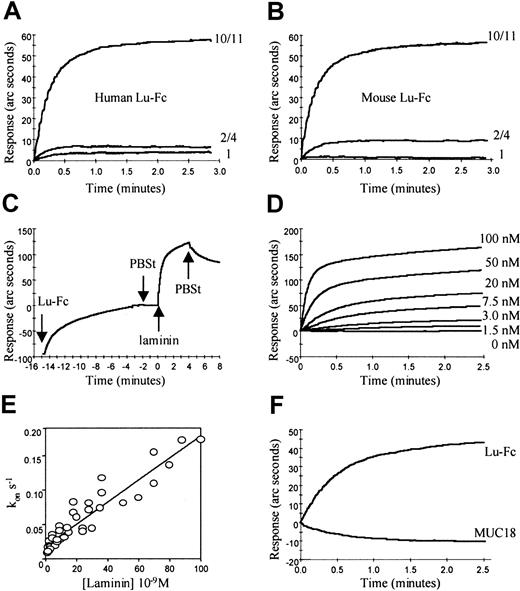
To define the laminin isoform(s) that associates with Lu gp, the optical biosensor assay was used. Recombinant human Lu-Fc was captured on the anti-Fc biosensor cuvette; then affinity purified human laminin 10/11, crude human placental laminin, or mouse laminin 1 was applied (each at a final concentration of 42 nM) and association was recorded. We observed that human Lu-Fc protein bound human laminin 10/11 (Figure4A). In marked contrast, a relatively small association response was observed with an equal molar concentration of the crude human placental laminin preparation (laminin 2/4) and no binding occurred to mouse laminin 1 (Figure 4A).
Human and mouse Lu-Fc bind laminin 10/11 specifically and with high affinity.
Binding of human laminin 10/11 to recombinant Lu-Fc proteins was examined in an optical biosensor. (A) Human Lu-Fc bound 42-nM purified human laminin 10/11 (10/11) but not 42-nM crude human, placental laminin 2/4 (2/4), or 42-nM mouse, EHS, laminin 1(1). (B) Mouse Lu-Fc bound 42-nM purified human laminin 10/11 (10/11) but not 42-nM crude human, placental laminin 2/4 (2/4), or 42-nM mouse laminin 1(1). (C) Lu-Fc was captured on the anti-Fc surface at time −15 minutes and allowed to associate until the biosensor recorded a response of 100 arc seconds. The Lu-Fc was removed and fresh buffer (PBSt) was added at time −2.5 minutes. Soluble, human laminin 10/11 was applied (final concentration 24 nM) at time 0 minute and association was recorded for 4 minutes before the laminin was exchanged for PBSt and dissociation rate (koff) of the complex was measured. In all experiments Fc fusion proteins were applied to the anti-Fc matrix to give an instrument response of 100 arc seconds before washing with PBSt, addition of laminin at time 0 minute and measurement of laminin binding. (D) Association rates (kon) for the interaction of human Lu-Fc with human laminin 10/11 were measured at laminin concentrations in the range 0.1 to 100 nM by curve fitting using the biosensor software; representative data are shown. (E) The measured values for kon were plotted against laminin concentration and the association rate constant (kass) was derived from the gradient of this plot using the biosensor software. (F) Human Lu-Fc but not the structural analogue MUC 18-Fc (MUC18) bound 10 nM laminin 10/11.
Human and mouse Lu-Fc bind laminin 10/11 specifically and with high affinity.
Binding of human laminin 10/11 to recombinant Lu-Fc proteins was examined in an optical biosensor. (A) Human Lu-Fc bound 42-nM purified human laminin 10/11 (10/11) but not 42-nM crude human, placental laminin 2/4 (2/4), or 42-nM mouse, EHS, laminin 1(1). (B) Mouse Lu-Fc bound 42-nM purified human laminin 10/11 (10/11) but not 42-nM crude human, placental laminin 2/4 (2/4), or 42-nM mouse laminin 1(1). (C) Lu-Fc was captured on the anti-Fc surface at time −15 minutes and allowed to associate until the biosensor recorded a response of 100 arc seconds. The Lu-Fc was removed and fresh buffer (PBSt) was added at time −2.5 minutes. Soluble, human laminin 10/11 was applied (final concentration 24 nM) at time 0 minute and association was recorded for 4 minutes before the laminin was exchanged for PBSt and dissociation rate (koff) of the complex was measured. In all experiments Fc fusion proteins were applied to the anti-Fc matrix to give an instrument response of 100 arc seconds before washing with PBSt, addition of laminin at time 0 minute and measurement of laminin binding. (D) Association rates (kon) for the interaction of human Lu-Fc with human laminin 10/11 were measured at laminin concentrations in the range 0.1 to 100 nM by curve fitting using the biosensor software; representative data are shown. (E) The measured values for kon were plotted against laminin concentration and the association rate constant (kass) was derived from the gradient of this plot using the biosensor software. (F) Human Lu-Fc but not the structural analogue MUC 18-Fc (MUC18) bound 10 nM laminin 10/11.
Having obtained the predicted sequence of the murine homologue of Lu gp, we were also able to determine whether murine Lu gp interacts with the same laminin isoforms as human Lu gp. After expressing the predicted extracellular domains of murine Lu as an Fc fusion protein, we tested this recombinant polypeptide in the biosensor assay. Mouse Lu-Fc also bound human laminin isoforms 10 and 11, a small proportion of crude human placental laminin but not mouse laminin 1 (Figure 4B). These data clearly show that both human and mouse Lu are specific membrane receptors for laminin isoform(s) containing the α5 polypeptide chain.
To characterize the kinetics of the human Lu gp/laminin interaction, soluble recombinant, full-length Lu-Fc was added to PBSt buffer (final concentration 10 μg/mL) and captured on the anti-Fc surface (Figure 4C) until the biosensor recorded a response of 100 arc seconds and then the buffer was changed. Dissociation of Lu-Fc from the anti-Fc was negligible (Figure 4C). Soluble, human laminin 10/11 was then applied (final concentration 24 nM) and kinetics of the protein-protein interaction was recorded for 4 minutes before the laminin was exchanged for PBSt and dissociation of the protein complex recorded (Figure 4C).
The association rates (kon) and dissociation rates (koff) for the interaction with human laminin 10/11 were measured at laminin concentrations in the range 0.1 to 100 nM. Values for kon and koff were estimated by curve fitting using biosensor software assuming simple, monophasic interaction (A+B↔AB, representative data for the association of laminin 10/11 with human Lu-Fc are shown in Figure 4D). Measured values for kon were plotted against laminin concentration and the association rate constant (kass) was derived from the gradient of this plot using biosensor software. Data for the association of laminin 10/11 with human Lu-Fc are shown in Figure 4E. The dissociation rate constant (kdiss) was taken as the mean value for koff. The dissociation equilibrium constant (KD) was calculated (KD = Kdiss/Kass). Errors for kass were obtained using the biosensor software and errors for kdissand KD were calculated using the sum of squares method. The results of kinetic analysis for the interaction of human laminin 10/11 with human Lu-Fc revealed high-affinity binding (kass = 1.6 ± 0.09 × 106M−1s−1, kdiss = 1.26 ± 0.01 × 10−2 s−1, KD = 7.9 ± 0.47 nM).
The interaction of human laminin 10/11 with mouse Lu-Fc was examined in a similar manner (data not shown). Compared with human Lu-Fc the affinity of the binding of laminin 10/11 to mouse Lu-Fc was found to be approximately an order of magnitude lower (kass = 3.5 ± 0.7 × 105M−1s−1, kdiss = 2.62 ± 0.01 × 10−2s−1, KD = 74.5 ± 14.5 nM).
To determine the specificity of the binding interaction, we tested other cell adhesion molecules expressed as soluble Fc fusion proteins. We observed that the extracellular region of the human structural analogue MUC 18-Fc failed to bind 10 nM laminin (Figure 4F) as did LW-Fc, NCAM-Fc, and CD47-Fc (data not shown). Soluble human thrombospondin (final concentration 2.9 nM) was also applied to captured human Lu-Fc and failed to bind (data not shown).
K562 transfectants expressing human Lu gp specifically bind human laminin 10/11
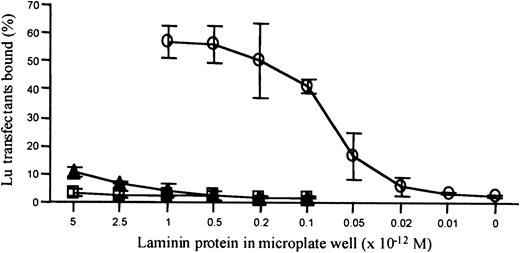
As an independent and complementary approach to identifying the laminin ligand, K562 transfectants were tested in a microplate cell adhesion assay. Laminin preparations were titrated and added to microplates in the range 5 × 10−12 to 5 × 10−16 mole per microplate well. Fluorescently labeled K562 transfectants expressing full-length human Lu gp were allowed to adhere to the coated microplates, fluorescence was measured before and after washing, and the percentage bound cells was calculated. The titration results clearly show that in this static adhesion assay system the human Lutheran transfectant adhered in microplate wells coated with small quantities of human laminin 10/11 (37% cell adhesion at 5 × 10−13 mole per well added laminin, Figure 5). In contrast, the transfectants showed poor adhesion in wells containing 10-fold greater quantities of mouse laminin 1 or crude human placental laminin (3% and 11% cell adhesion, respectively, at 5 × 10−12 mole per well added laminin, Figure 5). The data from this series of experiments, therefore, confirmed the results of the biosensor assays and clearly demonstrated that human Lutheran Fc fusion proteins specifically bind human laminins containing the α5 polypeptide chain.
Human Lu gp transfectants specifically adhere to immobilized laminin 10/11.
Laminin preparations were titrated and tested for binding of fluorescently labeled K562 transfectants expressing human Lu gp in a microplate adhesion assay. The transfectants adhered to purified laminin 10/11 in a concentration-dependent manner (open circles, 55% binding at 1 × 10−12 mole added protein/well). The transfectants adhered only poorly to crude human, placental laminin 2/4 (closed triangles, 11% binding at 5 × 10−12 mole added protein/well) and to mouse laminin 1 (open squares, 3% binding at 5 × 10−12 mole added protein/well). Results are the mean percentage binding from 3 experiments ± 1 SD.
Human Lu gp transfectants specifically adhere to immobilized laminin 10/11.
Laminin preparations were titrated and tested for binding of fluorescently labeled K562 transfectants expressing human Lu gp in a microplate adhesion assay. The transfectants adhered to purified laminin 10/11 in a concentration-dependent manner (open circles, 55% binding at 1 × 10−12 mole added protein/well). The transfectants adhered only poorly to crude human, placental laminin 2/4 (closed triangles, 11% binding at 5 × 10−12 mole added protein/well) and to mouse laminin 1 (open squares, 3% binding at 5 × 10−12 mole added protein/well). Results are the mean percentage binding from 3 experiments ± 1 SD.
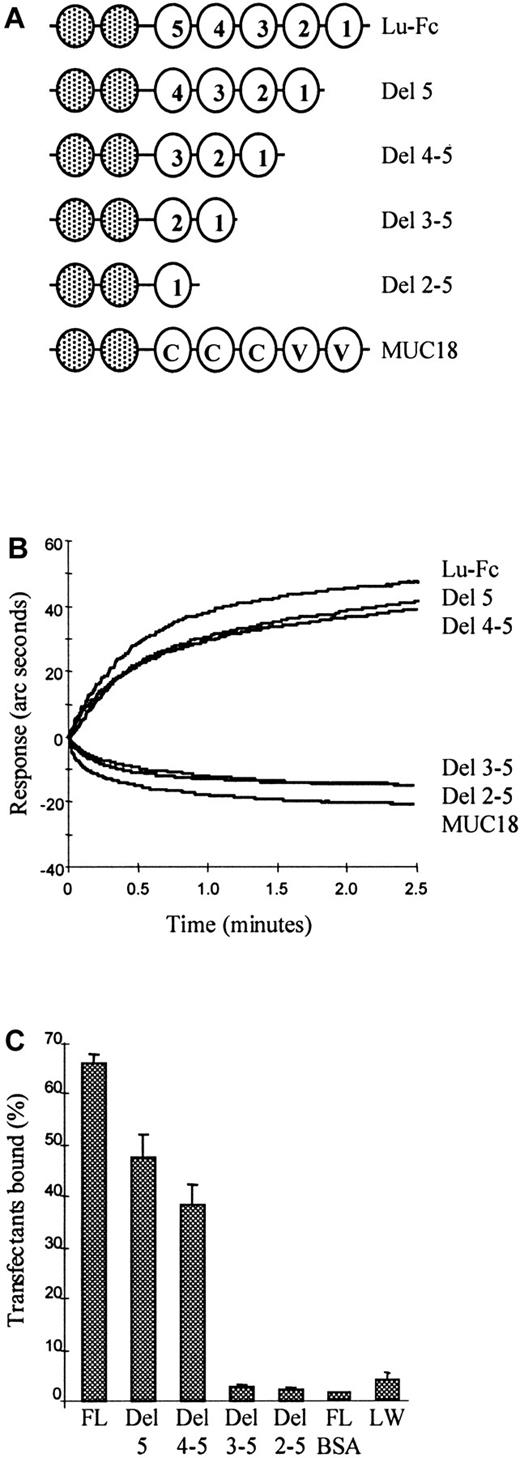
The first 3, N-terminal IgSF domains of human Lu gp bind human laminin 10/11
To identify the laminin-binding domain(s) on Lu gp, Lu deletion mutants, lacking one or more IgSF domains (Figure6A) were expressed as Fc fusion proteins and assayed for laminin binding. Each Fc fusion protein was captured on the anti-Fc biosensor cuvette to give an instrument response of 100 arc seconds. Human laminin 10/11 (final concentration 10 nM) was added to the cuvette and the binding response was recorded. Full-length Lu-Fc, the mutant lacking Lu domain 5 (Del 5), and the mutant lacking domains 4 and 5 (Del 4-5) gave a positive binding curve, whereas the mutant Lu-Fc fusion proteins lacking domains 3, 4, and 5 (Del 3-5); the mutant lacking domains 2, 3, 4, and 5 (Del 2-5); and MUC18-Fc (MUC18) control failed to bind laminin 10/11 (Figure 6B). These data imply that the first 3, N-terminal IgSF domains are critical for laminin binding to Lu gp.
The first 3, N-terminal IgSF domains of human Lutheran bind human laminin 10/11.
(A) Cartoons depicting the structures of full-length Lu-Fc, Lu deletion mutants lacking one or more IgSF domains also expressed as Fc fusion proteins, and the structural analogue MUC18-Fc. (B) These constructs were captured on the anti-Fc biosensor cuvette, then purified human laminin 10/11 (final concentration 10 nM) was added at time 0 minute and association was recorded. Lu-Fc and the mutants Del 5 and Del 4-5 gave a positive binding curve, whereas the mutants Del 3-5 and Del 2-5 and the MUC18-Fc (MUC18) control failed to bind laminin 10/11. (C) Fluorescently labeled K562 transfectants expressing full-length Lu gp or domain deleted Lu gp were tested for adhesion in microplates coated with 10−12 mole purified human laminin 10/11 per well. The extracellular IgSF domain organization of the Lu gp constructs expressed by these transfectants is the same as that depicted for the Fc fusion protein constructs in panel A.22 Transfectants expressing full-length Lu (FL) and mutants Del 5 and Del 4-5 adhered to laminin 10/11 coated plastic, whereas transfectants expressing mutants Del 3-5 or Del 2-5 did not adhere. Negative controls were a K562 transfectant expressing the blood group active LW glycoprotein tested as above (LW) and the K562 transfectant expressing full length Lu gp applied to immobilized bovine albumin (FL BSA).
The first 3, N-terminal IgSF domains of human Lutheran bind human laminin 10/11.
(A) Cartoons depicting the structures of full-length Lu-Fc, Lu deletion mutants lacking one or more IgSF domains also expressed as Fc fusion proteins, and the structural analogue MUC18-Fc. (B) These constructs were captured on the anti-Fc biosensor cuvette, then purified human laminin 10/11 (final concentration 10 nM) was added at time 0 minute and association was recorded. Lu-Fc and the mutants Del 5 and Del 4-5 gave a positive binding curve, whereas the mutants Del 3-5 and Del 2-5 and the MUC18-Fc (MUC18) control failed to bind laminin 10/11. (C) Fluorescently labeled K562 transfectants expressing full-length Lu gp or domain deleted Lu gp were tested for adhesion in microplates coated with 10−12 mole purified human laminin 10/11 per well. The extracellular IgSF domain organization of the Lu gp constructs expressed by these transfectants is the same as that depicted for the Fc fusion protein constructs in panel A.22 Transfectants expressing full-length Lu (FL) and mutants Del 5 and Del 4-5 adhered to laminin 10/11 coated plastic, whereas transfectants expressing mutants Del 3-5 or Del 2-5 did not adhere. Negative controls were a K562 transfectant expressing the blood group active LW glycoprotein tested as above (LW) and the K562 transfectant expressing full length Lu gp applied to immobilized bovine albumin (FL BSA).
As an independent and complementary approach to defining the ligand binding domain on the Lu gp receptor, fluorescently labeled K562 transfectants expressing full-length or domain-deleted Lu gp were tested in the microplate adhesion assay in microplates coated using 10−12 mole human laminin 10/11 per well. Fluorescence was measured before and after washing and the percentage bound cells quantitated. K562 transfectants expressing full-length Lu and mutants Del 5 and Del 4-5 adhered to laminin 10/11, whereas transfectants expressing mutants Del 3-5 and Del 2-5 did not adhere (Figure 6C). Negative controls were K562 transfectants expressing LW glycoprotein and full-length Lu gp applied to laminin and immobilized bovine albumin, respectively. Considered together these data clearly show that a laminin-binding site(s) is present in the 3 membrane distal IgSF domains.
Lu interacts with the spectrin-based membrane skeleton
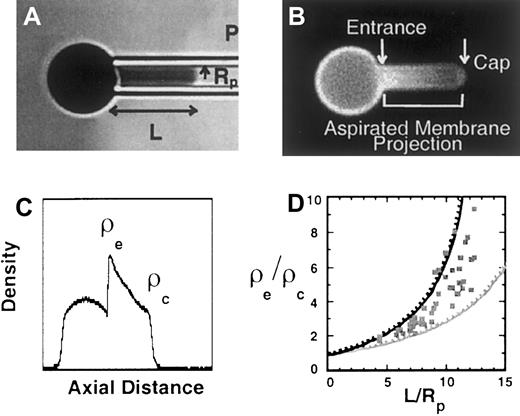
To begin to characterize the binding interactions of the cytoplasmic domain of Lu gp we determined whether Lu gp interacts with the spectrin-based skeleton. For these studies, Lu gp in human erythrocytes was specifically labeled with FITC-conjugated monoclonal anti-Lu gp BRIC 221 and analyzed by fluorescence imaged microdeformation. The redistribution of Lu in response to microdeformation was mapped by quantitating the ratio of entrance to cap density (ρε/ρc) as a function of aspiration length (L/Rp). We observed that FITC-labeled anti-Lu gp exhibited a steep gradient in concentration along the projection, which was almost as steep as that of skeletal-actin (Figure7). This is in contrast to findings with glycophorin A or band 3, which exhibit less steep gradients reflecting the presence of both skeleton-associated and freely diffusing populations of these molecules. Based on these findings we suggest that BRIC 221-labeled Lu is predominantly linked to the membrane skeleton and we speculate that the interaction of its cytoplasmic tail may be critical for laminin-induced signal transduction.
Redistribution of Lu gp after microdeformation.
(A) Bright field image of red cell being aspirated into micropipette. (B) Fluorescence micrograph and (C) corresponding intensity profile of Lu gp labeled with fluorescent antibody BRIC 221 in the partially aspirated red cell. The intensity profile indicated that the density of FITC-labeled anti-Lu gp increased markedly at the pipette entrance (ρε) and subsequently decreased toward the aspiration cap (ρc). (D) The ratio of entrance to cap density (ρε/ρc) of FITC-labeled anti-Lu gp (squares) plotted as a function of aspiration length (L/Rp). For comparison, the upper dashed black line shows prior data of rhodamine phalloidin-labeled actin, indicating the dependence of ρε/ρc versus L/Rp for molecules tightly associated with the membrane skeleton. The lower dashed gray line shows prior data of eosin-5-maleimide (EMA)-labeled band 3, and indicates the presence of both skeleton-associated and freely diffusing populations of band 3. The interpretation of the density gradient obtained with the fluorescence-imaged microdeformation technique is based on accumulated data from FIMD experiments on red cell membranes with well-defined membrane protein abnormalities. We know that the steepness of the density gradient of a particular protein is determined by the extent of its linkage to the cytoskeleton by analyzing the density distribution of integral proteins in cells in which the skeletal proteins linking these integral proteins to the cytoskeleton are lacking. Specifically, although glycophorin C in normal cells exhibits a steep gradient, it is freely diffusable in protein 4.1-deficient cells because in these cells it is not linked to the membrane skeleton due to absence of protein 4.1.26,27 Similarly, in ankyrin-deficient cells, the density of band 3 fails to exhibit a gradient40 because ankyrin molecules are not available to link band 3 to spectrin.
Redistribution of Lu gp after microdeformation.
(A) Bright field image of red cell being aspirated into micropipette. (B) Fluorescence micrograph and (C) corresponding intensity profile of Lu gp labeled with fluorescent antibody BRIC 221 in the partially aspirated red cell. The intensity profile indicated that the density of FITC-labeled anti-Lu gp increased markedly at the pipette entrance (ρε) and subsequently decreased toward the aspiration cap (ρc). (D) The ratio of entrance to cap density (ρε/ρc) of FITC-labeled anti-Lu gp (squares) plotted as a function of aspiration length (L/Rp). For comparison, the upper dashed black line shows prior data of rhodamine phalloidin-labeled actin, indicating the dependence of ρε/ρc versus L/Rp for molecules tightly associated with the membrane skeleton. The lower dashed gray line shows prior data of eosin-5-maleimide (EMA)-labeled band 3, and indicates the presence of both skeleton-associated and freely diffusing populations of band 3. The interpretation of the density gradient obtained with the fluorescence-imaged microdeformation technique is based on accumulated data from FIMD experiments on red cell membranes with well-defined membrane protein abnormalities. We know that the steepness of the density gradient of a particular protein is determined by the extent of its linkage to the cytoskeleton by analyzing the density distribution of integral proteins in cells in which the skeletal proteins linking these integral proteins to the cytoskeleton are lacking. Specifically, although glycophorin C in normal cells exhibits a steep gradient, it is freely diffusable in protein 4.1-deficient cells because in these cells it is not linked to the membrane skeleton due to absence of protein 4.1.26,27 Similarly, in ankyrin-deficient cells, the density of band 3 fails to exhibit a gradient40 because ankyrin molecules are not available to link band 3 to spectrin.
Discussion
A major finding of the current study is that Lu gp is a specific, high-affinity receptor for human laminins containing the α5 polypeptide chain. Of the 10 laminin isoforms identified to date, 2 isoforms are in this category—laminin 10 (α5β1γ1) and laminin 11 (α5β2γ1). Our data further suggest that Lu gp may bind laminins 10/11 via the α5 chain and not via the β or γ chain, because we clearly showed that human laminins 2 (α2β1γ1) and 4 (α2β2γ1), which share common β and γ chains with laminins 10/11, do not associate with Lu gp. This report, therefore, extends the findings that Lu gp(s) is a membrane laminin receptor of erythroid cells9 10 by characterizing the affinity of the Lu gp/laminin interaction and by identifying the specific laminin-binding partner.
We and others have previously shown that Lu gp first appears in erythroid differentiation at the stage of orthochromatic erythroblasts.32,33 These findings suggest that Lu gp/laminin binding may have a role in the later stages of erythroid maturation, specifically in the migration of maturing erythrocytes through the subendothelium of bone marrow sinusoids. Recently, a study of human and murine bone marrow demonstrated that laminin 10/11 is expressed in sinusoidal ECM.34 Our current data showing that laminin 10/11 is the specific Lu gp binding partner strengthens the hypothesis that Lu gp may mediate erythroid adhesion with bone marrow sinusoidal ECM and play a functional role in marrow egress.
Our identification of α5 chain-containing laminins as the Lu gp specific ligand is also consistent with accumulating evidence suggesting that the Lu gp/laminin interaction contributes to the abnormal adhesion of sickle red blood cells to vascular endothelium.9-11,15-17,35 Sickle cells show abnormal adhesion to laminin and it is now clear that an increased expression of Lu gp found on unfractionated red blood cells from patients with sickle cell disease may be the direct causal mechanism of this phenomenon.10,11,36 Indeed, we have found that the Lu gp copy number on sickle cells may be as much as 50% higher than on control red cells from healthy, Afro-Caribbean volunteers (Gardner, Spring, Parsons, Anstee, unpublished observations). It has recently been reported that sickle cells bind to human laminin isoforms 10 and 11 but not to human laminin 2, human laminin 4, or mouse laminin 1.17 Strikingly, our present data indicate that human Lu gp demonstrates a similar adherence pattern and also binds to human laminin-isoforms 10 and 11 but not to human laminin 2, human laminin 4, or mouse laminin 1. These data thus imply that the observed binding interaction between sickle cells and laminins 10 and 11 involves the Lu gp laminin receptor.
To begin to understand the precise molecular mechanisms involved in Lu gp receptor function we characterized in more detail the protein-protein interactions of the extracellular and cytoplasmic domains of this adhesion molecule. Using domain-deleted Lu gp constructs expressed both as soluble fusion proteins and on the membranes of stable transfectants, we have located a laminin binding region within the first 3, N-terminal IgSF domains. Independently, another group has reported that cells transfected with a domain-deleted Lu gp construct containing only the membrane proximal, fifth IgSF domain adhered to laminin.11 In contrast to the data presented here, these investigators found that deletion of domain 5 abolished laminin binding. We have shown, using 2 different techniques, that domain 5 is not critical for laminin binding. Of note, however, in our experiments with transfected K562 cells we did observe that fewer cells expressing mutants with deletions of domain 5 or 4-5 bound laminin compared to cells expressing full-length Lu gp. Hence our current data, coupled with that of Zen and colleagues,11raises the intriguing possibility that there may be 2 laminin-binding regions on the Lu gp structure: one on domains 1-3 and the other on domain 5. There is precedence for such a hypothesis since VCAM-1, which has 7 IgSF domains, contains 2 distinct binding sites for α4β1 integrin: one on the membrane distal IgSF domain and the other on the fourth IgSF domain.37 38
The protein-protein interactions of the cytoplasmic domain of a receptor molecule can play critical roles in regulating receptor function. To determine whether Lu interacts with the spectrin-based skeleton we used a molecular mapping technique that is a powerful means of providing insight into in situ protein linkages. Our findings that BRIC 221-labeled Lu exhibited a steep gradient in concentration along the membrane projection, which was almost as steep as that of skeletal actin, implies that Lu is predominantly skeleton associated. We postulate that this interaction of the Lu gp cytoplasmic tail with the spectrin-based skeleton may also be critical for laminin-induced signal transduction. In future studies it will be interesting to determine whether laminin binding alters the protein interactions of the Lu cytoplasmic domain.
Because we believe that the development of a suitable animal model is a key requirement for obtaining a detailed understanding of the role of Lu in normal erythropoiesis and in sickle cell adhesion, we cloned and sequenced the mouse gene encoding the murine homologue to human Lu gp. Our characterization of mouse Lu confirms and extends the observations very recently reported by Rahuel and colleagues.39 These investigators described the intron/exon boundaries, sequenced 7 of the 14 introns, and estimated the sizes of the remaining 7 introns. We have obtained and submitted to GenBank the complete sequence of the 15 exons and all 14 introns. The splice acceptor and donor site sequences obtained by our 2 groups are very similar with the differences being primarily at the single nucleotide level. With the exception of introns 1, 2, and 8 earlier estimates of intron size39 are within 4 to 94 nucleotides of our complete sequence data. In the case of introns 1, 2, and 8, Rahuel and colleagues predicted sizes of more than 2000, ∼1600, and ∼4200 nucleotides, respectively, whereas our genomic sequence data have shown that intron 1 is 1834 nt, intron 2 is 1365, and intron 8 is 3671. The exonic sequences derived from our genomic sequence were completely homologous with the cDNA sequence we obtained of the EST clones with the exception of single nucleotide variations in exons 5, 6, 8, 9, and 13. The only nucleotide differences that would result in changes in amino acid sequence occurred in exons 6, 8, and 9 where the genomic sequence would encode an aspartic acid, glycine, and alanine, respectively, whereas the cDNA sequence would encode an asparagine, arginine, and valine, respectively. Interestingly in human Lu gp, blood group single nucleotide polymorphisms have been mapped to each of the 5 IgSF domains22 and it is possible that a similar situation may exist for mouse.
From our characterization of the murine homologue we discovered that human and mouse Lu gps are highly conserved at the amino acid sequence level (72% overall identity). This conservation was reflected within individual IgSF domains but, among candidate ligand-binding domains, the first domains showed least identity (65%), whereas the second and third domains were most conserved (82% and 75%, respectively). The fifth domain showed 69% identity. Strikingly consistent with these findings, the predicted extracellular IgSF domains of mouse Lu gp, when expressed as a fusion protein, displayed the same binding pattern as human Lu gp and specifically bound human laminin 10/11 but not human laminin 2/4 or mouse laminin 1. However, the affinity of the interaction of human laminin 10/11 with mouse Lu gp, while still in the nanomolar range, was an order of magnitude lower than that observed for human Lu gp. Examination of published amino acid sequences of various laminin α chains revealed that while only a partial sequence is published for the human α5 chain, it shows 68% identity with mouse α5. We speculate that the observed differences in affinities might reflect minor divergences at the primary sequence or functional levels during evolution. In contrast to our results, Rahuel and coworkers reported that transfectants expressing a fusion protein comprising N-terminal c-myc and full length mouse Lu gp bound mouse laminin 1.39 One possible explanation for this is that the presence of N-terminal c-myc at or close to the laminin-binding region may have caused partial loss of ligand specificity in this construct. However, our finding that the murine Lu gp has conserved the laminin-binding specificity of the human receptor molecule strongly suggests that the Lu gp/laminin 10/11 interaction may have a key biologic function. Ahead lies the challenge of defining the precise role of Lu gp in normal erythropoiesis and in sickle cell disease.
Acknowledgments
We would like to thank Dr Marja Ekblom for advice on the nature of commercial laminin preparations. We are very grateful to Dr Uli Weier and Dr Robert Lersch (LBNL, Berkeley, CA) for their invaluable help in FISH analysis and to Mr Michael Patterson for his expert technical assistance.
Supported in part by National Institutes of Health grants DK56267, DK26263, DK32094, and HL31579 and by the Director, Office of Health and Environment Research Division, US Department of Energy, under Contract DE-AC03- 76SF00098.
D.J.A. and J.A.C. contributed equally to the direction of this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dr Joel Anne Chasis, Lawrence Berkeley National Laboratory, Bldg 74, 1 Cyclotron Rd, Berkeley, CA 94720; e-mail:jachasis@lbl.gov; David J. Anstee, Bristol Institute for Transfusion Sciences, Southmead Road, Bristol, BS10 5ND, United Kingdom; e-mail:david.anstee@nbs.nhs.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal