Abstract
Cytokine receptors activate signals that regulate the transcription factor E2F-1, which then coordinates the expression of genes essential for DNA synthesis and cell cycle progression. Overexpression of E2F-1 most often induces S-phase entry followed by apoptosis, but in some cell types it leads to continuous proliferation and transformation. Here, it is shown that constitutive expression of E2F-1 promotes cytokine-independent proliferation in the murine pro-B cell line BaF-B03. There was no enhancement of apoptosis following cytokine withdrawal in these cells, despite the presence of intact p53-dependent apoptotic pathways. Notwithstanding the continuous presence of E2F-1, the cell cycle–dependent expression of cyclin A, cyclin B1, cyclin D1, cyclin E, and proliferating-cell nuclear antigen was restored with a pattern equivalent to that associated with cytokine stimulation. These findings provide evidence that, in the absence of cytokine, constitutive expression of E2F-1 can promote cell cycle progression and prevent apoptosis.
Introduction
The balance between cell proliferation and apoptosis is essential to immune development and plays a pivotal role in immunoregulatory processes. Many of these processes are mediated by cytokine receptors, which control divergent signal transduction pathways that regulate cell survival, differentiation, and proliferation of hematopoietic cells. For receptors belonging to the cytokine receptor superfamily, the initiating events in these pathways involve Jak kinases and lead to recruitment of phosphoinositide second messengers and activation of the GTPase, Ras.1,2The transcription factor E2F-1 is a primary target of cytokine receptor signals and a key regulator of the cell cycle machinery. Recent developments have identified a more complex biology of E2F-1 and have found that it acts as a bimodal switch that has the capacity to control both proliferation and cell survival.3 4
Together with its heterodimeric partner DP-1, E2F-1 promotes progression of the cell cycle beyond the G1 restriction point by binding to promoter regions of genes encoding for factors essential for DNA synthesis and cell cycle control and by inducing their transcription prior to and during S phase.5 Activity of E2F-1 is negatively regulated in G1 phase by interaction with the retinoblastoma protein (pRB), which, upon hyperphosphorylation by cyclin D or cyclin E kinase complexes, releases E2F-1, thereby allowing E2F-1 target gene activation. During progression of cells through S phase, E2F-1 activity is regulated by cyclin A:CDK2 complexes that phosphorylate E2F-1:DP-1 complexes and inhibit binding to DNA target sequences.6-9 A key regulatory event in the control of E2F-1 activity is growth factor–dependent expression and activation of D-type cyclins in G1 phase of the cell cycle.10 Several signaling pathways have been implicated in this process, including phosphatidylinositol 3-kinase–dependent pathways,10 JAK-STAT pathways,11 and Ras-MAP kinase pathways.12 13
The pivotal role of E2F-1 in regulating cell proliferation requires very strict control, and overexpression leads to DNA synthesis and S-phase entry in some cells.14-16 However, these cells mostly progress to apoptosis following S-phase entry, and transformation with oncogenic potential has been demonstrated only in a small number of cell types, including rat embryo fibroblasts,17,18 mouse fibroblasts,19chicken embryo fibroblasts,20 and astrocytes.21 In order to understand the role of this pathway in cell cycle regulation by cytokine growth factor receptors, we have overexpressed E2F-1 in the murine pro-B cell line BaF-B03. The BaF-B03 cell line is dependent on interleukin (IL)–3 for proliferation and survival and has been widely used for cytokine research. The results of this study demonstrate that E2F-1 overexpression leads to cytokine-independent proliferation of BaF-B03 cells and associated restoration of cell cycle progression in an orderly fashion. No aberration of DNA synthesis or dysregulation of the cell cycle occurred in this model. Using the technique of bivariate flow-cytometric analysis, we demonstrate that a cytokine mitogenic stimulus induces cell cycle–dependent expression of cyclin and proliferating-cell nuclear antigen (PCNA) proteins, but not CDK proteins, in the BaF-B03 cell line, and all of these events are coordinately regulated by E2F-1. The results highlight the role of E2F-1 as an integration point for cytokine-dependent proliferative signaling in the BaF-B03 model and demonstrate that under appropriate conditions, E2F-1 can replace cytokine for continuous cell proliferation.
Materials and methods
Cell culture and proliferation assays
Unless indicated otherwise, BaF-B03–derived cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 10% WEHI supernatant (a source of IL-3 from WEHI 3B[d−] cell cultures), 50 μM glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin, and 2 μM 2-mercaptoethanol. For the generation of E2F-1–overexpressing cells, BaF-B03 cells were transfected by electroporation with an expression vector (pRcCMV) containing a wild-type hemagglutinin-tagged E2F-1 (HA-E2F1) complementary DNA (cDNA) inserted at the HindIII and EcoRI restriction sites (a kind gift of Dr W. Krek6). Following electroporation and selection in the presence of growth factor and geneticin (500 μg/mL), cells were cultured in media without cytokine (RPMI supplemented with 10% FBS). Cell proliferation assays were performed in triplicate in flat-bottomed 96-well plates. Cells were seeded at an initial concentration of 1 × 105 cells per milliliter in RPMI containing a final concentration either of 10% FBS or of 10% FBS and 10% WEHI supernatant. Cells were harvested and stained with 0.4% trypan blue and then enumerated by light microscopy.
5-bromo-2′-deoxyuridine (BrdU)–incorporation assays
Cells were initially cultured in IL-3 media. Prior to assay, cells were washed 3 times in Hanks balanced salt solution (HBSS) and then seeded into 24-well plates at an initial concentration of 1 × 105 cells per milliliter in RPMI supplemented with either 10% FBS or 10% FBS plus 10% WEHI supernatant. After an 11-hour incubation, cells were pulsed with 25 mM BrdU for 1 hour. BrdU incorporation was determined by an indirect immunofluorescence microscopy kit according to the manufacturer's protocol (Boehringer Mannheim, Sydney, Australia).
Immunoblot analysis
We analyzed p53 following exposure of cells to various DNA-damaging agents (25 J/m2 UV irradiation; 1-hour pulse with 100 μM camptothecin [Sigma, St Louis, MO]) in the presence of IL-3 medium or, alternatively, following withdrawal of cytokine. Whole-cell lysates were prepared by boiling in denaturing sample buffer (2% sodium dodecyl sulfate [SDS], 62.5 mM Tris-HCl pH 6.8, 10% glycerol, 5% 2-mercaptoethanol). Equivalent amounts of protein were resolved by SDS–polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes with the use of semidry transfer in buffer consisting of 0.7% acetic acid with 10% methanol. Membranes were incubated with 5% bovine serum albumin (BSA) diluted with TBST (Tris-buffered saline with 0.05% Tween-20) to block nonspecific binding sites and then probed with antimouse p53 antibody (0.5 μg/mL) (Zymed, San Francisco, CA) for 2 hours. Membranes were then washed with TBST followed by a 30-minute incubation with antimouse immunoglobulin (Ig)–G horseradish peroxidase–conjugated antibody (Amersham, Sydney, Australia) and then washed further with TBST and TBS. We detected p53 by enhanced chemiluminescence detection reagents according to the manufacturer's recommendations.
DNA laddering apoptosis assay
Cells were seeded into 24-well plates at an initial concentration of 1 × 106 cells per milliliter in RPMI containing either 10% FBS or 10% FBS supplemented with 10% WEHI supernatant. Following incubation periods of 24 and 48 hours at 37°C, cells were harvested and fixed in 70% ethanol and then stored at −20°C for 24 hours. We extracted low molecular weight DNA from each sample by incubating cell pellets in a phosphate-citrate buffer (0.192 M sodium hydrogen phosphate and 4 mM citric acid pH 7.8) for 30 minutes at room temperature. Following centrifugation, NP40 (0.25%) (Calbiochem, San Diego, CA) and RNase A (65 μg/mL) were added to the recovered supernatants prior to a 30-minute incubation at 37°C. DNA samples were subsequently treated with proteinase K (65 μg/mL) and incubated at 37°C overnight. Fragmented DNA was resolved by electrophoresis on a 1.5% TreviGel 500 gel (Trevigen, Gaithersburg, MD), stained with 0.5 μg/mL ethidium bromide, and visualized with a UV transilluminator.
Cell cycle analysis and quantitation of apoptosis
Control and E2F-1–overexpressing cells were cultured in complete medium (RPMI supplemented with 10% FBS and 10% WEHI supernatant) and then washed 3 times in HBSS prior to being seeded into 24-well plates at a concentration of 1 × 106 cells per milliliter in 10% FBS/phenol-free RPMI. Cells were UV-irradiated immediately (10 J/cm2) and then supplemented with 10% WEHI supernatant (final concentration) and cultured for a further 24 hours at 37°C. At this time, cells were fixed in 70% ethanol and stored at −20°C for 24 hours. As a mock treatment, an identical plate of control and E2F-1–overexpressing cells was prepared but was not UV-irradiated. Fixed cells were centrifuged, and the ethanol was removed, stained with propidium iodide containing RNase A (50 U/mL), and allowed to stand at room temperature for 18 to 30 hours to enable extraction of low molecular weight DNA. Cell cycle analysis was performed by flow cytometry by means of CellFIT software (Becton Dickinson, CA). All experiments for analysis of apoptosis were performed in triplicate, with 20 000 events acquired for each experiment. Apoptosis was quantitated as the proportion of cells with subdiploid DNA content.
Bivariate analysis of cyclin/PCNA/CDK expression and DNA content
Cells (1 × 106) were washed with phosphate-buffered saline (PBS) and then fixed in 90% methanol at −20°C for 24 hours. Following centrifugation, cells were washed and then resuspended in a permeabilization solution (PBS, 0.25% Triton X-100, 1% BSA) at 4°C for 5 minutes. After further washes, staining for specific intracellular proteins was performed with the use of rabbit polyclonal anti–cyclin A (C-19), rabbit polyclonal anti–cyclin B1 (M-20), mouse monoclonal anti–cyclin D1 (72-13G), rabbit polyclonal anti–cyclin E (M-20), mouse monoclonal anti-PCNA (PC10), rabbit polyclonal anti-CDK4 (C-22), or mouse monoclonal anti-p34cdc217 antibodies (all antibodies purchased from Santa Cruz Biotechnology, Santa Cruz, CA) diluted in 1% BSA in PBS for 1 hour at room temperature. Purified rabbit IgG antibody or mouse myeloma IgG2b antibody was used as control (both antibodies purchased from Zymed). Following a wash with 1% BSA in PBS, cells were incubated with either goat antirabbit IgG fluorescein-conjugated antibody (Sigma) or sheep antimouse IgG fluorescein-conjugated F(ab′)2 fraction (Silenus, Melbourne, Australia) for 30 minutes at room temperature. After further washing, DNA was stained with 1 μg/mL propidium iodide in PBS containing 200 μg/mL RNase A for 30 minutes at room temperature. Flow cytometric analysis was performed as described above.
Results
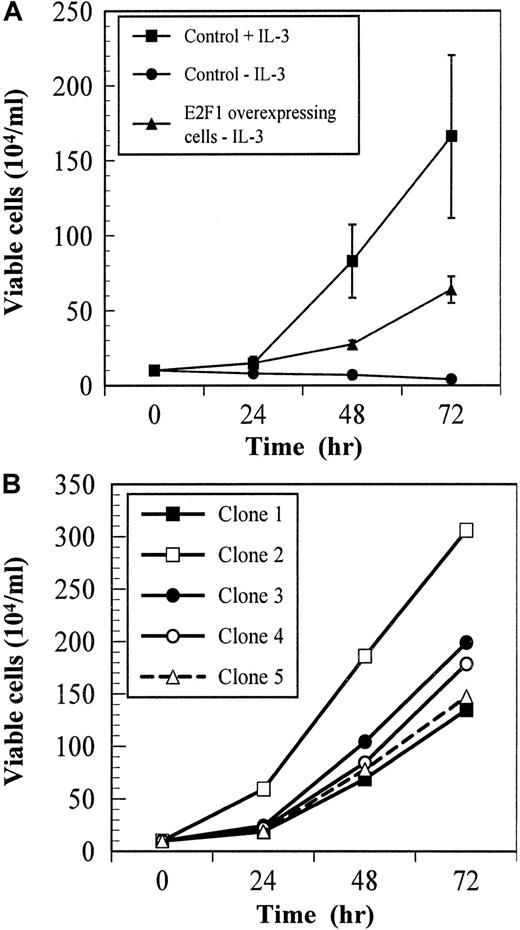
The IL-3–dependent cell line BaF/3 and the subclone BaF-B03 have been widely used for studies of cytokine-mediated proliferation. To examine the role of E2F-1 in controlling cytokine-dependent proliferation in these cells, we transfected BaF-B03 cells with an expression construct containing a human E2F-1 cDNA under the control of a constitutively active CMV promoter.8 The proliferative response of control and E2F-1–overexpressing BaF-B03 cells to IL-3 was assessed in short-term cultures. As expected, control cells proliferated in response to IL-3, but died rapidly when IL-3 was withdrawn (Figure1A). In contrast, E2F-1–overexpressing cells continued to proliferate and expand indefinitely following withdrawal of cytokine growth factor stimulation. We examined 5 clones of E2F-1–overexpressing cells for cytokine growth factor–independent proliferation; all were found to proliferate in the absence of cytokine growth factor (Figure 1B). The proliferative response of individual clones exceeded that of uncloned cells by as much as 5 times that observed in Figure 1A. These data indicate that overexpression of E2F-1 leads to cytokine independence in BaF-B03 cells and suggests that activation of the cyclin:CDK-pRB:E2F-1 pathway is sufficient to support proliferation in this cell line.
Cell proliferation assays.
Overexpression of E2F-1 confers cytokine growth factor independence to the IL-3–dependent cell line BaF-B03. Proliferation was assessed at the indicated times by staining with trypan blue. (A) Cells were cultured with or without IL-3. (B) Clones of E2F-1–overexpressing cells were cultured without IL-3.
Cell proliferation assays.
Overexpression of E2F-1 confers cytokine growth factor independence to the IL-3–dependent cell line BaF-B03. Proliferation was assessed at the indicated times by staining with trypan blue. (A) Cells were cultured with or without IL-3. (B) Clones of E2F-1–overexpressing cells were cultured without IL-3.
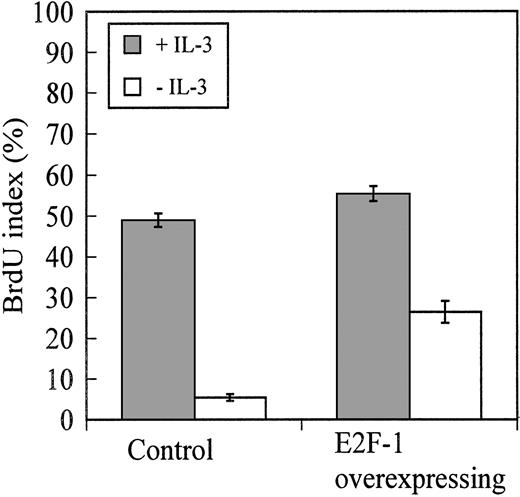
Consistent with the proliferation assays, BrdU-incorporation assays showed that after 12 hours of culture in cytokine-deficient medium, BaF-B03 cells overexpressing E2F-1 were being driven into S phase and were actively synthesizing DNA (BrdU index, 28%) whereas control cells incorporated only a minimal amount of BrdU (BrdU index, 5%) (Figure2). At the time BrdU incorporation was measured, control and E2F-1–overexpressing cells had a BrdU index of 50% and 55%, respectively, when stimulated with IL-3. Cell cycle analysis of control and E2F-1–overexpressing cells cultured without IL-3 for 24 hours reveals that 62.6% of E2F-1–overexpressing cells were present in S phase despite the absence of cytokine growth factor, in contrast to only 7.5% of control cells (Table1). When stimulated with IL-3, 58.5% and 72.1% of control and E2F-1–overexpressing cells, respectively, were present in S phase. Collectively, these results indicate that control cells were arrested prior to S-phase entry because mitogenic stimulation normally provided by IL-3 was not available to drive the cells into S phase. However, overexpression of E2F-1 was sufficient to induce S-phase entry and DNA synthesis. This is consistent with the role identified in other cell-types in which availability of “free” E2F-1 drives the G1/S-phase transition.
BrdU-incorporation assay.
Overexpression of E2F-1 promotes cell cycle progression in BaF-B03 cells following cytokine growth factor withdrawal. Both cell populations were cultured with or without IL-3 for 11 hours and then pulsed with 25 mM BrdU for 1 hour. BrdU incorporation was determined by indirect immunofluorescence microscopy.
BrdU-incorporation assay.
Overexpression of E2F-1 promotes cell cycle progression in BaF-B03 cells following cytokine growth factor withdrawal. Both cell populations were cultured with or without IL-3 for 11 hours and then pulsed with 25 mM BrdU for 1 hour. BrdU incorporation was determined by indirect immunofluorescence microscopy.
Effects of E2F-1 overexpression on cell cycle S phase in the presence and absence of cytokine growth factor
| . | Cells in S phase at 24 hours (%) . | |
|---|---|---|
| IL-3 . | No cytokine . | |
| Control cells | 58.5 | 7.5 |
| E2F-1–overexpressing cells | 72.1 | 62.6 |
| . | Cells in S phase at 24 hours (%) . | |
|---|---|---|
| IL-3 . | No cytokine . | |
| Control cells | 58.5 | 7.5 |
| E2F-1–overexpressing cells | 72.1 | 62.6 |
Cells were cultured with or without interleukin (IL)–3 for 24 hours and then fixed in 70% ethanol prior to staining with propidium iodide. Cell cycle analysis was performed on the nonapoptotic (viable) cell population by flow cytometry. Shown are the mean results for the percentage of cells in S phase, as determined from 3 separate experiments.
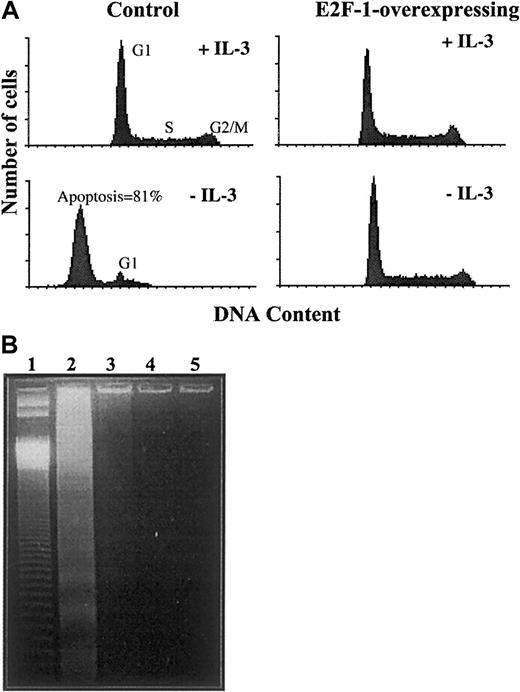
To date, only a small number of immortalized cell lines that overexpress E2F-1 has been shown to sustain continuous proliferation, whereas most proceed to apoptosis via a p53-dependent mechanism following entry into S phase.15,16 22 Having established that E2F-1 was promoting S-phase entry in BaF-B03 cells despite the absence of cytokine growth factor, we examined the effect on the rate of apoptosis. Both control cells and E2F-1–overexpressing cells were cultured with or without cytokine for 24 hours, and cell cycle profiles were then analyzed by flow cytometry. Cell cycle profiles reflected the results of the proliferation assays. Both cell populations, when cultured in the presence of IL-3, displayed normal DNA profiles (Figure3A); all phases of the cell cycle are evident and no abnormal peaks are seen. Cytokine withdrawal arrested control cells in G1 phase, and no cells were present in either S or G2/M phases. Of those cells blocked in G1 phase, 81% proceeded to apoptosis, as determined from the proportion of cells with a subdiploid content of DNA. In contrast, removal of IL-3 did not result in apoptosis in E2F-1–overexpressing cells, which continued to progress through the cell cycle normally and were not arrested at any point.
Effect of overexpression of E2F-1 on apoptosis following cytokine growth factor withdrawal.
Apoptosis following cytokine growth factor withdrawal is prevented by overexpression of E2F-1. Cells were cultured with or without IL-3 for 24 hours. (A) Cell cycle analysis was assessed by flow cytometry after staining with propidium iodide. Histograms show DNA content (x-axis) and cell number (y-axis). (B) DNA laddering apoptosis assay. Low molecular weight DNA was isolated and resolved on a 1.5% TreviGel 500 gel. Lane 1 shows 50 base-pair ladder; lane 2, control cells without IL-3; lane 3, control cells with IL-3; lane 4, E2F-1–overexpressing cells without IL-3; and lane 5, E2F-1–overexpressing cells with IL-3.
Effect of overexpression of E2F-1 on apoptosis following cytokine growth factor withdrawal.
Apoptosis following cytokine growth factor withdrawal is prevented by overexpression of E2F-1. Cells were cultured with or without IL-3 for 24 hours. (A) Cell cycle analysis was assessed by flow cytometry after staining with propidium iodide. Histograms show DNA content (x-axis) and cell number (y-axis). (B) DNA laddering apoptosis assay. Low molecular weight DNA was isolated and resolved on a 1.5% TreviGel 500 gel. Lane 1 shows 50 base-pair ladder; lane 2, control cells without IL-3; lane 3, control cells with IL-3; lane 4, E2F-1–overexpressing cells without IL-3; and lane 5, E2F-1–overexpressing cells with IL-3.
Next, low molecular weight DNA was isolated from both cell populations for assessment of DNA laddering produced by nuclease-fragmented DNA. In accordance with the results of cytofluorometric analysis, DNA laddering was observed in DNA extracted from control cells deprived of IL-3 for 24 hours, but was not detected in IL-3–deprived E2F-1–overexpressing cells or in either cell population cultured with IL-3 (Figure 3B). The data demonstrate that overexpression of E2F-1 does not promote apoptosis in BaF-B03 cells, but rather is able to substitute for the role performed by IL-3 with regard to both cell cycle progression and cell survival.
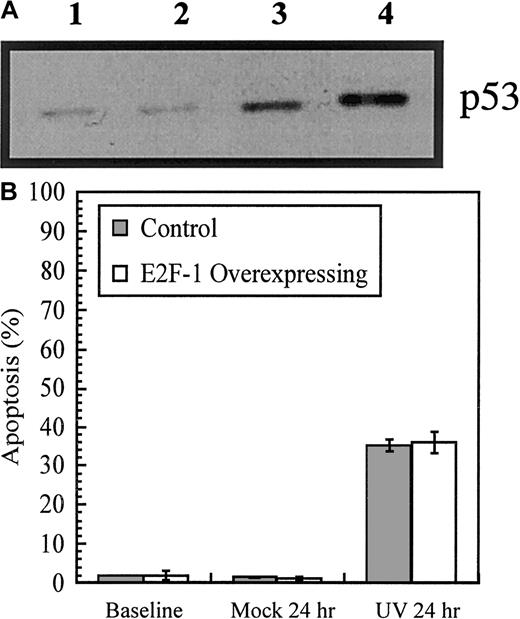
One mechanism by which overexpression of E2F-1 drives cells into apoptosis is p53-dependent pathways. To examine the role played by p53 in the response of BaF-B03 cells, we sequenced p53 cDNA and assessed the response to agents that cause DNA damage, namely, UV irradiation and camptothecin. UV irradiation induces a p53-mediated apoptosis following strand breakage via effects exerted upon the mechanisms of DNA repair and replication,23 and camptothecin causes breaks in double-stranded DNA during DNA synthesis by inhibiting the activity of topoisomerase-I.24
Immunoblot analysis revealed that low levels of p53 are present in the E2F-1–overexpressing cells prior to the induction of DNA damage and that, following exposure to DNA-damaging agents, the levels increased (Figure 4A). Previous studies have shown that BaF/3 cells stimulated by IL-3 are arrested at the G1checkpoint following exposure to ionizing radiation and that, in the absence of IL-3, p53-mediated apoptosis results.25Analysis of cell cycle profiles of UV-irradiated control and E2F-1–overexpressing cells revealed that, 24 hours postirradiation, the level of apoptosis in both cell populations was approximately 35% (Figure 4B). Overexpression of E2F-1 failed to prevent the onset of apoptosis in response to DNA damage. Further, cytokine growth factor stimulation provided by IL-3 did not rescue cells from apoptosis.
Effect of DNA damage on p53 and apoptosis in E2F-1–overexpressing cells.
DNA damage upregulates p53 and induces apoptosis in E2F-1–overexpressing cells. (A) p53 immunoblot. Cells were exposed to UV irradiation (25 J/m2) or camptothecin (1-hour pulse, 100 mM) in the presence of IL-3 and then washed extensively and cultured for a further 11 hours in IL-3 medium. Lane 1 shows cells at baseline; lane 2, cells after 12 hours' culture in IL-3 with 0.05% dimethyl sulfoxide, 3 to 12 hours after UV irradiation and 4 to 11 hours after being pulsed with camptothecin. (B) Cells were cultured in IL-3 medium, washed, and resuspended in 10% FBS/phenol-free RPMI for irradiation (10 J/cm2). Cells were then cultured for a further 24 hours in IL-3 medium. As a mock treatment, cells were not irradiated. Cell cycle analysis was performed by flow cytometry.
Effect of DNA damage on p53 and apoptosis in E2F-1–overexpressing cells.
DNA damage upregulates p53 and induces apoptosis in E2F-1–overexpressing cells. (A) p53 immunoblot. Cells were exposed to UV irradiation (25 J/m2) or camptothecin (1-hour pulse, 100 mM) in the presence of IL-3 and then washed extensively and cultured for a further 11 hours in IL-3 medium. Lane 1 shows cells at baseline; lane 2, cells after 12 hours' culture in IL-3 with 0.05% dimethyl sulfoxide, 3 to 12 hours after UV irradiation and 4 to 11 hours after being pulsed with camptothecin. (B) Cells were cultured in IL-3 medium, washed, and resuspended in 10% FBS/phenol-free RPMI for irradiation (10 J/cm2). Cells were then cultured for a further 24 hours in IL-3 medium. As a mock treatment, cells were not irradiated. Cell cycle analysis was performed by flow cytometry.
Cytokine receptor–mediated control of cell cycle progression is tightly regulated by the availability of E2F-1 at specific points in the cell cycle. E2F-1 coordinates the induction of genes required for G1/S-phase transition, including cyclins A and E, and cyclins closely regulate the activity of pRB.5 To examine the effects of overexpression of E2F-1 and its availability throughout the cell cycle on cyclin expression profiles, we performed bivariate analysis of cyclin and DNA content. With this technique, it is possible to measure intracellular levels of cyclin proteins at the single-cell level and correlate their expression to cell cycle position.26 This multiparameter method of analysis facilitates measurement of cell cycle–dependent intracellular cyclin levels in asynchronous cell culture populations and thus has the advantage of avoiding cell cycle disturbances that commonly accompany attempts at synchronizing cells by chemical inhibitors or growth factor withdrawal. Additionally, variations in cyclin expression can be detected within populations of cells occupying the same phase of the cell cycle.
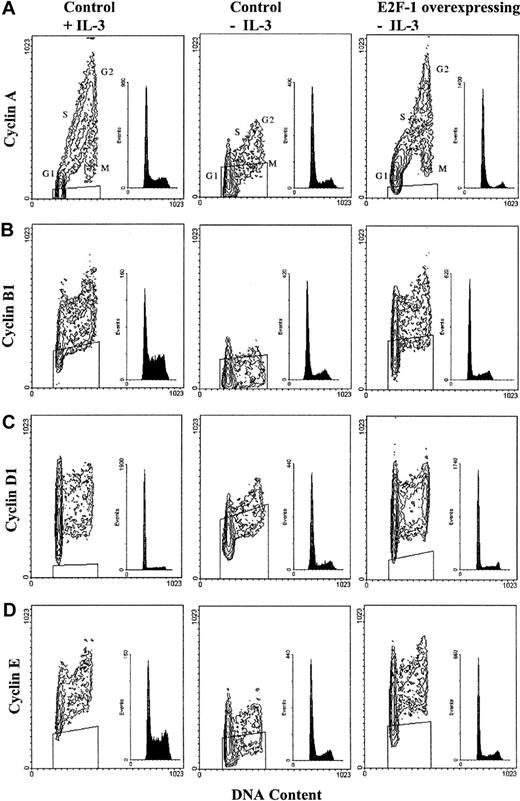
The pattern of cyclin A expression in control cells stimulated with IL-3 is presented in Figure 5A; low expression in G1 phase is followed by increased expression throughout S phase and peak levels in G2 phase. Cyclin A expression then decreases in M phase to levels slightly higher than those observed in G1 phase. In the absence of IL-3, cyclin A continued to be expressed in G1 phase, but expression was substantially diminished in all other cell cycle phases. In contrast, the pattern of cyclin A expression in E2F-1–overexpressing cells following cytokine withdrawal resembled the pattern established by IL-3 in control cells. These observations were consistent for the expression of cyclins B1, D1, and E and demonstrate that E2F-1 overexpression mimics the effects of IL-3 without leading to deregulated cyclin profiles.
E2F-1 and cell cycle–dependent expression of cyclins A, B1, D1, and E after removal of cytokine growth factor.
E2F-1 restores cell cycle–dependent expression of cyclins A, B1, D1, and E following removal of cytokine growth factor. Cells were cultured in the presence or absence of IL-3 for 24 hours and then fixed in 90% methanol prior to staining with the appropriate cyclin antibodies and propidium iodide. Bivariate analysis was performed by flow cytometry; results are represented as contour plots, with DNA content corresponding to the x-axis and intracellular cyclin content corresponding to the y-axis. The corresponding DNA histogram for each study is shown as an inset, and the boxed region indicates the outer limits of the signal obtained with the use of a control primary antibody.
E2F-1 and cell cycle–dependent expression of cyclins A, B1, D1, and E after removal of cytokine growth factor.
E2F-1 restores cell cycle–dependent expression of cyclins A, B1, D1, and E following removal of cytokine growth factor. Cells were cultured in the presence or absence of IL-3 for 24 hours and then fixed in 90% methanol prior to staining with the appropriate cyclin antibodies and propidium iodide. Bivariate analysis was performed by flow cytometry; results are represented as contour plots, with DNA content corresponding to the x-axis and intracellular cyclin content corresponding to the y-axis. The corresponding DNA histogram for each study is shown as an inset, and the boxed region indicates the outer limits of the signal obtained with the use of a control primary antibody.
Also of interest was the pattern of cyclin B1 expression. In lymphocytes, cyclin B1 is commonly expressed at very low levels throughout G1 phase and at slightly higher levels in S phase, with peak levels of this cyclin observed at G2/M phases.26 In BaF-B03 cells, cyclin B1 expression increased throughout G1 phase and was maintained in S phase, before being maximally expressed in G2/M phases of both control cells stimulated with IL-3 and E2F-1–overexpressing cells in the absence of IL-3 (Figure 5B). Cyclin D1 is a regulator of G1-phase events, and its expression and interaction with CDKs are dependent upon extracellular growth factor stimulation.5 The expression of this cyclin typically increases consistently throughout G1 phase and peaks late in G1 phase, and this was observed in IL-3–stimulated control cells as well as IL-3–deprived E2F-1–overexpressing cells (Figure 5C). During S phase, cyclin D1 expression was lower and then elevated in G2/M phases. Interestingly, cytokine withdrawal from control BaF-B03 cells appeared to attenuate cyclin D1 expression in G1 and S phases, yet expression remained high in G2/M phases. Cyclin E is a regulator of G1/S-phase transition and in other cell types is normally expressed during late G1 and early S phase. This pattern of expression was observed in IL-3–stimulated control cells and IL-3–deprived E2F-1–overexpressing cells (Figure 5D). In contrast to other cell types, where expression of cyclin E decreases in M phase, cyclin E expression increased slightly during S phase and peaked at G2/M phase, where it was maintained; this variant cyclin E expression profile has been reported in certain tumor cell lines.26 These data demonstrate that E2F-1 restores cell cycle–dependent expression of cyclin proteins in the absence of cytokine growth factor stimulation and provides a mechanism by which cell proliferation is sustained.
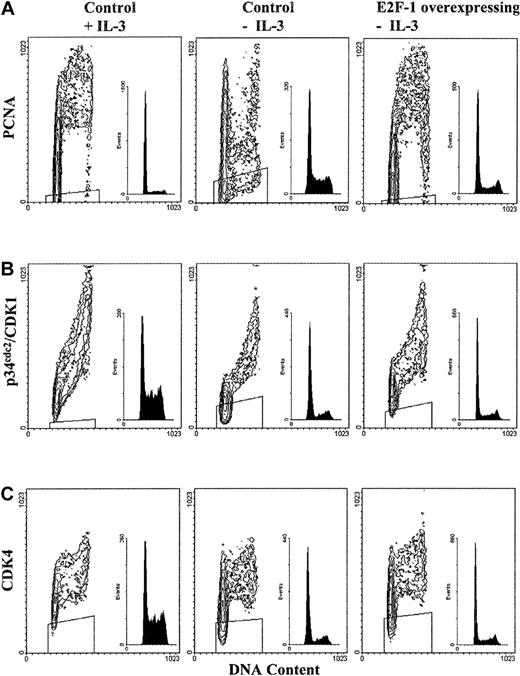
Additional cell cycle regulatory molecules such as PCNA or CDKs, which tightly regulate cyclin activity, were examined to assess whether they were deregulated in the presence of overexpressed E2F-1. PCNA plays an essential role in DNA replication and regulation of cyclins and CDK inhibitors (CDKIs), in particular, the D-type cyclins and the CDKIs p21 and p57.27 In IL-3–stimulated control cells and IL-3–deprived E2F-1–overexpressing cells, PCNA expression increased throughout G1 phase, with maximal levels expressed during S phase (Figure 6A). In IL-3–deprived control cells, PCNA expression dropped to basal levels in S phase prior to an increase during G2/M phase, where levels were maximal. This demonstrates the importance of cytokine growth factor stimulation in maintaining PCNA expression during DNA synthesis and shows that E2F-1 overexpression restores normal PCNA expression in the absence of cytokine growth factor stimulation.
Effect of cytokine withdrawal on PCNA and CDK expression.
PCNA expression is restored by E2F-1 following cytokine withdrawal; however, cytokine withdrawal does not alter CDK expression. Bivariate analysis of DNA content and PCNA or CDK expression is shown; experimental details are similar to those for Figure 5.
Effect of cytokine withdrawal on PCNA and CDK expression.
PCNA expression is restored by E2F-1 following cytokine withdrawal; however, cytokine withdrawal does not alter CDK expression. Bivariate analysis of DNA content and PCNA or CDK expression is shown; experimental details are similar to those for Figure 5.
Next, the expression of the CDKs p34cdc2 (CDK1) and CDK4 were analyzed; p34cdc2 complexes with both cyclins A and B1 during G2 phase and is required for entrance into M phase. CDK4 complexes D-type cyclins during G1 phase, where it is required for the phosphorylation of pRB. In addition, cyclin D:CDK4 complex formation commits cells to S-phase entry.28 29Neither cytokine withdrawal nor overexpression of E2F-1 resulted in the deregulated expression of p34cdc2 or CDK4 in BaF-B03 cells (Figures 6A-C), suggesting that expression of CDKs is independent of E2F-1 effects.
Discussion
Progression of cells through G1 phase relies upon stimulation by growth factors, and passage beyond the restriction point in late G1 phase commits cells to a cell cycle progression program. The D-type cyclins, together with their dimerization partners CDK4 and CDK6, are important mediators of growth factor receptors and ultimately lead to phosphorylation of pRB and uninhibited activity of E2F-1. The results of the present study demonstrate that E2F-1 and its downstream effects are sufficient for S-phase entry and subsequent completion of the cell cycle in the cytokine growth factor–dependent cell line BaF-B03. This cell line has been widely used for studying proliferative responses to a range of cytokines, including IL-2, IL-3, IL-4, erythropoietin, and granulocyte-macrophage colony-stimulating factor.30-36 Previous studies with BaF-B03 cells have shown that there are at least 3 pathways that may cooperate to produce a proliferative signal.37 The results of the present study emphasize the pivotal role of the cyclin:CDK-pRB:E2F-1 pathway in mediating the cellular proliferative response in BaF-B03 cells and suggest that multiple pathways converge at E2F-1. Indeed, recent evidence from studies in other lymphoid cells have shown a convergence of p70 S6 kinase–mediated signals and PKB/Akt-dependent activation of E2F-1.10 38
The ability of constitutive E2F-1 expression not only to initiate DNA synthesis and S-phase entry but also to sustain cell proliferation with orderly progression of cells through all cell cycle phases in BaF-B03 cells is consistent with a role for E2F-1 in cell cycle–related functions additional to S-phase entry. E2F-target genes include not only a limited number required for DNA synthesis, but also genes encoding for other proteins that contribute to cell proliferation. One such role may be related to timely accumulation of cyclin B1, which precedes G2/M transition.39 In our study, cyclin B1 accumulation was clearly induced by E2F-1 overexpression. Taken together, the results demonstrate that E2F-1 can provide all of the necessary cell cycle–regulatory events to support continuous proliferation of cells in the absence of cytokine growth factor, with some cell-type–specific requirements. Although the factors that predispose to this permissive phenotype are not known, they may reflect a genetic background associated with a stage of cell maturation.
E2F-1 is recognized not only as a regulator of cell cycle progression but also, in more recent studies, for its role in controlling apoptosis. In a range of cell lines, E2F-1 overexpression leads to apoptosis following entry into S phase.15,22,40,41 In addition, mice with a homozygous deficiency of E2F-1 show abnormal T-lymphocyte development with an intrathymic excess of mature T lymphocytes owing to a stage-specific defect in thymocyte apoptosis.42 Additionally, as E2F-1−/− mice age, a second phenotype characterized by aberrant cell proliferation and tumorigenesis becomes evident, with hyperproliferation of cortical thymocytes and formation of reproductive tract sarcomas, lung adenocarcinomas, lymphomas, and various other solid tissue tumors.42 43 In the present study, E2F-1 overexpression inhibited apoptosis following cytokine growth factor withdrawal. Withdrawal of IL-3 in cultures of BaF-B03 control cells resulted in a demonstrable induction of apoptosis within 24 hours, whereas E2F-1–overexpressing cells showed no evidence of apoptosis even in long-term cultures in the absence of cytokine growth factor. The mechanism underlying this failure of E2F-1 dysregulation to promote apoptosis in BaF-B03 cells is currently unknown, but further investigation of the intracellular signaling pathways that mediate E2F-1–induced apoptosis may ultimately provide some further clues.
The tumor-suppressor protein p53 plays an important but not exclusive role in E2F-1–induced apoptosis.15,22,44 Depending upon the cell line studied, other additional p53-independent pathways may also mediate apoptosis resulting from E2F-1 dysregulation15,16 and may be inhibited by Bcl-2.45,46 Sequence and immunoblot analysis of p53 in E2F-1–overexpressing BaF-B03 cells confirmed the presence of wild-type p53 and demonstrated an increase of p53 activity in response to DNA-damaging agents. These data demonstrate that p53 dysfunction was not responsible for the phenotype induced by E2F-1. This is supported by the findings that there was no aberrant DNA synthesis or cell cycle progression, which are also associated with recruitment of p53-dependent pathways.18 Our results also demonstrate that overexpression of E2F-1 is not sufficient to overcome DNA damage in this cell line and hence they do not conflict with evidence that E2F-1 has a direct role in apoptosis following DNA damage.47 Given the evidence for a normally functioning p53 pathway in response to DNA-damaging agents, a mutation of thep19ARF gene in this cell line may be predicted as a possible explanation for the absence of apoptosis in response to E2F-1 overexpression. Expression of certain oncogenes, including E2F-1, c-myc, E1A, and oncogenic Ras mutants, results in accumulation of p19ARF, which then acts to promote p53 stabilization and function.48 However, p19ARFmediates induction of p53 via a pathway that is separable from the pathway following DNA damage, and further characterization of the p19ARF-p53 pathway in this model should contribute to understanding of the pathways involved in E2F-1–mediated apoptosis.
E2F-1 transcriptional activity mediating cytokine-independent cell proliferation was sufficient for complete restoration of the cell cycle–dependent expression of cyclins A, B1, D1, and E, as well as of PCNA. Degradation of E2F-1 protein via the ubiquitin-proteasome pathway,49 may play a role in controlling the levels of E2F-1 during various phases of cell cycle in the BaF-B03 system, but whether this is essential for appropriate cyclin expression and cell cycle progression is unknown. Transcription of E2F target genes beyond the G1/S transition is necessary for coordinated S-phase progression and cell division, and continuing E2F-1–mediated regulatory activity may be essential for the timely expression of important cell cycle regulatory proteins.39 Our results are consistent with a model in which deregulated E2F-1 expression does not lead to abnormal cyclin expression, but rather promotes the orderly expression of cyclins during all phases of the cell cycle. This provides the fundamental mechanism by which E2F-1 allows continued growth and cytokine-independent proliferation in this model.
Supported in part by grants from the National Health and Medical Research Council of Australia, and New South Wales Cancer Council.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Peter Williamson, Dept of Medicine, University of Sydney, Westmead Hospital, NSW 2145, Australia; e-mail: peterw@westgate.wh.usyd.edu.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal