Abstract
Protein synthesis in reticulocytes depends on the availability of heme. In heme deficiency, inhibition of protein synthesis correlates with the activation of heme-regulated eIF-2α kinase (HRI), which blocks the initiation of protein synthesis by phosphorylating eIF-2α. HRI is a hemoprotein with 2 distinct heme-binding domains. Heme negatively regulates HRI activity by binding directly to HRI. To further study the physiological function of HRI, the wild-type (Wt) HRI and dominant-negative inactive mutants of HRI were expressed by retrovirus-mediated transfer in both non-erythroid NIH 3T3 and mouse erythroleukemic (MEL) cells. Expression of Wt HRI in 3T3 cells resulted in the inhibition of protein synthesis, a loss of proliferation, and eventually cell death. Expression of the inactive HRI mutants had no apparent effect on the growth characteristics or morphology of NIH 3T3 cells. In contrast, expression of 3 dominant-negative inactive mutants of HRI in MEL cells resulted in increased hemoglobin production and increased proliferative capacity of these cells upon dimethyl-sulfoxide induction of erythroid differentiation. These results directly demonstrate the importance of HRI in the regulation of protein synthesis in immature erythroid cells and suggest a role of HRI in the regulation of the numbers of matured erythroid cells.
Introduction
Phosphorylation of the α-subunit of translational initiation factor 2 (eIF-2α) is important in the regulation of protein synthesis in mammalian cells under various conditions of stress such as heme-deficiency,1 viral infection (reviewed in Proud2), nutrient starvation,3-5 heat shock,3,5,6 and stress in endoplasmic reticulum (ER) (reviewed in Brostrom and Brostrom7). Phosphorylation of the α-subunit of eIF-2 at the serine 51 residue by activated eIF-2α kinases results in the formation of an eIF-2(αP)/eIF-2B complex that renders eIF-2B nonfunctional. eIF-2B is required for the exchange of guanosine triphosphate for guanosine diphosphate bound to eIF-2 in the recycling of eIF-2 for another round of initiation. Since eIF-2B is limiting, the phosphorylation of only a fraction of the total eIF-2α present in the cell is sufficient to shut off protein synthesis (reviewed in Jackson8 and Clemens9). Overexpression of either nonphosphorylatable eIF-2αAla51,10 inactive mutant forms of double-stranded RNA (dsRNA)–dependent eIF-2α kinase (PKR),11-13 or the p58 inhibitor of PKR14 has been shown to result in malignant transformation of NIH 3T3 cells. On the other hand, overexpression of PKR resulted in apoptosis.15-18 These studies underscore the importance of eIF-2α phosphorylation by eIF-2α kinases with respect to regulation of protein synthesis and cell growth.
In addition to the 2 extensively studied mammalian heme-regulated eIF-2α kinases (HRIs) and the PKR, there are also the GCN2 protein kinase and the recently identified mammalian ER resident eIF-2α kinase (PERK),19 which is identical to the pancreatic eIF-2α kinase (PEK) highly expressed in the pancreas.20 These eIF-2α kinases share extensive homology in the kinase catalytic domains19-25 and phosphorylate eIF-2α at the serine 51 residue.19,20,26,27 However, the regulatory domains and the mechanism of each of these eIF-2α kinases are very different. HRI is regulated by heme, the prosthetic group of hemoglobin.1,28,29 PKR is regulated by dsRNA through the 2 N-terminal dsRNA-binding domains (reviewed in Proud2). GCN2 is activated under condition of amino acid starvation through the C-terminal domain, which contains a His-transfer-RNA-synthase–like sequence (reviewed in Hinnebusch30). PERK is activated by ER stress and contains a lumenal domain that is similar to the sensor domain of the ER-stress kinase, Irel.19
It is well documented that protein synthesis in intact reticulocytes and their lysates is dependent upon the availability of heme. In heme deficiency, inhibition of protein synthesis correlates with the activation of HRI (reviewed in Chen and London,1Jackson,8 Clemens,9 and Hershey31). Hemin has been shown to inhibit both the autokinase and eIF-2α kinase of HRI by inhibiting adenosine triphosphate binding to HRI.32,33 We have shown previously that expression of Wt HRI in insect Sf9 cells causes global inhibition of protein synthesis and that baculovirus-expressed Wt HRI is a hemoprotein and is regulated by micromolar concentrations of hemin both in vitro and in vivo.28,29 Furthermore, there are 2 distinct heme-binding sites in HRI. The heme binding to the N-terminal domain of HRI is stable and copurified with HRI to homogeneity. The heme binding to the kinase-insertion domain is reversible and is responsible for sensing the heme and regulates HRI activity.34
We have reported earlier that the protein and messenger RNA (mRNA) of HRI are expressed predominantly in erythroid cells of adult rabbits. In addition, the level of HRI mRNA is increased during erythroid differentiation of mouse erythroleukemic (MEL) cells, and this increase in HRI mRNA is dependent on the presence of heme.35 Since then, 2 reports have described the presence of small amounts of HRI mRNA in non-erythroid rat and mouse tissues and the presence of HRI-like activity in mouse liver and NIH 3T3 cells.25 36These reports suggested a possible role of HRI in translational regulation in non-erythroid cells in addition to its role in erythroid cells. It is to be noted that no HRI protein was reported in either of these 2 studies.
To directly examine the physiological function of HRI, we have expressed Wt and inactive mutants of HRI in both non-erythroid 3T3 cells and erythroid MEL cells. We report here that expression of Wt HRI in NIH 3T3 cells results in severe inhibition of cell growth and ultimately cell death, while cells that express inactive HRI mutants appear to grow normally. The expression of inactive HRI mutants in 3T3 cells resulted in no detectable change in morphology and did not cause oncogenic transformation. In contrast, the expression of these inactive HRI mutants in MEL cells significantly increased the hemoglobin content and proliferative capacity of the differentiating MEL cells. These results demonstrate a functional role of HRI in erythroid MEL cells, but not in non-erythroid 3T3 cells.
Materials and methods
Plasmid constructions
All HRI constructs contained the 5′ untranslated leader of tobacco mosaic virus and the entire 3′ untranslated region as described previously.21 K199R (single-letter amino acid codes) HRI complementary DNA (cDNA) was prepared and reported previously.28 The internal deletion of amino acids 375 through 384 of HRI (Δ10 HRI) and of amino acids 375 through 394 of HRI (Δ20 HRI) was prepared by recombinant polymerase chain reaction (PCR) as described.37 Briefly, the primers used for deletions were 30 nucleotides long, flanking the desired deleted sequence with 15 nucleotides each on both the 5′ and 3′ ends. The fragments of Dra III to Hpa I sites (nucleotides 805 through 1875) with the desired deletions were amplified as 2 separate fragments by 2 separate PCR reactions. The first half was from the Dra III site to the start site of the deletion. The second half was from 3′ terminal to the site of deletion to Hpa I site. These PCR reactions and the following recombinant PCR for the production of the Dra III to Hpa I fragment were as described previously.28 The Dra III to Hpa I fragment with Δ10 or Δ20 deletion was digested with Dra III and Hpa I and then subcloned to TMV-HRI vector.21 The deletions were confirmed by DNA sequencing. In vitro transcription and translation of Δ10 and Δ20 HRI in the TMV-HRI plasmid were also performed to ensure that protein products with the expected sizes were made. The Δ10 and Δ20 HRI cDNAs were then excised from the TMV-HRI plasmid and subcloned into 1392 baculovirus-transferring vector as described previously.28 Production of recombinant HRI baculoviruses and the expression of HRI were as described.28
Wt, K199R, Δ10, and Δ20 HRI retroviral constructs were prepared by the use of pLXSN retroviral vector38 (provided by D. Miller, Fred Hutchinson Cancer Research Center, Seattle, WA) with neomycin- or puromycin-resistant gene. The HRI cDNA inserts of Wt, K199R, Δ10, and Δ20 were excised by digestion with Bgl II and Eco RI. These inserts were blunt-ended by Klenow fragment and ligated to the pLXSN vector, which was digested with Eco RI and blunt-ended.
Transfection, infection, and selection of cells
NIH 3T3 cells were obtained from American Type Culture Collection (Manassas, VA); BOSC23 and Bing cells39 were provided by W. Pear and D. Baltimore (Massachusetts Institute of Technology, Cambridge, MA); and Ψcrip and Ψcre cells40 were provided by R. Mulligan (Harvard Medical School, Boston, MA). NIH 3T3/eIF-4E and PKR Δ6-12 cells were provided by N. Sonenberg (McGill University, Montreal, Quebec, Canada). NIH 3T3 cells, Ψcrip, and Ψcre cells were grown at 37°C in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated calf serum, 4.5 mg/mL glucose, 2 mmol/L glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. For BOSC23, Bing, and MEL cells, 10% heat-inactivated fetal calf serum was used.
The packaging cell lines BOSC23, Bing, Ψcrip, and Ψcre were grown and transfected as described.39,40 Plasmid DNAs (20 μg) prepared by the Qiagen, Valencia, CA procedure were transfected into these cells by means of a calcium phosphate precipitation method (Ependorf-5 Prime Inc, Boulder, CO). The recombinant retroviruses produced were harvested from the culture supernatants by filtering through the 0.22 μm filter units as described.39 40 All infections were carried out in the presence of 8 μg/mL polybrene (Sigma; St Louis, MO). The recombinant retroviruses were used to infect NIH 3T3 or MEL cells. For cells infected with Wt HRI, 5 μmol/L hemin was added to the culture media to maintain cell viability. Retrovirally transduced cells were selected 2 days after infection with 500 μg/mL G418 (Gibco/BRL, Rockville, MD) or 1.5 μg/mL puromycin (Sigma). For experiments, these transduced cells were cultured in the absence of selection agents.
Lysate preparations and Western blot analysis
Preparations of cell lysates and Western blot analysis of lysate proteins were performed as previously described.35 Mouse antirabbit HRI monoclonal antibody was used to detect HRI protein by means of either ECL (Amersham, Piscataway, NJ) or NBT-BCIP color development system (Promega, Madison, WI).
Results
Inhibition of the Wt HRI activity by inactive mutant HRI
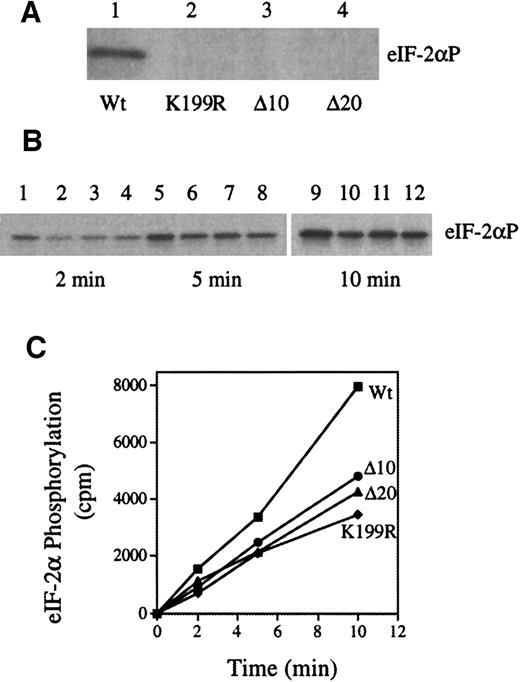
Since we planned to use the inactive HRI mutants to diminish endogenous HRI activity, we examined the abilities of inactive HRI mutants to inhibit the activity of Wt HRI by coexpression of Wt HRI with 3 HRI mutants in Sf9 cells using recombinant baculoviruses. The expression of large quantities of HRI in this system readily permits the biochemical analysis of HRI kinase activity.28 The K199R mutant HRI has a mutation of K199 in catalytic domain II to R. K199R HRI is inactive when expressed in insect cells28 and in yeast cells.27 The Δ10 and Δ20 HRIs are the internal deletions of amino acids 375 through 384 and amino acids 375 through 394, respectively, which are located in kinase domain VI and highly conserved among all eIF-2α kinase. These 2 deletion mutants are similar to PKR Δ6 mutation.
As shown in Figure 1A, in contrast to Wt HRI (lane 1), K199R, Δ10, and Δ20 HRI lacked eIF-2α kinase activity (lanes 2, 3, and 4). In addition, co-expressions of these inactive mutants with Wt HRI decreased the rate of eIF-2α phosphorylation by 40% to 50% (Figure 1B-C). We did the co-expressions 4 times with similar results. We have shown previously that co-expression of Wt HRI with interleukin 1-β does not affect HRI activity.28 Thus, these observations indicate that inhibition of theWt HRI activity by these 3 inactive mutant HRIs is specific and is not due to the general effect of co-expression. Similarly, co-expression of the mutant HRI with Wt HRI in NIH 3T3 cells relieved the inhibitory effect of Wt HRI (data not shown). Previous work by others and by us has demonstrated that HRI forms homodimers,29 41-43 It is most likely that inactive HRI mutants inhibit the activity of Wt HRI by forming heterodimers with Wt HRI. Consistent with this interpretation, it is important to note that during co-expression there is always a fraction of active Wt HRI dimer present, at least 25%. Therefore, one would not expect complete inhibition of the HRI activity by co-expression.
Inhibition of the protein kinase activity of Wt HRI by co-expression with the HRI mutants.
Wt and mutant HRI (K199R, Δ10, and Δ20) were expressed individually or co-expressed in Sf9 cells as described previously.28Protein kinase assays were performed with the use of cytoplasmic cell extracts and purified eIF-2 as described previously.28 (A) eIF-2α kinase activity of Wt, K199R, Δ10, or Δ20 HRI. (B) eIF-2α kinase activity of co-expressed HRI. Wt HRI alone (lanes 1, 5, and 9); co-expression of Wt HRI with K199R (lanes 2, 6, and 10), with Δ10 (lanes 3, 7, and 11), or with Δ20 (lanes, 4, 8, and 12). The time intervals of protein kinase assays are 2, 5, and 10 minutes as indicated. (C) The extent of eIF-2α phosphorylation was quantified by scintilation counting of the gel slices containing eIF-2α. ■, wt; ●, Δ10/wt; ▴, Δ20/wt; ⧫, K199R/wt.
Inhibition of the protein kinase activity of Wt HRI by co-expression with the HRI mutants.
Wt and mutant HRI (K199R, Δ10, and Δ20) were expressed individually or co-expressed in Sf9 cells as described previously.28Protein kinase assays were performed with the use of cytoplasmic cell extracts and purified eIF-2 as described previously.28 (A) eIF-2α kinase activity of Wt, K199R, Δ10, or Δ20 HRI. (B) eIF-2α kinase activity of co-expressed HRI. Wt HRI alone (lanes 1, 5, and 9); co-expression of Wt HRI with K199R (lanes 2, 6, and 10), with Δ10 (lanes 3, 7, and 11), or with Δ20 (lanes, 4, 8, and 12). The time intervals of protein kinase assays are 2, 5, and 10 minutes as indicated. (C) The extent of eIF-2α phosphorylation was quantified by scintilation counting of the gel slices containing eIF-2α. ■, wt; ●, Δ10/wt; ▴, Δ20/wt; ⧫, K199R/wt.
Expression of Wt HRI in NIH 3T3 cells inhibits their growth
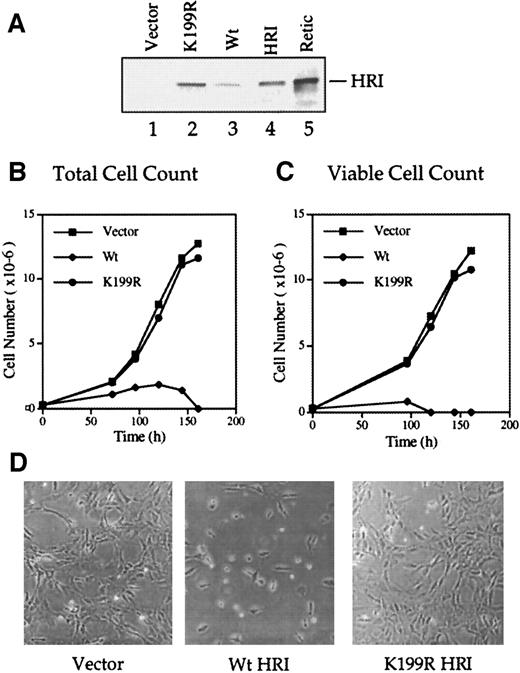
We produced the Wt and inactive K199R HRI retroviruses and used them to infect NIH 3T3 cells as described in “Materials and methods.” Western blot analyses of the retrovirally transduced cells with the anti-HRI monoclonal antibody showed that full-length Wt and K199R HRI protein were synthesized in these cells (Figure2A, lanes 2 and 3). The level of Wt HRI protein expression in these cells was much lower than that of the inactive K199R HRI (lanes 2 and 3). Since Wt HRI expressed in 3T3 cells was an active eIF-2α kinase when analyzed by in vitro protein kinase assays (data not shown), it is likely that Wt HRI inhibited its own synthesis as well as protein synthesis generally in NIH 3T3 cells. Similar observations were made when HRI was expressed in Sf9 cells by recombinant baculoviruses.28
Growth characteristics of NIH 3T3 cells overexpressing Wt and K199R HRI.
(A) Levels of HRI protein expression. NIH 3T3 cells overexpressing vector alone (lane 1), Wt HRI (lane 3), or K199R HRI (lane 2) were generated by infections with retroviruses as described in “Materials and methods.” Cytoplasmic extracts (20 μg) from these cells, as indicated in the Figure, were separated by 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed for HRI by Western blot analysis. Purified rabbit reticulocyte HRI (lane 4) and reticulocyte lysates (lane 5) were used as positive controls. (B) Growth curve using total cell numbers. NIH 3T3 cells expressing either the pLXSN retroviral vector alone, Wt HRI, or K199R HRI were seeded at equal densities of (2.5 × 105) per 100 mm. Cells expressing Wt HRI were washed 3 times with DMEM to remove the hemin supplemented in the medium and maintained in the culture medium without hemin supplement for 24 hours prior to seeding. Total numbers of cells at different times of culture were plotted. (C) Growth curve of viable cells. Cells were grown as described in panel B, and the cell viability was determined by staining with trypan blue. The number of trypan blue–negative cells was plotted relative to time in culture. (D) Phase contrast photographs. NIH 3T3 cells expressing vector, Wt, or K199R HRI at day 3 of growth as shown in panels B and C were photographed at 200 × magnification.
Growth characteristics of NIH 3T3 cells overexpressing Wt and K199R HRI.
(A) Levels of HRI protein expression. NIH 3T3 cells overexpressing vector alone (lane 1), Wt HRI (lane 3), or K199R HRI (lane 2) were generated by infections with retroviruses as described in “Materials and methods.” Cytoplasmic extracts (20 μg) from these cells, as indicated in the Figure, were separated by 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed for HRI by Western blot analysis. Purified rabbit reticulocyte HRI (lane 4) and reticulocyte lysates (lane 5) were used as positive controls. (B) Growth curve using total cell numbers. NIH 3T3 cells expressing either the pLXSN retroviral vector alone, Wt HRI, or K199R HRI were seeded at equal densities of (2.5 × 105) per 100 mm. Cells expressing Wt HRI were washed 3 times with DMEM to remove the hemin supplemented in the medium and maintained in the culture medium without hemin supplement for 24 hours prior to seeding. Total numbers of cells at different times of culture were plotted. (C) Growth curve of viable cells. Cells were grown as described in panel B, and the cell viability was determined by staining with trypan blue. The number of trypan blue–negative cells was plotted relative to time in culture. (D) Phase contrast photographs. NIH 3T3 cells expressing vector, Wt, or K199R HRI at day 3 of growth as shown in panels B and C were photographed at 200 × magnification.
Forced expression of Wt HRI in NIH 3T3 cells resulted in a severe inhibition of cell growth and ultimately in cell death (Figure 2B-D). The addition of a low concentration (5 μmol/L) of hemin to the culture medium enabled us to maintain Wt HRI-expressing 3T3 cells. All the experiments described here used pooled cells and were, therefore, not subject to clonal variation. For characterizing the growth curve of cells expressing Wt HRI, these cells were cultured for 24 hours in the absence of hemin and plated in the absence of hemin. As shown in Figures 2B and 1C, the initial growth of Wt HRI was much slower than that of cells expressing vector alone or K199R HRI, and after 4 days in culture most of the cells were detached. These cells appeared condensed and round and displayed an increased refractivity similar to that of apoptotic cells. There were few trypan blue–negative cells remaining by day 4 and none by day 5 (Figure 2C). In contrast, expression of the K199R HRI had no apparent effect on the morphology or growth rate of 3T3 cells (Figure 2B-D). Similar results were obtained with overexpression of Δ10 or Δ20 HRI (data not shown).
Overexpression of inactive K199R, Δ10, or Δ20 HRI does not cause malignant transformation of NIH 3T3 cells
It has been shown that overexpression of the K296R or Δ6 inactive mutants of PKR in 3T3 cells results in malignant transformation.11,13 44 We examined the growth in monolayer, cloning efficiency, and tumorigenicity of 3 K199R HRI clones. Each of our K199R HRI-expressing clones exhibited a normal morphology and had doubling times and saturation densities unchanged from those of uninfected or vector-only control cells (Table1). We observed similar results using pooled K199R, Δ10, or Δ20 HRI-expressing 3T3 cells (data not shown). Cells expressing K199R, Δ10, or Δ20 HRI failed to grow on soft agar (Tables 1 and 2), in contrast to NIH 3T3 cells expressing the inactive mutant PKR Δ6, which exhibited cloning efficiencies of 19.8% ± 0.9% (Table 1). None of our K199R HRI 3T3 clones developed tumors in athymic (nu/nu) nude mice within the 10-week observation period, while all 3 nude mice injected with PKR Δ6-12 cells developed tumors within 28 to 34 days. The inability of inactive HRI to transform 3T3 cells is consistent with our earlier finding that HRI is expressed predominantly in eythroid cells in contrast to the ubiquitous expression of PKR.
Growth characteristics of K199R HRI NIH 3T3 cells
| NIH 3T3 cells . | Growth in monolayers* . | Tumorigenicity† (animals with tumors/animals injected) . | ||
|---|---|---|---|---|
| Doubling times (h) . | Saturation density (106 cells) . | Cloning efficiency (%)1-153 . | ||
| pLXSN | 28.5 ± 1.0 | 3.4 ± 0.4 | 0 | 0/3 |
| K199R HRI.1 | 28.8 ± 1.3 | 3.1 ± 0.3 | 0 | 0/3 |
| K199R HRI.2 | 28.5 ± 0.5 | 3.3 ± 0.3 | 0 | 0/3 |
| K199R HRI.3 | 29.3 ± 0.9 | 3.3 ± 0.5 | 0 | 0/3 |
| PKRΔ6-12 | 24.2 ± 1.1 | 4.5 ± 0.5 | 19.8 ± 0.9 | 3/3 (28-34 days) |
| NIH 3T3 cells . | Growth in monolayers* . | Tumorigenicity† (animals with tumors/animals injected) . | ||
|---|---|---|---|---|
| Doubling times (h) . | Saturation density (106 cells) . | Cloning efficiency (%)1-153 . | ||
| pLXSN | 28.5 ± 1.0 | 3.4 ± 0.4 | 0 | 0/3 |
| K199R HRI.1 | 28.8 ± 1.3 | 3.1 ± 0.3 | 0 | 0/3 |
| K199R HRI.2 | 28.5 ± 0.5 | 3.3 ± 0.3 | 0 | 0/3 |
| K199R HRI.3 | 29.3 ± 0.9 | 3.3 ± 0.5 | 0 | 0/3 |
| PKRΔ6-12 | 24.2 ± 1.1 | 4.5 ± 0.5 | 19.8 ± 0.9 | 3/3 (28-34 days) |
Cell growth in monolayers was measured by seeding 5 × 104 cells in 100-mm dishes in Dulbecco's modified Eagle's medium (DMEM) + 10% calf serum. The medium was changed every 3 days. The doubling times were determined by counting the cells every day for a 6-day period and calculating the growth rate of exponentially growing cells. The saturation density was determined by counting the number of cells in culture at 3 days post-confluence.
The ability of these cells to form tumors was examined in 6-week-old athymic mice (nu/nu); 1 × 106 cells in 100 μL of phosphate-buffered saline were injected subcutaneously.
For studies on cloning efficiency, cells (1 × 104) were suspended in a 0.35% agar solution in DMEM + 20% fetal calf serum (FCS) and overlaid onto a 0.5% agar matrix in DMEM + 20% FCS in 35-mm plates. One day after incubation at 37°C in 5% CO2, 2 mL DMEM + 20% FCS was added. Cells grown in soft agar were counted 10 days after plating. Cloning efficiency was determined by the number of colonies divided by the number of cells plated. The figures presented are the averages of 3 experiments with SD.
Cloning efficiency of NIH 3T3 cells overexpressing Wt and mutant HRI
| Cell lines . | Cloning efficiency (%) . |
|---|---|
| Vector | 0 |
| Wt HRI | 0 |
| K199R HRI | 0 |
| Δ20 HRI | 0 |
| Bcl-2 | 0 |
| 4E | 11.0 ± 0.3 |
| 4E/Wt HRI | 2.8 ± 0.2 |
| 4E/K199R HRI | 12.2 ± 0.7 |
| 4E/Δ20 HRI | 12.8 ± 0.7 |
| PKRΔ6/Wt HRI | 5.6 ± 0.3 |
| Cell lines . | Cloning efficiency (%) . |
|---|---|
| Vector | 0 |
| Wt HRI | 0 |
| K199R HRI | 0 |
| Δ20 HRI | 0 |
| Bcl-2 | 0 |
| 4E | 11.0 ± 0.3 |
| 4E/Wt HRI | 2.8 ± 0.2 |
| 4E/K199R HRI | 12.2 ± 0.7 |
| 4E/Δ20 HRI | 12.8 ± 0.7 |
| PKRΔ6/Wt HRI | 5.6 ± 0.3 |
The cloning efficiency of various cell lines was determined as described in Table 1.
It has been reported that 3T3 cells that overexpressed eIF-4E exhibit a morphology indicative of a transformed phenotype and grew in soft agar.45-47 We examined the ability of Wt HRI to inhibit the transformation by eIF-4E and PKR Δ6. Interestingly, we found that the cloning efficiency of eIF-4E cells was dramatically reduced by co-expression with Wt HRI, but not by co-expression with K199R or Δ20 HRI. Similarly, the cloning efficiency of PKR Δ6 was also reduced significantly by co-expression of Wt HRI. These results indicated that HRI could inhibit protein synthesis in cells transformed by eIF-4E or PKR Δ6. The ability of HRI to inhibit protein synthesis in eIF-4E–overexpressing cells is consistent with the current knowledge of independent regulation of protein synthesis by eIF-4E and eIF-2. The ability of Wt HRI to inhibit PKR Δ6 transformation suggests that PKR and HRI do not form a heterodimer and that both PKR and HRI function as homodimers.
Expression of inactive mutant HRI increases hemoglobin production and proliferative capacity of differentiating MEL cells
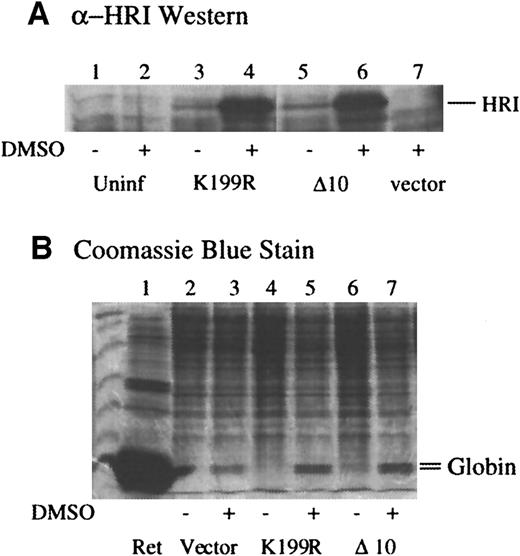
Next, we examined the effects of expression of these mutants in MEL cells in which endogenous HRI resides.35 MEL cells were infected with amphotropic retroviruses containing mutant HRI cDNA and/or puromycin selection marker. Pooled selected cells were plated in the absence of puromycin, and erythroid differentiation was induced by dimethyl sulfoxide (DMSO). The levels of HRI expression before and after DMSO induction were determined by Western blot analysis. As shown in Figure 3A, the levels of K199R and Δ10 HRI expression were increased substantially upon DMSO induction of MEL cells (lanes 4 and 8 vs lanes 3 and 7). It has been shown previously that the promoter activity of Moloney retrovirus LTR is enhanced by DMSO induction.48
Expression of inactive mutant HRI increases the α and β globin contents of differentiating MEL cells.
(A) Expression of retrovirally transduced HRI in MEL cells. MEL cells expressing vector alone, K199R HRI, or Δ10 HRI were plated at 1 × 107 cells per 100-mm plate the night before treatment with 2% DMSO as indicated. Cells were counted and harvested on day 5. Cells were lysed at 1 × 108 cells/mL. Cytoplasmic extracts, equivalent to 2 × 106 cells, were used for anti-HRI Western blot analysis. Lanes 1 and 2 are uninfected MEL cell controls; lanes 3 and 4 are K199R-infected cell extracts; lanes 5 and 6 are Δ10 HRI-infected cell extracts; lane 7, vector-infected cell extracts. (B) α and β globin contents of MEL cells expressing mutant HRI. Lanes 2, 4, and 6 are induced with DMSO. Cytoplasmic extracts, equivalent to 5 × 105 cells, were separated by 15% SDS-PAGE and stained with Coomassie blue. The positions of α and β globin were marked. Lane 1, rabbit reticulocyte lysates. Lanes 2 and 3, cell extracts; lanes 4 and 5, K199R HRI-expressing cell extracts; lanes 6 and 7, Δ10 HRI-expressing cell extracts; lanes 3, 5, and 7 are treated with DMSO.
Expression of inactive mutant HRI increases the α and β globin contents of differentiating MEL cells.
(A) Expression of retrovirally transduced HRI in MEL cells. MEL cells expressing vector alone, K199R HRI, or Δ10 HRI were plated at 1 × 107 cells per 100-mm plate the night before treatment with 2% DMSO as indicated. Cells were counted and harvested on day 5. Cells were lysed at 1 × 108 cells/mL. Cytoplasmic extracts, equivalent to 2 × 106 cells, were used for anti-HRI Western blot analysis. Lanes 1 and 2 are uninfected MEL cell controls; lanes 3 and 4 are K199R-infected cell extracts; lanes 5 and 6 are Δ10 HRI-infected cell extracts; lane 7, vector-infected cell extracts. (B) α and β globin contents of MEL cells expressing mutant HRI. Lanes 2, 4, and 6 are induced with DMSO. Cytoplasmic extracts, equivalent to 5 × 105 cells, were separated by 15% SDS-PAGE and stained with Coomassie blue. The positions of α and β globin were marked. Lane 1, rabbit reticulocyte lysates. Lanes 2 and 3, cell extracts; lanes 4 and 5, K199R HRI-expressing cell extracts; lanes 6 and 7, Δ10 HRI-expressing cell extracts; lanes 3, 5, and 7 are treated with DMSO.
Since hemoglobin is the major protein in erythroid cells, the effects of forced expression of the dominant-negative HRI mutants on the levels of α and β globin chains in differentiating MEL cells were examined. The contents of α and β globin were determined by SDS-PAGE and Coomassie blue staining of the cell extracts from equal numbers of cells. As shown in Figure 3B, DMSO induction of MEL cells induced the globin expression in the cells expressing puromycin-resistant gene alone as expected (lane 3 vs 2). Upon DMSO induction, MEL cells overexpressing K199R or Δ10 HRI had significantly higher levels of globin chains (lanes 5 and 7 vs lane 3). Before DMSO induction, there were no apparent changes in the profiles of protein expressed in MEL cells expressing inactive mutants. This is perhaps to be expected since the levels of the expression of the mutant HRI were low before induction.
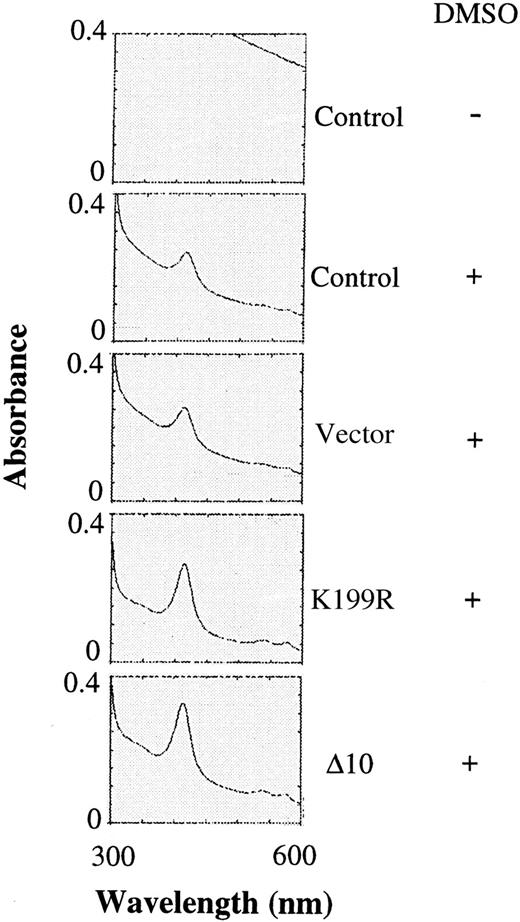
Since there was an increase in the α and β globin chains upon expression of K199R and Δ10 HRI, the amounts of hemoglobin in differentiating MEL cells were also determined by scanning the visible spectra of the cell extracts for the characteristic Soret band of hemoglobin. There was no detectable signal of Soret band without DMSO induction (Figure 4). Upon DMSO induction, the peak of Soret band was visible. Overexpression of the K199R or Δ10 HRI in MEL cells resulted in a 2- to 3-fold increase in the hemoglobin content of these cells upon DMSO induction as compared with uninfected MEL cells or MEL cells expressing the puromycin-resistant vector control (Figure 4) or Bcl-2 (data not shown). This increase in hemoglobin synthesis is most likely the result of the inhibition of the endogenous HRI activity by these inactive HRI mutants. Thus, the increase of hemoglobin synthesis is the consequence of increase of general protein synthesis.
Hemoglobin contents of differentiating MEL cells expressing inactive mutant HRI.
The same cytoplasmic extracts described in Figure 3 were diluted 1:5 and then analyzed for the presence of the Soret band of hemoglobin at 414 nm by scanning from 300 to 600 nm as described previously.29
It is well established that upon terminal differentiation, cells lose their ability to divide (proliferative capacity). We have shown above that expression of Wt HRI in 3T3 cells resulted in the inhibition of the proliferation of these cells. We therefore asked whether HRI was involved in the loss of the proliferative capacity of differentiating MEL cells. The cell numbers of differentiating MEL cells expressing mutant HRI at days 4 and 5 after DMSO induction were determined and are shown in Table 3. The cell pellets of K199R and Δ10 HRI-expressing cells were visibly larger and redder than those of uninfected cells, cells expressing vector alone, or cells expressing Bcl-2. At both 4 and 5 days after DMSO induction, there was a significant increase in the number of K199R or Δ10 HRI-expressing cells as compared with the number of control uninfected cells or cells expressing the puromycin-resistant gene only or Bcl-2 (Table 3). In addition, cell numbers of K199R and Δ10 HRI-expressing cells continued to increase substantially from day 4 to day 5 while the cell number of vector control cells, Bcl-2–expressing cells, or uninfected cells did not increase much. These observations suggested that K199R and Δ10 HRI cells continued to proliferate from day 4 to day 5 after DMSO induction. Therefore, it appears that HRI may play a role in the loss of proliferative capacity of differentiating erythroid cells. In the absence of DMSO, there was no significant change in cell number of K199R or Δ10 HRI-expressing cells. This may be due to the low-level expression of this mutant HRI without DMSO induction as shown in Figure 3A. Thus, these results demonstrate that overexpression of K199R or Δ10 HRI in DMSO-induced MEL cells affects not only the production of hemoglobin, but also the proliferative capacity of these cells.
Effect of expression of inactive mutant HRI on the growth of MEL cells
| Cell lines . | Cell number (×106) . | ||||
|---|---|---|---|---|---|
| DMSO, day 4 . | DMSO, day 5 . | ||||
| − . | + . | − . | + . | % . | |
| Uninfected | 63.5 | 65.0 | 90.5 | 71.5 | 100 |
| Vector control | 67.2 | 80.0 | 3-150 | 74.0 | 103.5 |
| K199R HRI | 60.8 | 105.0 | 78.0 | 123.0 | 172.0 |
| Δ10 HRI | 72.0 | 83.0 | 77.0 | 98.8 | 138.2 |
| Bcl-2 | 70.5 | 59.5 | 82.5 | 74.5 | 104.2 |
| Cell lines . | Cell number (×106) . | ||||
|---|---|---|---|---|---|
| DMSO, day 4 . | DMSO, day 5 . | ||||
| − . | + . | − . | + . | % . | |
| Uninfected | 63.5 | 65.0 | 90.5 | 71.5 | 100 |
| Vector control | 67.2 | 80.0 | 3-150 | 74.0 | 103.5 |
| K199R HRI | 60.8 | 105.0 | 78.0 | 123.0 | 172.0 |
| Δ10 HRI | 72.0 | 83.0 | 77.0 | 98.8 | 138.2 |
| Bcl-2 | 70.5 | 59.5 | 82.5 | 74.5 | 104.2 |
MEL cells with expression vector, K199R HRI, Δ10 HRI, and Bcl-2, and the uninfected MEL cells were cultured and induced for erythroid differentiation as described in the Figure 3 legend. At days 4 and 5, both control and induced cells were harvested and counted. The percentage indicates the increase in cell number upon expression of inactive mutant HRI, Bcl-2, or vector control after DMSO inductin at day 5. The cell number of uninfected cells at day 5 is defined as 100%.
Cell number was not determined.
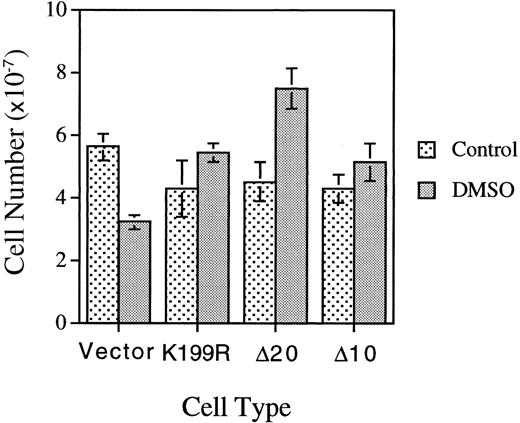
We repeated the experiments described above in Figures 3 and 4 and Table 3 with a separate batch of MEL cells infected with another batch of HRI retroviruses. In this experiment, MEL cells were plated at one fifth of the cell density employed in the previous experiment presented in Table 3. Under this condition, the loss of the proliferative capacity of DMSO-treated differentiating MEL cells was observed more clearly; ie, the cell number of DMSO-treated MEL cells expressing vector is 55.5% of that of the untreated cells (Figure5). In contrast to the loss of proliferative capacity of the control vector-differentiating cells, MEL cells expressing K199R, Δ10, or Δ20 HRI had increased proliferative capacity, with higher cell numbers upon DMSO-induction as compared with the untreated cells. The cell-cycle status of these cells was also analyzed by flow cytometry and is presented in Table4. At 4.5 days after subculture and in the absence of DMSO, approximately 20% to 30% of cells were in S+G2/M phase. Upon DMSO induction for 4.5 days, the percentage of cells in S+G2/M decreased substantially to 6% to 8%, a 3-fold drop as compared with untreated cells. This decrease was indicative of loss of proliferative capacity of differentiating cells. Significantly, the cells expressing K199R or Δ10 HRI had a higher percentage of cells in S+G2/M. The Δ20 HRI-expressing cells had only a slightly higher percentage of cells in S+G2/M, perhaps because these cells had already reached the saturation density since the number of Δ20 HRI-expressing cells was considerably higher than the number of K199R or Δ10 HRI-expressing cells. Collectively, these results from Tables 3 and 4and Figure 5 indicate that HRI also regulates the proliferation of differentiating erythroid cells.
Effects of expression of inactive mutant HRI on the proliferative capacities of differentiating MEL cells.
Cells expressing vector or mutant HRI as indicated were plated at 2 × 106 cells per 100-mm plate and induced as described in the Figure 4 legend. Cell numbers were counted on day 4.5 after induction.
Effects of expression of inactive mutant HRI on the proliferative capacities of differentiating MEL cells.
Cells expressing vector or mutant HRI as indicated were plated at 2 × 106 cells per 100-mm plate and induced as described in the Figure 4 legend. Cell numbers were counted on day 4.5 after induction.
Effect of expression of mutant HRI on the cell-cycle status in MEL cells
| Cell lines . | Without DMSO . | With DMSO . | ||
|---|---|---|---|---|
| G0/G1 . | S + G2/M . | G0/G1 . | S + G2/M . | |
| Uninfected | 76.51 | 23.49 | 93.88 | 6.12 |
| Vector | 78.44 | 21.56 | 91.58 | 8.42 |
| K199R HRI | 80.37 | 19.63 | 84.37 | 15.63 |
| Δ10 HRI | 72.62 | 27.38 | 87.56 | 12.44 |
| Δ20 HRI | 63.39 | 36.61 | 90.46 | 9.54 |
| Cell lines . | Without DMSO . | With DMSO . | ||
|---|---|---|---|---|
| G0/G1 . | S + G2/M . | G0/G1 . | S + G2/M . | |
| Uninfected | 76.51 | 23.49 | 93.88 | 6.12 |
| Vector | 78.44 | 21.56 | 91.58 | 8.42 |
| K199R HRI | 80.37 | 19.63 | 84.37 | 15.63 |
| Δ10 HRI | 72.62 | 27.38 | 87.56 | 12.44 |
| Δ20 HRI | 63.39 | 36.61 | 90.46 | 9.54 |
MEL cells expressing vector and HRI as indicated were cultured and induced for erythroid differentiation as described in the Figure 5legend. At day 4.5, both control and induced cells were harvested and stained with propidium iodide for cell-cycle analysis as described previously.53 The number represents the percentage of cells in each phase of cell cycle.
Recently, it has been reported that there is HRI-like activity in NIH 3T3 cells.25 As shown in Figure 2, a very low-level expression of Wt HRI in NIH 3T3 cells resulted in cell death. However, expression of inactive mutant HRI alone in nonerythroid 3T3 cells did not result in any enhancement in cell growth or changes in cell morphology (Figure 2; Tables 1 and 2). These results strongly suggest that if there is HRI protein present in 3T3 cells, it is not active under our experimental conditions. Most important, we observed that expression of inactive mutant HRI in erythroid MEL cells resulted in phenotypical changes in these cells, ie, increased hemoglobin content and proliferative capacity. The levels of expression of these dominant-negative mutants were comparable in 3T3 and MEL cells (data not shown). Thus, the most plausible explanation for the observed phenotypic changes upon expression of dominant-negative mutants in erythroid cells, but not in non-erythroid cells, is that active HRI is present in MEL cells but not in 3T3 cells.
Discussion
In this study, we have examined the consequence of overexpression of Wt and inactive mutant HRI in nonerythroid and erythroid cells. We have shown here that forced expression of Wt HRI in 3T3 cells caused an inhibition of cell growth and ultimately cell death (Figure 2). We were able to maintain the growth of Wt HRI-expressing 3T3 cells by the addition of hemin to the culture medium. Upon removal of hemin, Wt HRI became active, shut off protein synthesis by phosphorylating eIF-2α (data not shown), and killed cells. This lethal effect of overexpression of Wt HRI in 3T3 cells is similar to that of the overexpression of PKR in these cells.16 Interestingly, we found that expression of Wt HRI can inhibit the transformation phenotype of PKR Δ6-overexpressing 3T3 cells (Table 2). This finding indicates that HRI and PKR do not form heterodimers in vivo. Thus, HRI and PKR act independently of each other and represent different targets of regulation of protein synthesis in response to different kinds of stress.
It has been demonstrated that overexpression of inactive mutants of PKR in NIH 3T3 cells results in the malignant transformation of those cells.11,13,44 Because PKR is a dimer, it is believed that inactive PKR protein acts in a transdominant-negative manner to form an inactive heterodimer with the endogenous PKR.11 However, we found that overexpression of the inactive K199R, Δ10, or Δ20 HRI did not result in any change in the morphology that would be indicative of malignant transformation. These cells did not grow in soft agar or form tumors in nude mice. We have previously shown that co-expression of Wt HRI with K199R HRI in baculovirus-infected insect cells leads to diminished kinase activity of the Wt HRI.28 This reduced activity is thought to be the result of the formation of an inactive HRI heterodimer. We showed similar results here using the inactive HRI deletion mutants Δ10 and Δ20 (Figure 1). Our observation that forced expression of K199R, Δ10, or Δ20 HRI fails to confer upon NIH 3T3 cells a transformed phenotype is consistent with there being no active HRI in these cells. These results are in agreement with our earlier finding that HRI is expressed predominantly in erythroid cells.35
Recently, it has been reported that HRI mRNA is expressed in NIH 3T3 cells and in nonerythroid tissues.25 They have also partially purified HRI-like activity from young mouse liver (6 to 8 weeks old) and NIH 3T3 cells. It is important to note that fetal liver is a site of erythropoiesis during development and that erythropoiesis persists in the liver of the mouse after birth. In addition, NIH 3T3 cells are embryonic in origin. It is to be noted that in these 2 reports no Western blot analysis of HRI in tissues or 3T3 cells has been performed to establish that HRI protein is indeed present in nonerythroid cells. We show here that a very low-level expression of Wt HRI in NIH 3T3 cells (1/100 to 1/500 of rabbit reticulocytes) is sufficient to inhibit cell growth (Figure 2). These results indicate that if there is HRI protein present in NIH 3T3 cells, it is probably not active under our experimental conditions. Recently, 2 more mammalian eIF-2α kinases have been cloned, the PEK (PERK)19,20 and the GCN2.49 50 Both of these eIF-2α kinases are, like PKR, universally expressed.
In contrast to no phenotypical change in 3T3 cells, forced expression of the inactive mutant HRI in differentiating MEL cells results in increased hemoglobin synthesis (Figures 3 and 4) and proliferative capacity of these cells (Tables 3 and 4; Figure 5). These phenotypical changes not only provide the direct in vivo evidence for the function of HRI in the regulation of hemoglobin synthesis in erythroid cells, but also suggest a role of HRI in the regulation of the proliferation of the differentiating erythroid cells through, most likely, the inhibition of protein synthesis. However, it is possible that HRI may have an as yet unidentified substrate other than eIF-2α, and the phosphorylation of this substrate might be involved in the regulation of cell proliferation. The regulation of hemoglobin synthesis by HRI in erythroid cells is the result of its regulation of general protein synthesis. We have shown recently that HRI is a hemoprotein with 2 distinct heme-binding sites.29, 34 Thus, HRI serves as a sensor of heme and provides a feedback mechanism to coordinate the synthesis of globins according to heme concentration in erythroid cells.
In addition to regulating HRI activity, heme also regulates its own synthesis in reticulocytes at one or more steps prior to the formation of δ-aminolevulinic acid, the first intermediate in the heme biosynthetic pathway.51 Furthermore, inhibition of protein synthesis in reticulocytes by the addition of cycloheximide or puromycin also results in the inhibition of heme biosynthesis at the formation of δ-aminolevulinic acid.52 These earlier studies indicate that in the absence of globin synthesis, the accumulation of heme inhibits heme biosynthesis. Thus, heme coordinates the synthesis of heme and globins in erythroid cells by regulating HRI and its own synthesis.
Acknowledgments
We thank all investigators who provided materials as indicated in “Materials and methods” and Dr Karen Westerman for providing expertise in the production of retroviruses.
Supported by National Institutes of Health grants DK-16272 (J.-J.C.) and HL-55435 (P.L.B).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jane-Jane Chen, E25-545, Massachusetts Institute of Technology, Cambridge, MA 02139; e-mail: j-jchen@mit.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal