Abstract
The purposes of this study were to determine the frequency of mutations in SH2D1A in X-linked lymphoproliferative disease (XLP) and the role of SH2D1A mutations and Epstein-Barr virus (EBV) infection in determining the phenotype and outcome of patients with XLP. Analysis of 35 families from the XLP Registry revealed 28 different mutations in 34 families—large genomic deletions (n = 3), small intragenic deletions (n = 10), splice-site (n = 3), nonsense (n = 3), and missense (n = 9) mutations. No mutations were found in 25 males, so-called sporadic XLP (males with an XLP phenotype after EBV infection but no family history of XLP) or in 9 patients with chronic active EBV syndrome. Of 304 symptomatic males in the XLP Registry, 38 had no evidence of EBV infection at first clinical manifestation. When fulminant infectious mononucleosis (FIM) was excluded, there was no statistical difference in the frequency of EBV infectivity in the other XLP phenotypes. Furthermore, there was no difference at age of first clinical manifestation between EBV+ and EBV−males or in survival when patients with FIM were excluded. In conclusion, it was found that mutations in the SH2D1A gene are responsible for XLP but that there is no correlation between genotype and phenotype or outcome. It was also found that though EBV infection often results in FIM, it is unnecessary for the expression of other manifestations of XLP, and it correlates poorly with outcome. These results suggest that unidentified factors, either environmental or genetic (eg, modifier genes), contribute to the pathogenesis of XLP.
Introduction
X-linked lymphoproliferative disease (XLP) (Duncan disease; MIM 308 240) is a congenital immunodeficiency disease estimated to affect approximately 1 in 1 × 106males.1 Although more than 98% of persons in the general population experience little or no clinical manifestations after Epstein-Barr virus (EBV) infection, XLP has been characterized by exquisite vulnerability to EBV infection. The spectrum of phenotypes seen in XLP is varied. The most common phenotype is fulminant infectious mononucleosis (FIM), which is often fatal and clinicopathologically identical to the virus-associated hemophagocytic syndrome. Lymphoproliferative disorders (LPD) are the next most often observed. These may manifest as polyclonal or monoclonal proliferations and may range from lymphoid vasculitides to malignant lymphoma. Both B-cell and T-cell non-Hodgkin lymphoma and Hodgkin disease have been observed. Dysgammaglobulinemia (dys-γ) is also often observed. Usually this manifests as hypogammaglobulinemia or agammaglobulinemia, but abnormalities in one or more immunoglobulin isotypes have been observed (eg, IgA deficiency, hyper-IgM). Cytopenia is rare (3% of affected persons) and usually manifests as aplastic anemia (AA), but isolated anemias, autoimmune aplasia, and pure red cell aplasia have been observed.1,2 Males often develop more than one phenotype over time. Most often one of these phenotypes appears after exposure to EBV; however, the XLP phenotype can be observed in affected persons without serologic or molecular evidence of prior EBV infection.2-4
The gene mutated in XLP has been mapped to the interval between DXS1001 and DXS8057.5 This chromosomal region contains the tenascin-m gene, a human homologue of the Drosophilaextracellular protein coding gene and SH2D1A.6 The latter was found to be mutated in 9 of 16 unrelated patients with XLP.6 The gene spans 40 kb of genomic sequence and contains 4 exons. The human SH2D1A gene gives rise to a transcript of 2.5 kb expressed in CD4+ and CD8+ T lymphocytes, the thymus, fetal liver, colon, and spleen, which encodes a protein with a src homology 2 (SH2) domain.6-8 The SH2 domain is a highly conserved structure containing approximately 100 amino acid residues and is found in cytosolic nonreceptor tyrosine kinases and in a number of proteins that play key roles in signal transduction that modulate enzyme activity or target proteins to certain cellular locations. The core element of the structure is an antiparallel β-sheet that is sandwiched between 2 α-helices.9 The SH2D1A protein interacts with SLAM (signaling lymphocyte-activating molecule), 2B4, and presumably other molecules required for a controlled T-cell response during EBV infection.7,10,11 Germ-line mutations in the SH2D1A gene have been identified in approximately 55% to 60% of the patients tested manifesting one of the XLP phenotypes.6-8 It has been suggested that patients with XLP without SH2D1A mutations in the coding region of the gene might harbor mutations in the intronic sequences, the regulatory region of the SH2D1A gene, or in another gene such as tenascin-m.6-8 Alternatively, the low frequency of mutations in the SH2D1A gene in the analyzed families could stem from a misdiagnosis of XLP. Indeed, the diagnosis of XLP is difficult given its broad clinical heterogeneity and the lack of a diagnostic laboratory test, particularly in the setting of a single male manifesting a phenotype consistent with XLP but without a family history supporting an X-linked inheritance. This group has been designated, sporadic XLP, an inappropriate term, as this study demonstrates.
This study is a comprehensive analysis of the correlation of SH2D1A mutations and EBV infection with clinical phenotype and outcome in XLP. Data and material from the David T. Purtilo International XLP Registry were used for these studies. SH2D1A mutational analysis was performed on 35 families with a confirmed X-linked inheritance (ie, 2 or more maternally related affected males from the XLP Registry). We also studied 25 unrelated patients with sporadic XLP, a phenotype reminiscent of XLP but without the corroborating family history and 9 persons with chronic, active EBV (CAEBV) syndrome.12 Using data from the XLP Registry, the EBV status of boys with XLP was determined at the time of their first clinical manifestation to determine what effect EBV infection has on clinical phenotype and outcome. We provide evidence that mutations in the SH2D1A gene are responsible for XLP. However, this study illustrates that EBV infection is not the only trigger for the clinical manifestations of XLP, and that neither EBV infections nor mutations in the SH2D1A gene are predictive of clinical phenotype or outcome.
Patients and methods
Patients
Subjects studied were collected as part of a long-standing study on the genetics of X-linked lymphoproliferative disease through the David T. Purtilo International XLP Registry and in accordance with institutional review board guidelines. The study was composed of members of families with XLP and of male patients with phenotypes reminiscent of XLP (mostly FIM) but without family histories of the disease, who were listed with the Department of Pathology and Microbiology before June 1999 and for whom samples were available for comprehensive analysis of the SH2D1A gene. Patients were considered affected by XLP if they had the following: 2 or more maternally related males manifesting an XLP phenotype (FIM, dys-γ, LPD or AA), and strong linkage to locus-specific markers. In this regard, 33 of 35 families were demonstrated by RFLP linkage analysis to be linked near the SH2D1A locus. Genetic diagnosis was not possible for the patients with sporadic XLP (males with clinical phenotypes reminiscent of XLP (FIM, dys-γ, LPD) without a family history of the disease. Patients with chronic, active Epstein-Barr virus (CAEBV) syndrome were from Japan, and no family history of the disease was reported. CAEBV is a severe illness with unusual EBV activation that persists for years, and its pathogenesis is largely unknown.12
Linkage analysis
Haplotype analysis.
Haplotypes for the RFLP markers DXS37, DXS6, DXS100, and DXS42 were constructed, as described earlier.13 Labeled microsatellite markers DXS8088, DXS1220, DXS8055, DXS8053, DXS8081, DXS8064, DXS8067, DXS1001, DXS8057, AFM240xb10, DXS8093, DXS8009, DXS1206, DXS8078, DXS1046, and DXS1047 were also used in some cases. Primers and annealing temperatures used to amplify the microsatellite markers were as previously described.14
Mutation detection.
Peripheral blood lymphocytes from family members with XLP, patients with so-called sporadic XLP, and patients with CAEBV were collected throughout North and South America, Europe, the Middle East, Australia, and Japan. Genomic DNA was isolated from blood samples using standard techniques. When lymphoblasts were available, they were grown in RPMI 1640 supplemented with 10% fetal bovine serum under standard culture conditions. DNA and RNA were isolated using standard techniques and Qiagen (Valencia, CA) RNA isolation kits, respectively. Coding sequences, 5′ regulatory region (−300 nucleotides from the transcription initiation site), and intronic splice-site sequences were amplified by the polymerase chain reaction (PCR). From each XLP family, at least 2 affected members, 2 carriers, and 2 normal members were analyzed for mutations in the SH2D1A gene. Amplified exons from patients and control subjects were purified by the Qiaquick PCR purification kit (Qiagen) and sequenced using appropriate end-labeled primers. Labeled products were separated by a LI-COR 4000L automated DNA sequencer (Li-Cor; Lincoln, NE). Exon deletions, initially detected by a failure of the exons to amplify in PCR assays, were confirmed by Southern blotting and hybridization with DNA probes derived from the cDNA fragments of the SH2D1A gene. To verify that similar changes were not present in healthy persons (ie, they represent polymorphic variants), the 5′ regulatory, coding, and splice-site sequences were also analyzed in 100 additional normal X-chromosomes.
Reverse transcriptase–PCR
Total RNA was isolated from EBV-immortalized peripheral blood lymphocytes of patients, carriers, and healthy persons by TRIzol Reagent (BRL, Gaithersburg, MA). One microgram total RNA was reverse-transcribed with Moloney murine leukemia virus reverse transcriptase (RT) of the Gene Amp RNA PCR kit from PerkinElmer (Roche, Branchburg, NJ) according to the supplier's recommendation. PCR was performed in 100 μL with the primer pair XL37RTU (CGCAGTGGCTGTGTATCAT) and XL37RTL (ACTTCTAGCTGAGGACTTCTTCTC), giving a 325-nucleotide–long product of normal SH2D1A transcript. As a control, actin was used (actin F, TGAAGTGTGACGTGGACATC; actin R, ACTCGTCATACTCCTGCTTG), producing a 246-nucleotide product. The annealing temperature for both primer pairs was 60°C.
EBV status
Males who underwent EBV testing at time of their first clinical manifestation were analyzed for age, phenotype of first manifestation, and survival. Evidence of EBV infection included one or more of the following: positive heterophile antibody test result, positive EBV titer to any EBV antigen, or evidence of EBV by in situ hybridization in tissue or by PCR in tissue or blood. Patients who did not undergo testing or who previously received gamma-globulin supplementation or blood products before serologic testing of EBV status were considered indeterminable for EBV status.
Statistical analysis
Median age of first clinical presentation and survival calculated for the EBV+ and EBV− groups were compared using the Mann-Whitney U test. Differences in phenotypes between the groups were compared by the χ2 test. Statistical analysis and calculations were performed using software from Statview SE+Graphics, version 1.02 (Abacus Concepts, Berkeley, CA).
Results
We previously reported that a definitive diagnosis of XLP can be made if 2 or more maternally related males manifest an XLP phenotype.2 To determine the frequency of mutations of the SH2D1A gene in XLP, 35 families with definitive diagnoses of XLP were analyzed. Thirty-three of the 35 underwent previous RFLP linkage analysis studies that demonstrated a strong association with the SH2D1A locus. The families are of diverse ethnic origins, from the United States, Canada, the United Kingdom, Australia, The Netherlands, New Zealand, Israel, Finland, Turkey, and Argentina. Twenty-five males with sporadic XLP who manifested a phenotype consistent with XLP (FIM, LPD or dys-γ after EBV infection) but without a family history supporting an X-linked disorder, and 9 patients with CAEBV syndrome were included in this study. All available affected subjects were examined, or the clinical and laboratory reports of the patients were reviewed by one of the authors.
To identify the molecular alterations in the SH2D1A gene responsible for the pathologic phenotype(s) of XLP, we designed 5 primer pair sets based on the genomic sequence of the SH2D1A gene to amplify the 4 exons, adjacent intron regions, and 300 nucleotides of the 5′ regulatory region of the SH2D1A gene (Table1). This analysis revealed 3 different types of mutations in 34 of 35 (97%) families with a definitive diagnosis of XLP: (1) deletion of the entire SH2D1A gene or part of the coding region, (2) splice-site mutations, and (3) nucleotide substitutions, including nonsense and missense mutations. In the 35 families surveyed for mutations, 28 different mutations were found. Table 2 summarizes the mutations observed. Codon numbering starts with the first in-frame methionine of the SH2D1A gene.
Sequence of the primers used to amplify exons, exon/intron junctions, and presumed regulatory region of the SH201A
| . | Forward primer . | Reverse primer . | bp . | Temp (°C) . |
|---|---|---|---|---|
| XLP.1 | cacgacgttgtaaaacgacacagaagcattactaagc | ggataacaatttcacacaggccagctgctgttgcccac | 608 | 55 |
| XLP.2 | cacgacgttgtaaaacgaccaatgacaccatatacgtgt | ggataacaatttcacacagggctttcttaatgatccatga | 380 | 55 |
| XLP.3 | cacgacgttgtaaaacgaccaagttacacaaatgtta | ggataacaatttcacacaggcttggactcataactctg | 348 | 55 |
| XLP.4 | cacgacgttgtaaaacgactcattgtgagttttatgcag | ggataacaatttcacacagggctcaccgaactgtatta | 272 | 55 |
| XLP.A | cacgacgttgtaaaacgacaaagcaccagttaaatgattc | ggataacaatttcacacaggtcacacttcagttaaaacgag | 718 | 55 |
| . | Forward primer . | Reverse primer . | bp . | Temp (°C) . |
|---|---|---|---|---|
| XLP.1 | cacgacgttgtaaaacgacacagaagcattactaagc | ggataacaatttcacacaggccagctgctgttgcccac | 608 | 55 |
| XLP.2 | cacgacgttgtaaaacgaccaatgacaccatatacgtgt | ggataacaatttcacacagggctttcttaatgatccatga | 380 | 55 |
| XLP.3 | cacgacgttgtaaaacgaccaagttacacaaatgtta | ggataacaatttcacacaggcttggactcataactctg | 348 | 55 |
| XLP.4 | cacgacgttgtaaaacgactcattgtgagttttatgcag | ggataacaatttcacacagggctcaccgaactgtatta | 272 | 55 |
| XLP.A | cacgacgttgtaaaacgacaaagcaccagttaaatgattc | ggataacaatttcacacaggtcacacttcagttaaaacgag | 718 | 55 |
The intronic primers were tagged at the 5′ end by oligos complementary to M13 forward and reverse primers (italics) to be able to sequence the amplified products using IR 800 M13 forward and reverse primers from Li-Cor.
SH2D1A mutations in families with definitive diagnosis of XLP
| No. . | Family . | Affected males in family . | Clinical phenotypes of affected males . | Detection . | Type of mutation . | Nucleotides affected . | Predicted effect . |
|---|---|---|---|---|---|---|---|
| 1 | K043 | 2 | Dys, g | S, PCR | Deletion | 3.5 Mb DNA deleted at Xq25 | Neither SH2D1A nor tenascin-m proteins |
| 2 | K063 | 4 | FIM, LPD | S, PCR | Deletion | 3.5 Mb DNA deleted at Xq25 | Neither SH2D1A nor tenascin-m proteins |
| 3 | K073 | 4 | FIM, LPD | S, PCR | Deletion | 3.5 Mb DNA deleted at Xq25 | Neither SH2D1A nor tenascin-m proteins |
| 4 | Ado1 | 3 | FIM | S, PCR | Deletion | Exons 1-4 | No SH2D1A Protein |
| 5 | K051 | 3 | Dys-g | S, PCR | Deletion | Exons 1-4 | No SH2D1A Protein |
| 6 | K005 | 5 | FIM, LPD | S, PCR | Deletion | Exons 2-4 | Truncated SH2D1A protein |
| 7 | K083 | 2 | FIM | S, PCR | Deletion | Exons 2-4 | Truncated SH2D1A protein |
| 8 | K001 | 11 | FIM, LPD dys-g | S, PCR | Deletion | Exon 2 | Truncated SH2D1A protein |
| 9 | K032 | 3 | FIM, dys-g | PCR.2 | Deletion | Exon 2 | Truncated SH2D1A protein |
| 10 | K055 | 5 | FIM, AA LPD, dys-g | SPCR.2 | Deletion | 89 bp of intron 1 + exon 2 + intron 2 | Truncated SH2D1A protein |
| 11 | K060 | 4 | LPD, dys-g | PCR.2 | Deletion | 159 bp of exon 2 from nucleotide 448 + intron 2 | Truncated SH2D1A protein |
| 12 | K061 | 3 | FIM, LPD | PCR.2 | Deletion | 439A-485T | Frameshift, truncated SH2D1A protein |
| 13 | K008 | 7 | FIM, LPD | PCR.2 | Splice | ACAGgtACCGgt intron 1 3′ splice site | Exon 2 skipping |
| 14 | K048 | 2 | LPD | PCR.1 | Splice | ctGTGA > ctGCGA intron 1 5′ splice site | Intron retention truncated SH2D1A protein |
| 15 | K065 | 7 | FIM, dys-g | PCR.2 | Splice | agGTAT > agATAT intron 2 5′ splice site | Intron retention truncated SH2D1A protein |
| 16 | K003 | 16 | LPD | PCR.2 | Nonsense | 462C → T | Arg55stop truncated SH2D1A protein |
| 17 | K004 | 6 | Dys-g | PCR.2 | Nonsense | 462C → T | Arg55stop truncated SH2D1A protein |
| 18 | K019 | 2 | FIM, LPD | PCR.2 | Nonsense | 462C → T | Arg55stop truncated SH2D1A protein |
| 19 | K030 | 2 | LPD | PCR.2 | Nonsense | 462C → T | Arg55stop truncated SH2D1A protein |
| 20 | K069 | 2 | FIM, LPD | PCR.2 | Nonsense | 462C → T | Arg55stop truncated SH2D1A protein |
| 21 | K081 | 3 | FIM, LPD | PCR.2 | Nonsense | 462C → T | Arg55stop |
| 22 | K006 | 8 | FIM, LPD | PCR.2 | Nonsense | 462C → T | Gln58stop truncated SH2D1A protein |
| 23 | K054 | 3 | LPD | PCR.2 | Nonsense | 490G → A | Trp64stop, truncated SH2D1A protein |
| 24 | K062 | 2 | FIM | PCR.3 | Missense | 604T → G | Val102Gly |
| 25 | K046 | 3 | FIM, AA | PCR.1 | Missense | 319A → G | Tyr7Cys |
| 26 | K068 | 6 | LPD, dys-g | PCR.1 | Missense | 383C → G | Ser28Arg |
| 27 | K037 | 5 | FIM, dys-g | PCR.1 | Missense | 425C → G | Cys42Trp |
| 28 | K053 | 14 | FIM, LPD, dys-g | PCR.2 | Missense | 457C → G | Thr53Ile |
| 29 | K082 | 3 | FIM, dys-g | PCR.3 | Missense | 559T → C | Phe87Ser |
| 30 | K002 | 17 | FIM, LPS, dys-g | PCR.3 | Missense | 595A → C | Gln99Pro |
| 31 | K084 | 6 | FIM | PCR.2 | Nonsense | 490G → A | Trp64stop truncated SH2D1A protein |
| 32 | K042 | 2 | FIM dys-g | ? | No mutation found in the SHD1A gene | N/A | |
| 33 | K080 | 2 | FIM, dys-g | PCR.2 | Intron | 7-bp deletion in the first intron | Truncated SH2D1A transcript |
| 34 | K089 | 3 | ML | PCR.1 | Missense | 391T → C | Leu31Pro |
| 35 | K038 | 2 | LPD, FIM | PCR.2 | Missense | 460A → G | Tyr54Cys |
| No. . | Family . | Affected males in family . | Clinical phenotypes of affected males . | Detection . | Type of mutation . | Nucleotides affected . | Predicted effect . |
|---|---|---|---|---|---|---|---|
| 1 | K043 | 2 | Dys, g | S, PCR | Deletion | 3.5 Mb DNA deleted at Xq25 | Neither SH2D1A nor tenascin-m proteins |
| 2 | K063 | 4 | FIM, LPD | S, PCR | Deletion | 3.5 Mb DNA deleted at Xq25 | Neither SH2D1A nor tenascin-m proteins |
| 3 | K073 | 4 | FIM, LPD | S, PCR | Deletion | 3.5 Mb DNA deleted at Xq25 | Neither SH2D1A nor tenascin-m proteins |
| 4 | Ado1 | 3 | FIM | S, PCR | Deletion | Exons 1-4 | No SH2D1A Protein |
| 5 | K051 | 3 | Dys-g | S, PCR | Deletion | Exons 1-4 | No SH2D1A Protein |
| 6 | K005 | 5 | FIM, LPD | S, PCR | Deletion | Exons 2-4 | Truncated SH2D1A protein |
| 7 | K083 | 2 | FIM | S, PCR | Deletion | Exons 2-4 | Truncated SH2D1A protein |
| 8 | K001 | 11 | FIM, LPD dys-g | S, PCR | Deletion | Exon 2 | Truncated SH2D1A protein |
| 9 | K032 | 3 | FIM, dys-g | PCR.2 | Deletion | Exon 2 | Truncated SH2D1A protein |
| 10 | K055 | 5 | FIM, AA LPD, dys-g | SPCR.2 | Deletion | 89 bp of intron 1 + exon 2 + intron 2 | Truncated SH2D1A protein |
| 11 | K060 | 4 | LPD, dys-g | PCR.2 | Deletion | 159 bp of exon 2 from nucleotide 448 + intron 2 | Truncated SH2D1A protein |
| 12 | K061 | 3 | FIM, LPD | PCR.2 | Deletion | 439A-485T | Frameshift, truncated SH2D1A protein |
| 13 | K008 | 7 | FIM, LPD | PCR.2 | Splice | ACAGgtACCGgt intron 1 3′ splice site | Exon 2 skipping |
| 14 | K048 | 2 | LPD | PCR.1 | Splice | ctGTGA > ctGCGA intron 1 5′ splice site | Intron retention truncated SH2D1A protein |
| 15 | K065 | 7 | FIM, dys-g | PCR.2 | Splice | agGTAT > agATAT intron 2 5′ splice site | Intron retention truncated SH2D1A protein |
| 16 | K003 | 16 | LPD | PCR.2 | Nonsense | 462C → T | Arg55stop truncated SH2D1A protein |
| 17 | K004 | 6 | Dys-g | PCR.2 | Nonsense | 462C → T | Arg55stop truncated SH2D1A protein |
| 18 | K019 | 2 | FIM, LPD | PCR.2 | Nonsense | 462C → T | Arg55stop truncated SH2D1A protein |
| 19 | K030 | 2 | LPD | PCR.2 | Nonsense | 462C → T | Arg55stop truncated SH2D1A protein |
| 20 | K069 | 2 | FIM, LPD | PCR.2 | Nonsense | 462C → T | Arg55stop truncated SH2D1A protein |
| 21 | K081 | 3 | FIM, LPD | PCR.2 | Nonsense | 462C → T | Arg55stop |
| 22 | K006 | 8 | FIM, LPD | PCR.2 | Nonsense | 462C → T | Gln58stop truncated SH2D1A protein |
| 23 | K054 | 3 | LPD | PCR.2 | Nonsense | 490G → A | Trp64stop, truncated SH2D1A protein |
| 24 | K062 | 2 | FIM | PCR.3 | Missense | 604T → G | Val102Gly |
| 25 | K046 | 3 | FIM, AA | PCR.1 | Missense | 319A → G | Tyr7Cys |
| 26 | K068 | 6 | LPD, dys-g | PCR.1 | Missense | 383C → G | Ser28Arg |
| 27 | K037 | 5 | FIM, dys-g | PCR.1 | Missense | 425C → G | Cys42Trp |
| 28 | K053 | 14 | FIM, LPD, dys-g | PCR.2 | Missense | 457C → G | Thr53Ile |
| 29 | K082 | 3 | FIM, dys-g | PCR.3 | Missense | 559T → C | Phe87Ser |
| 30 | K002 | 17 | FIM, LPS, dys-g | PCR.3 | Missense | 595A → C | Gln99Pro |
| 31 | K084 | 6 | FIM | PCR.2 | Nonsense | 490G → A | Trp64stop truncated SH2D1A protein |
| 32 | K042 | 2 | FIM dys-g | ? | No mutation found in the SHD1A gene | N/A | |
| 33 | K080 | 2 | FIM, dys-g | PCR.2 | Intron | 7-bp deletion in the first intron | Truncated SH2D1A transcript |
| 34 | K089 | 3 | ML | PCR.1 | Missense | 391T → C | Leu31Pro |
| 35 | K038 | 2 | LPD, FIM | PCR.2 | Missense | 460A → G | Tyr54Cys |
When analyzed under the same conditions, no mutations in the SH2D1A gene were found in any of the 25 males with sporadic XLP, suggesting that they do not have XLP. Similarly, no mutations of the SH2D1A gene were observed among the 9 patients with CAEBV (data not shown).
Deletion mutations
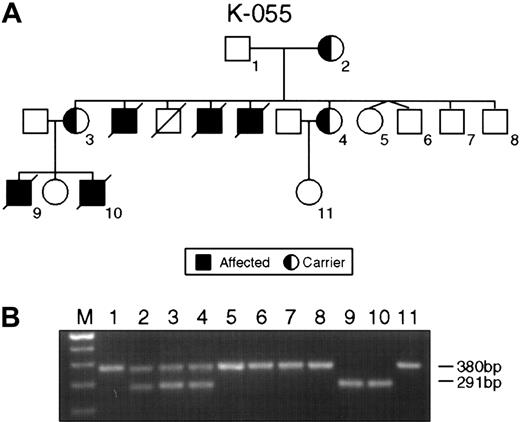
The single most common mutation was represented by deletions ranging from several megabase pairs of DNA in families K043, K063, and K073 to 47 nucleotides in family K061, leading to the complete or partial loss of the SH2D1A gene (Table 2). The small interstitial deletion of 47 nucleotides in the second exon of family K061 caused a frameshift mutation and premature termination of translation at the TGA stop codon 11 nucleotides downstream of the 3′ breakpoint of the deletion. In family K055, a deletion of 89 nucleotides was detected. The deletion involved the splice acceptor site in intron 1 and the splice donor site in intron 2 and led to skipping exon 2 (Figure1).
Intragenic deletions in the SH2D1A gene in families K055 and K061.
(A) Pedigree K055 shows XLP female carriers (2-4) affected males (9, 10) and unaffected persons (1, 5-8). (B) PCR product amplified with XLP2 primer pairs. Consistent with the pedigree, lanes 1, 5, 6, 7, and 8 indicate the product of 380 bp of normal SH2D1A. Lanes 2, 3, and 4 show the result of the PCR on DNA obtained from the obligate carriers; normal (380 bp) and mutant (291 bp) alleles are present. Lanes 9 and 10 display the result of PCR on DNA from affected males.
Intragenic deletions in the SH2D1A gene in families K055 and K061.
(A) Pedigree K055 shows XLP female carriers (2-4) affected males (9, 10) and unaffected persons (1, 5-8). (B) PCR product amplified with XLP2 primer pairs. Consistent with the pedigree, lanes 1, 5, 6, 7, and 8 indicate the product of 380 bp of normal SH2D1A. Lanes 2, 3, and 4 show the result of the PCR on DNA obtained from the obligate carriers; normal (380 bp) and mutant (291 bp) alleles are present. Lanes 9 and 10 display the result of PCR on DNA from affected males.
Splice-site mutations
Splice-site mutations accounted for 11% of this series. Two mutations were found at the 5′ donor splice site, and one was found at the 3′ acceptor splice site. An A-C transversion in the conserved AG of the intronic splice-acceptor region preceding exon 2 was observed in family K008 (Table 2). Family K065 carried a G-A transition in the conserved GT dinucleotide sequence (Table 2).
In family K048, for whom lymphoblasts were available for RNA extraction, we performed RT-PCR analysis to determine the consequences of the genomic mutation on the splicing process. The mutation changed the ctGTGA → ctGCGA splice donor site. To assess the level of RNA expression, we performed RT-PCR analysis of the total RNA isolated from lymphoblasts established from the member of the family K048. The patients who carried the T-C transition in the splice donor site in the first intron showed no detectable SH2D1A transcript using the XL37U/L primer pair (Figure 2).
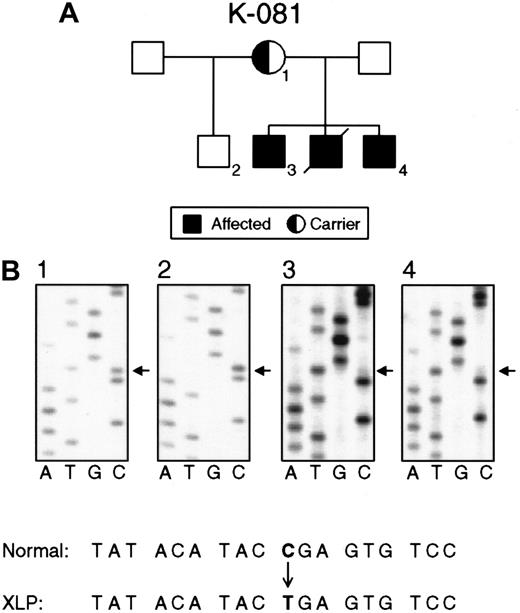
Nucleotide substitutions in the SH2D1A gene in families K081 and K089.
(A) Pedigree K081 shows XLP female carriers (1), normal persons (2), and affected males (3, 4). (B) Direct sequencing of PCR products demonstrating a nucleotide substitution (C462T) in exon 2 that causes a premature stop in the translation of SH2D1A. The status of the SH2D1A gene in an obligate carrier (1), unaffected person (2), and affected persons (3, 4).
Nucleotide substitutions in the SH2D1A gene in families K081 and K089.
(A) Pedigree K081 shows XLP female carriers (1), normal persons (2), and affected males (3, 4). (B) Direct sequencing of PCR products demonstrating a nucleotide substitution (C462T) in exon 2 that causes a premature stop in the translation of SH2D1A. The status of the SH2D1A gene in an obligate carrier (1), unaffected person (2), and affected persons (3, 4).
Nonsense mutations
Nine families with XLP showed 3 different nonsense mutations 462C → T (Arg55stop), 471C → T (Gln58stop), and 490G → A Trp64stop), resulting in the premature termination of polypeptide synthesis. Nonsense mutations were found solely in the second exon. The nonsense mutation 462C → T (Arg55stop) was detected in 6 unrelated families (Figure 2).
Missense mutations
Nine of the 28 mutations were missense mutations found in 9 XLP families. We screened 100 control X-chromosomes for the presence of missense substitutions identified in patients with XLP to exclude the possibility that such mutations represent rare polymorphisms. None of the missense mutations was found in the control chromosomes. All missense mutations resulted in the substitution of a conserved amino acid residue identical in the human and murine SH2D1A proteins.
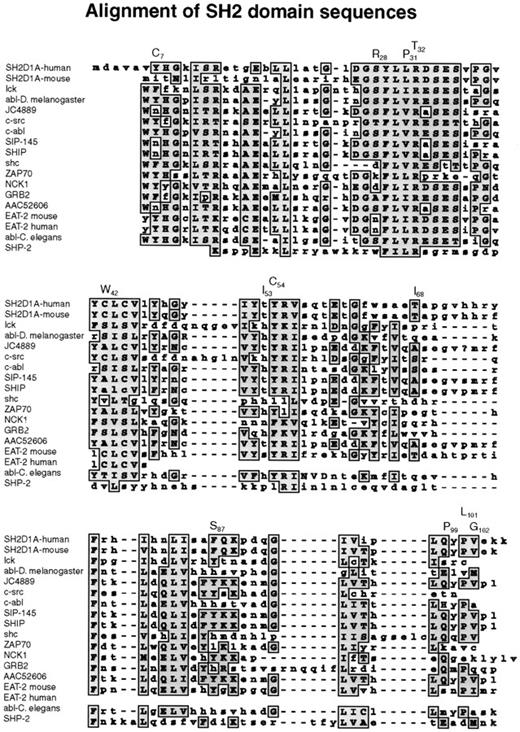
Missense mutations affected highly conserved amino acid residues in the SH2 domain, changing them to residues that are never found at this position in related SH2 domains (Figure3). The 319A → G transition replaced tyrosine 7 by a cysteine residue. Tyrosine 7 is in βA, though this region is not directly involved in phosphotyrosine binding; this residue is in a position where a mutation could affect other residues that interact with phosphotyrosine. The 383C → G missense mutation replaced a conserved serine at position 28 by arginine in the phosphotyrosine-binding site (Figures 3,4). The arginines are important because they are involved in the simultaneous recognition of phosphate groups and aromatic rings (Figure 5). Introducing another arginine into the phosphotyrosine binding site should have a significant impact on the structure and function of the protein. Mutations in the phosphotyrosine binding pocket of src,15 lck,16 vav,17 and p8518,19 have confirmed the importance of that group in the function of the SH2 domain. In family K037, the highly conserved hydrophilic cysteine in the βC chain was replaced by a hydrophobic tryptophan. In family K089, the extremely well-conserved leucine 31 in the FLVR motif was replaced by proline (Figures 3, 4). In the βD chain, threonine 53 in the conserved EF loop region of the hydrophobic binding pocket9 was replaced by isoleucine in family K053 (Figures 3, 4). A phenylalanine conserved between human and mouse SH2D1A protein in the α-helix B was replaced by serine in family K082. A valine residue of the βG structure was substituted by glycine in family K062. Glutamine 99 is very well conserved among SH2 domains; this region contributes to the formation of the hydrophobic binding pocket and was replaced by proline in family K002 (Figures 3, 4).
Alignment of SH2 domain sequences.
A subset of human SH2 domain–containing proteins was chosen using BLAST search from NCBI. Those that showed the highest homology to the human and murine SH2D1A proteins were selected, and their SH2 domains were aligned using the MacVector Clustal Program.
Alignment of SH2 domain sequences.
A subset of human SH2 domain–containing proteins was chosen using BLAST search from NCBI. Those that showed the highest homology to the human and murine SH2D1A proteins were selected, and their SH2 domains were aligned using the MacVector Clustal Program.
Schematic representation of the SH2D1A protein molecule showing the mutations found in the gene.
The boundaries of the secondary structural elements are indicated by solid boxes, and the notation for these elements is as described.9
Schematic representation of the SH2D1A protein molecule showing the mutations found in the gene.
The boundaries of the secondary structural elements are indicated by solid boxes, and the notation for these elements is as described.9
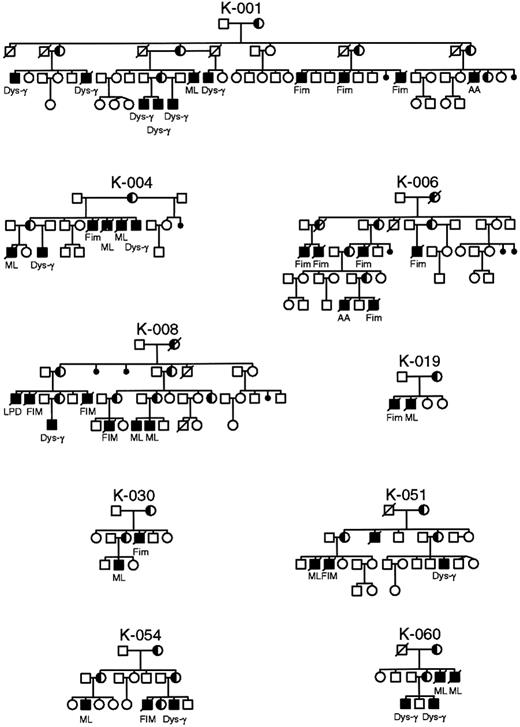
Pedigrees of XLP families in whom identical mutations in the SH2D1A gene manifest different phenotypes within the same family.
AA, aplastic anemia; Dys-γ, dysgammaglobulinemia; FIM, fulminant infectious mononucleosis; ML, malignant lymphoma.
Pedigrees of XLP families in whom identical mutations in the SH2D1A gene manifest different phenotypes within the same family.
AA, aplastic anemia; Dys-γ, dysgammaglobulinemia; FIM, fulminant infectious mononucleosis; ML, malignant lymphoma.
Specificity and sensitivity of SH2D1A mutation for XLP
In the 35 families with definitive diagnoses of XLP, we found 34 of 35 (97%) to have mutations in the SH2DA1 gene that should result in the abnormal expression of the protein. The one kindred in whom no mutation was found also did not undergo RFLP analysis. In 25 males with sporadic XLP and 9 with CAEBV, no mutation was found. Although we cannot exclude the possibility that mutations in the SH2D1A gene are involved in the pathogenesis of sporadic FIM and CAEBV, our failure to identify mutations in these patients makes it unlikely that a defect in the SH2D1A gene is the cause of sporadic XLP and CAEBV. Because there is no biochemical or laboratory test that is diagnostic for the defect in XLP, the gold standard must be stringent clinical criteria. Using the 35 families with definitive diagnoses as true positive and the other patients as true negatives for XLP, mutations in SH2D1A have a sensitivity of 97%, a specificity of 100%, a positive predictive value of 100%, and a negative predictive value of 97%.
Genotype-phenotype correlation
It was postulated that mutations that remove or truncate the SH2D1A protein are more often associated with a severe phenotype, whereas missense mutations occur preferentially in mildly affected patients.6 Not uncommonly, identical mutations manifest different phenotypes within the same family (Figure 5). To determine the functional significance of the mutations found in the SH2D1A gene, we undertook a careful evaluation of phenotype, age at first clinical manifestation, and survival (Table 3). No significant differences were observed in phenotypes or in severity of disease based on type (missense, nonsense, truncating) or localization of mutations (Table 3). The age of onset of clinical disease varied considerably, from younger than 1 year of age to 40 years, as did survival, but there was no correlation with the type of mutation. Indeed, such a variation was often seen between affected members of the same kindred, or even brothers. In addition, the large genomic deletion of 3.5 Mb in families K043 and K063 involving SH2D1A did not appear to have an impact on phenotype or severity of disease.
Correlation of mutation type, EBV status, clinical phenotype, and outcome in XLP
| Mutation type . | Kindreds . | Phenotypes . | EBV status at 1st manifestation . | Median age (y) of onset . | Median survival (y) . |
|---|---|---|---|---|---|
| Deletion (no SH2D1A) | K043, K051 | FIM (4), dys-(7) | EBV+ (5) | 5 | 7 |
| K063, K073, K088 | LPD (4), AA (0) | EBV− (5) | (0.5-26) | (1-41) | |
| Asx (1) | Und (6) | ||||
| Deletion (truncated SH2D1A) | K001, K005 | FIM (18), dys-(9) | EBV+ (16) | 4 | 9 |
| K032, K055, K060 | LPD (6), AA (2) | EBV− (5) | (0.5-34) | (1-41) | |
| K061, K083, K080 | Asx (0) | Und (14) | |||
| Splice defect | K008, K048 | FIM (11), dys-(1) | EBV+ (5) | 2 | 3 |
| K065 | LPD (3), AA (0) | EBV− (4) | (0.1-5) | (1-20) | |
| Asx (1) | Und (7) | ||||
| Nonsense | K003, K004, K006 | FIM (31), dys-(4) | EBV+ (24) | 3 | 6 |
| K019, K030, K054 | LPD (11), AA (1) | EBV− (8) | (0.5-23) | (0.5-38) | |
| K069, K081, K084 | Asx (1) | Und (17) | |||
| Missense | K002, K037, K038 | FIM (20), dys-(10) | EBV+ (14) | 4 | 7 |
| K046, K053, K062 | LPD (22), AA (2) | EBV− (7) | (1-33) | (1-45) | |
| K068, K082, K089 | Asx (2) | Und (35) |
| Mutation type . | Kindreds . | Phenotypes . | EBV status at 1st manifestation . | Median age (y) of onset . | Median survival (y) . |
|---|---|---|---|---|---|
| Deletion (no SH2D1A) | K043, K051 | FIM (4), dys-(7) | EBV+ (5) | 5 | 7 |
| K063, K073, K088 | LPD (4), AA (0) | EBV− (5) | (0.5-26) | (1-41) | |
| Asx (1) | Und (6) | ||||
| Deletion (truncated SH2D1A) | K001, K005 | FIM (18), dys-(9) | EBV+ (16) | 4 | 9 |
| K032, K055, K060 | LPD (6), AA (2) | EBV− (5) | (0.5-34) | (1-41) | |
| K061, K083, K080 | Asx (0) | Und (14) | |||
| Splice defect | K008, K048 | FIM (11), dys-(1) | EBV+ (5) | 2 | 3 |
| K065 | LPD (3), AA (0) | EBV− (4) | (0.1-5) | (1-20) | |
| Asx (1) | Und (7) | ||||
| Nonsense | K003, K004, K006 | FIM (31), dys-(4) | EBV+ (24) | 3 | 6 |
| K019, K030, K054 | LPD (11), AA (1) | EBV− (8) | (0.5-23) | (0.5-38) | |
| K069, K081, K084 | Asx (1) | Und (17) | |||
| Missense | K002, K037, K038 | FIM (20), dys-(10) | EBV+ (14) | 4 | 7 |
| K046, K053, K062 | LPD (22), AA (2) | EBV− (7) | (1-33) | (1-45) | |
| K068, K082, K089 | Asx (2) | Und (35) |
FIM, fulminant infectious mononucleosis; dys-γ, dysgammaglobulinemia; LPD, lyphoproliferative disease; AA, aplastic anemia; Asx, asymptomatic; Und, undeterminable. Median age of onset and survival are given in years, with ranges in parentheses below.
EBV-negative patients with XLP
Currently, there are 309 affected males in 89 kindreds in the XLP Registry. Five have been genetically diagnosed but, to date, have not had any clinical manifestations. Of the 304 in whom symptoms have developed, 38 (12.5%) had no evidence of EBV infection at time of their first clinical manifestation. The number may be higher than this because EBV status could not be determined at the time of first manifestation in 152 males. To determine the effect of EBV infection on clinical outcome, the 114 males with evidence of EBV infection at time of first clinical manifestation were compared with those without EBV infection (Table 4). FIM is the most common manifestation and is decidedly more frequent in the EBV+ group (P = .0001). However, when these patients are excluded, there was no statistical difference in the frequency of antecedent EBV infection when dys-γ and LPD were the presenting phenotypes (Table 4). AA appears to be more common in the EBV+ group, and this is not surprising because autoimmune cytopenias are a known complication of EBV infection, even in immunocompetent persons. An interesting finding is that 44 of 114 affected males had evidence of present or past EBV infection but that FIM never developed, suggesting factors other than EBV. In addition, the clinical outcomes of EBV+ and EBV−patients were evaluated. There was no difference in median time to first manifestation, but, again, the time of onset varied greatly for both groups and for all phenotypes, including FIM (ie, younger than 1 to 40 years of age). Survival for the EBV− group as a whole was significantly better (P = .0001), but this is entirely accounted for by the poor survival with FIM (Table5).
Effect of EBV infection on clinical phenotype in XLP
| Phenotype . | EBV+ (n = 114) . | EBV− (n = 38) . | P . |
|---|---|---|---|
| FIM | 70 | 0 | .0001 |
| Dysgammaglobulinemia | 19 | 15 | .30 |
| Lymphoproliferative disease | 20 | 21 | .24 |
| Aplastic anemia | 5 | 2 | .03 |
| Phenotype . | EBV+ (n = 114) . | EBV− (n = 38) . | P . |
|---|---|---|---|
| FIM | 70 | 0 | .0001 |
| Dysgammaglobulinemia | 19 | 15 | .30 |
| Lymphoproliferative disease | 20 | 21 | .24 |
| Aplastic anemia | 5 | 2 | .03 |
Effect of EBV infection on outcome in XLP
| . | Median age 1st manifestation (y) . | Median survival age (y) . |
|---|---|---|
| EBV+ | 4 (0.5-40) | 6 (0.5-40) |
| FIM | 3 (0.5-40) | 4 (0.5-40) |
| Dysgammaglobulinemia | 6 (0.5-32) | 14 (0.5-38) |
| Lymphoproliferative disease | 5 (2-19) | 6 (2-32) |
| Aplastic anemia | 6 (2-24) | 12 (2-27) |
| EBV− | 6 (0-34) | 6.55-150 (1-41) |
| Dysgammaglobulinemia | 4.5 (0.1-34) | 19 (4-41) |
| Lymphoproliferative disease | 8 (3-33) | 17 (4-39) |
| Aplastic anemia | 1, 11 | 7, 14 |
| . | Median age 1st manifestation (y) . | Median survival age (y) . |
|---|---|---|
| EBV+ | 4 (0.5-40) | 6 (0.5-40) |
| FIM | 3 (0.5-40) | 4 (0.5-40) |
| Dysgammaglobulinemia | 6 (0.5-32) | 14 (0.5-38) |
| Lymphoproliferative disease | 5 (2-19) | 6 (2-32) |
| Aplastic anemia | 6 (2-24) | 12 (2-27) |
| EBV− | 6 (0-34) | 6.55-150 (1-41) |
| Dysgammaglobulinemia | 4.5 (0.1-34) | 19 (4-41) |
| Lymphoproliferative disease | 8 (3-33) | 17 (4-39) |
| Aplastic anemia | 1, 11 | 7, 14 |
P < .05.
Discussion
This report provides a comprehensive analysis of the effects of SH2DA1 mutations and EBV infection on the clinical outcome of XLP. Previous studies demonstrated a relatively low frequency (approximately 62%) of mutations in a mixed population of familial (definitive) XLP and sporadic XLP (males with clinical phenotypes reminiscent of XLP but without a family history of the disease).6-8 It was postulated that XLP may be a polygenic disorder. In this study, we evaluated 35 families that met strict criteria for X-linked inheritance and symptoms consistent with XLP. We found 34 of 35 (97%) to have mutations that should result in the abnormal expression of the SH2D1A protein. In 25 males in whom sporadic XLP was diagnosed because FIM, dys-γ, or LPD developed after EBV infection, and in 9 patients with CAEBV, no mutations were found. The failure to identify mutations in these groups of patients makes it unlikely that defects in the SH2D1A gene are central to pathogenesis in patients with sporadic XLP or CAEBV. On the other hand, there is strong supporting evidence that the mutations identified in the SH2D1A gene are pathogenic in the XLP families studied. The data indicate that mutations segregate correctly in kindreds; mutations were not seen in 100 control chromosomes; large and small interstitial deletions and nonsense and splice-site mutations cause complete lack or truncation of the SH2D1A protein; and missense mutations should have dramatic effects on the conformation and function of the SH2 domain because they occur in highly conserved regions of the protein.20 Of note, the one kindred in whom no mutation was found also did not undergo RFLP analysis. Therefore, misdiagnosis cannot be ruled out because other inherited immunodeficiencies can manifest symptoms similar to XLP. These results strongly suggest that XLP is not a polygenic disease but is caused by a single gene, SH2D1A.
Before this report, 18 mutations in the SH2D1A gene had been reported in 29 unrelated patients with XLP disease or in a clinical phenotype reminiscent of XLP.4,6-8 In this study, 26 of 28 mutations have not been previously reported. The range of mutations identified spanned nearly all types of mechanisms by which genetic change can disrupt gene function. The most common changes consisted of deletions resulting in the complete loss of the SH2D1A gene and shorter intragenic deletions resulting in frameshifts and single-base changes leading to splice-site (3), nonsense (9), and missense (9) mutations. Of the nonsense mutations, more than two thirds were C-T transitions, which occurred in CpG dinucleotides. The mutations were distributed over the gene, though the number of mutations in the second exon appeared to be more frequent. One hot spot of mutations was detected at nucleotide position 462, resulting in an Arg55stop transition. The same mutation was also observed in unrelated families by Coffey et al6 and Nichols et al.8
Results from the current study, in addition to data previously reported, bring the total of different mutations to 56 in the SH2D1A gene.4,6-8 Most consist of point mutations, with only 10 occurring in CpG dinucleotides in the coding region of the gene; 10 amino acid substitutions may be of interest in functional studies. Mutations are evenly distributed throughout the gene, but there appears to be a clustering in the second exon. The mutations described herein and in 4 previous communications could shed light on the nature of the SH2D1A protein.20 Large genomic deletions result in the complete loss of the protein, whereas small intragenic deletions and splice-site mutations result in a nonfunctional protein and reveal only limited information about the protein.
In contrast, missense mutations that lead to disease can reveal significant information about the functionally important amino acid residues. Of the 56 mutations known, 11 are missense mutations that result in amino acid substitutions. As shown in Figure 5, all mutations leading to an amino acid change are located in the SH2 domain, but no mutations have been detected in the amino-terminal and carboxyl-terminal flanking sequences.
Coffey et al6 and Nichols et al8 have speculated about alternating mutations in SH2D1A and tenascin-m genes, accounting for the diverse XLP phenotypes. Yet, we could find no significant phenotypic differences among families with the loss of several megabase pairs of DNA (including SH2D1A and the adjacent tenascin-m gene) and families with missense mutations in the coding region of the SH2D1A gene (Tables 3, 4). The fact that identical mutations have been described in patients with greatly varying clinical phenotype suggests that additional factors are involved.
It has been a long-held doctrine that males affected with XLP will not manifest symptoms before EBV infection. Now we and others4provide data to the contrary. Although EBV infection is the most common trigger of symptoms and FIM is the most common phenotype, at least 12% of affected males have symptoms without evidence of a past or current EBV infection. We also demonstrate that when FIM is excluded, EBV infection is not a predictive factor for phenotype, age of onset of symptoms, or severity of disease (defined by survival). Therefore, there must be additional factors along with aberrant SH2D1A expression that contribute to the pathogenesis of XLP. Other elements that could be involved in shaping the clinical phenotypes include environmental factors, such as other viral triggers, or cellular factors, such as modifier genes. Modifier genes have been postulated for other diseases21 and are a particularly attractive hypothesis, given the structure and function of SH2D1A. Should such a gene (or genes) be found to be implicated, this could provide a mechanism to explain the diverse clinical phenotypes of XLP.
Acknowledgments
We thank Carolyn D. Hall for the excellent editorial assistance in the preparation of the manuscript and Joseph S. Edwards for the artwork.
Supported by the National Institutes of Health (grant 1 RO1 AI33532-OIA3), the Nebraska Department of Health LB506 (grant 93-07R), the William C. Havens Foundation, and the Lymphoproliferative Research Fund.
David T. Purtilo is deceased.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Janos Sumegi, Department of Pathology and Microbiology, University of Nebraska Medical Center, 985454 Nebraska Medical Center, Omaha, NE 68198-5454; e-mail: jsumegi@unmc.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal