Abstract
The platelet-derived neutrophil-activating peptide 2 (NAP-2, 70 amino acids) belongs to the ELR+ CXC subfamily of chemokines. Similar to other members of this group, such as IL-8, NAP-2 activates chemotaxis and degranulation in neutrophils (polymorphonuclear [PMN]) through chemokine receptors CXCR-1 and CXCR-2. However, platelets do not secrete NAP-2 as an active chemokine but as the C-terminal part of several precursors that lack PMN-stimulating capacity. As we have previously shown, PMN themselves may liberate NAP-2 from the precursor connective tissue-activating peptide III (CTAP-III, 85 amino acids) by proteolysis. Instead of inducing cell activation, continuous accumulation of the chemokine in the surroundings of the processing cells results in the down-regulation of specific surface-expressed NAP-2 binding sites and in the desensitization of chemokine-induced PMN degranulation. Thus, NAP-2 precursors may be regarded as indirect mediators of functional desensitization in neutrophils. In the current study we investigated the biologic impact of another major NAP-2 precursor, the platelet basic protein (PBP, 94 amino acids). We show that PBP is considerably more potent than CTAP-III to desensitize degranulation and chemotaxis in neutrophils. We present data suggesting that the high desensitizing capacity of PBP is based on its enhanced proteolytic cleavage into NAP-2 by neutrophil-expressed cathepsin G and that it involves efficient down-regulation of surface-expressed CXCR-2 while CXCR-1 is hardly affected. Correspondingly, we found PBP and, less potently, CTAP-III to inhibit CXCR-2– but not CXCR-1– dependent chemotaxis of neutrophils toward NAP-2. Altogether our findings demonstrate that the anti-inflammatory capacity of NAP-2 is governed by the species of its precursors.
Introduction
ELR+ CXC chemokines, such as IL-8 and neutrophil-activating peptide 2 (NAP-2), are well-known neutrophil (polymorphonuclear [PMN]) agonists that are considered important for PMN recruitment during many inflammatory processes. Bearing the functionally important amino acid motif consisting of glutamic acid (E), leucine (L), and arginine (R) directly in front of the first of their 4 conserved cysteines (C), ELR+ CXC chemokines specifically bind to receptors CXCR-1 and CXCR-2 on their target cells. This interaction can lead to subsequent activation of PMN functions such as chemotaxis and degranulation. To attenuate PMN-mediated deleterious effects during inflammation, considerable efforts have been focused on elucidating how ELR+ CXC chemokine activity is regulated.
In most cases, the generation of ELR+ CXC chemokines is controlled at the level of gene and subsequent protein expression after induction by inflammatory stimuli.1 The only ELR+ CXC chemokine regulated in a completely different manner is the 70 amino acids (aa) containing NAP-2 that originates essentially from platelets and megakaryocytes. The neutrophil-activating capacity of NAP-2 is strictly controlled by proteolytic processing of N-terminally extended NAP-2 precursor molecules (compare Figure 1), which themselves lack PMN-activating properties.2-4 These precursors, which together with NAP-2 are comprised under the term β-thromboglobulin antigen (β-TG Ag), are stored in the α-granules of platelets. On activation in the context of wounding, platelets degranulate leading to the immediate release of large quantities of β-TG Ag molecules, as apparent by serum concentrations that amount to 3 μmol/L.5 However, subsequent conversion of these precursors into mature NAP-2, which can activate PMN at nanomolar concentrations, is dictated by the presence of specific proteolytic activity required to remove the inhibitory N-terminus. As studies on the prevailing NAP-2 precursor in platelets—the connective tissue-activating peptide III (CTAP-III, 85 aa)—have shown, several serine proteases such as trypsin, chymotrypsin, and cathepsin G are capable of generating NAP-2 in vitro.2,6,7 In vivo, this proteolytic activity appears to be provided predominantly by a cell surface-bound, cathepsin G–like enzyme on PMN.8 In fact, in an earlier study, we could show that PMN generate their own stimulus NAP-2 from CTAP-III within minutes.3 However, these processing cells do not degranulate in response to the generating NAP-2 but become functionally desensitized toward subsequently administered higher dosages of NAP-2 and to other ELR+ CXC chemokines.3,9 Our previous findings that CTAP-III–mediated PMN desensitization strictly required proteolytic formation of NAP-23 and that this functional attenuation was associated with the reduction of specific NAP-2 binding sites on the cell surface3 9 indicated that desensitization might occur through down-regulation of chemokine receptors CXCR-2 and possibly CXCR-1 by the NAP-2 generated during processing. Through this mechanism, NAP-2 precursors could play an important role in regulating not only PMN responsiveness toward NAP-2 itself but toward ELR+ CXC chemokines in general.
Amino acid sequence of major β-TG Ag isoforms.
Numbers indicate residue positions relative to the N-terminus of NAP-2.
Amino acid sequence of major β-TG Ag isoforms.
Numbers indicate residue positions relative to the N-terminus of NAP-2.
In the current study we were interested in the role of the next most common NAP-2 precursor after CTAP-III, the platelet basic protein (PBP, 94 aa), which differs from CTAP-III by having an extended N-terminus (Figure 1). PBP reportedly not only contributes 11% to 49% of the total β-TG Ag found in platelets,10 but as the primeval gene product arising after the removal of a leader sequence, it is present at even higher proportions (more than 50%) in the progenitors of platelets, the megakaryocytes.11 We present data demonstrating that PBP acts as a desensitizing agent that is 6- to 7-fold more potent than CTAP-III and we discuss the mechanism underlying this enhanced effect. Moreover, we show that NAP-2 precursors affect not only the degranulation but also the chemotactic response of neutrophils. Altogether our data suggest that differential N-terminal processing of NAP-2 precursors could significantly contribute to the regulation of ELR+ CXC chemokine activity in inflammatory processes.
Materials and methods
Preparation of PBP, CTAP-III, and NAP-2
NAP-2 precursors were prepared from release-supernatants of thrombin-stimulated platelets, as described previously for CTAP-III.8 Briefly, total β-TG Ag was isolated from supernatants by immunoaffinity chromatography. The eluate obtained on acidification was split into fractions subsequently analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Western Blot analyses revealed PBP to preferentially elute in the later fractions. Therefore, early and late fractions were used for purification of CTAP-III and PBP, respectively. Fractions were adjusted to 10 mmol/L phosphate, pH 7.0, and separated on a cation-exchange column (MonoS HR5/5; Pharmacia/LKB, Freiburg, Germany) under a 0 to 0.5 mol/L NaCl-gradient. CTAP-III and PBP eluted at 0.23 to 0.29 and at 0.35 to 0.43 mol/L NaCl, respectively. Fractions were acidified with trifluoroacetic acid (TFA) and separated by reverse-phase high-performance liquid chromatography (HPLC) using a cyanopropyl column 4.6 × 250 mm, 5 μm (Baker Products, Phillipsburg, NJ). Under an n-propanol gradient (0%-35% in 0.1% TFA, 0.5 mL/min, 49 minutes), each precursor eluted at ≈20%. Protein concentrations were determined by the micro-bicinchoninic acid method using bovine serum albumin (BSA) as standard. On ≈500-fold overloaded silver-stained SDS-PAGE gels or immunostained blots, precursor preparations appeared homogeneous and devoid of proteins comigrating with NAP-2. Amino acid sequencing confirmed N-termini, whereas immunoreactivity to antiserum Rα-70/2 (see “Electrophoresis and immunoblotting”) confirmed C-termini in CTAP-III and PBP. Identity of precursors was further validated by mass spectrometry. NAP-2 was obtained on site-specific cleavage of CTAP-III with chymotrypsin, as described.12
Preparation of human neutrophils
PMN were isolated from the citrated blood of healthy donors by gradient centrifugation on Ficoll-Hypaque (Pharmacia/LKB) to a purity greater than 95%, as previously described.8
Degranulation assay
PMN (1 × 107/mL) suspended in D-PBS/BSA (Dulbecco-PBS/0.1% BSA) were preincubated for 10 minutes with 5 μg/mL cytochalasin B (Sigma, Deisenhofen, Germany) and supplemented with CaCl2 (1.8 mmol/L) and MgCl2 (1 mmol/L). Then 100 μL cells were added to 100 μL of samples preheated at 37°C. Samples acidified to stop cathepsin G activity were reneutralized with NaOH immediately before cells were added. After 30 minutes of incubation, cells were pelleted and supernatants were monitored for elastase enzymatic activity, as described elsewhere.9 Release rates were determined as percentage of total elastase activity in hexadecyl-trimethylammoniumbromide (0.1%)–treated PMN lysates. Backgrounds, as determined in the presence of buffer alone, were subtracted.
Desensitization of NAP-2–induced degranulation
To assess the effect of fixed concentrations of desensitizing agents on PMN degranulation in response to NAP-2, the degranulation assay was modified by pretreating PMN with the given agent 10 minutes before the stimulus was added. For analysis of various concentrations of desensitizing agent on the degranulation induced by 40 nmol/L NAP-2, PMN were suspended at 2 × 107 cells/mL and preincubated with cytochalasin-B. Then 50 μL of cell suspension was added to the same volume of 2-fold–concentrated desensitizing agent. After a 10-minute incubation, 100 μL of 80 nmol/L NAP-2 in D-PBS/BSA/CaCl2 (1.8 mmol/L/MgCl2(1 mmol/L)) was added. Subsequently, the degranulation assay was performed as described. Desensitization was expressed as percentage of release rates obtained with control PMN receiving no desensitizing agent.
Processing of NAP-2 precursors by neutrophils and cathepsin G and analyses of truncation products
NAP-2 precursors (1 μmol/L in 100 μL D-PBS/BSA) were mixed with either 100 μL PMN (1 × 107/mL) or cathepsin G (1 μg/mL) (Calbiochem, Frankfurt, Germany) in D-PBS/BSA and incubated for different time periods at 37°C (processing period). To terminate the enzyme reaction, samples were acidified with 0.1% TFA. Samples containing PMN were centrifuged and supernatants were analyzed for the presence of NAP-2 biologic activity. In the PMN degranulation assay, a standard of purified NAP-2 was run in parallel for the determination of NAP-2 activity equivalents. The initial velocity (V) of NAP-2 formation from precursors was determined and is given as an increase in NAP-2 activity equivalents per minute. The same samples were concomitantly monitored for the presence of NAP-2 by SDS-PAGE and Western blot analysis. Alternatively, supernatants were separated on a C2/C18 column (μRPC PC3.2/3; Amersham Pharmacia Biotech, Uppsala, Sweden) using a SMART chromatography unit (Amersham Pharmacia Biotech AB). Five hundred microliter volumes of samples acidified with TFA were loaded, and the column was developed with a linear acetonitrile-gradient (17.5%-37.5%). Peaks detected at 214 nm were collected and analyzed by mass spectroscopy (see “Mass spectrometry”).
Chemotaxis
Neutrophil chemotaxis was assayed in a modified 48-well Boyden chamber (Costar, Bodenheim, Germany), essentially as described.12
Flow cytometric analyses of CXCR-1 and CXCR-2 expression on neutrophils
Flow cytometric (FACS) analyses were performed as described.12 PMN (1 × 106/mL) suspended in D-PBS/BSA were incubated for 1 hour on ice with monoclonal antibodies RII 115 (2 μg/mL) and SE-2 (5 μg/mL) specific for CXCR-2 and CXCR-1, respectively. RII 115 was generated in our laboratory as described,12 and SE-2 was kindly provided by Dr O. Götze13 (University of Göttingen, Göttingen, Germany). Cell-bound antibodies were detected by fluorescein-conjugated goat α-mouse immunoglobulin (H+L) antibody (Dianova, Hamburg, Germany) (15 μg/mL), and analyses were performed on a flow cytometer (FACStar PLUS; Becton Dickinson, Heidelberg, Germany).
Electrophoresis and immunoblotting
SDS-PAGE was performed according to Schägger and von Jagow.14 Samples were reduced 10 minutes at 95°C, treated 30 minutes with iodoacetamide (2%) and loaded onto a 13% polyacrylamide gel with a 10% spacer gel and a 4% stacking gel on top. Rainbow Protein Markers (Amersham Buchler, Braunschweig, Germany) served as molecular mass markers. Western blotting was carried out as described previously.15 Rabbit antisera used for Western blot analyses were Rα-β-TG raised against a purified preparation of native β-TG Ag, reacting to all known variants of β-TG Ag and containing an antibody subpopulation interacting with residues 1 to 24 in PBP but not with residues 1 to 15 in CTAP-III (data not shown), and Rα-70/2 raised against the C-terminal octapeptide of NAP-2 (LAGDESAD). Rα-70/2 had characteristics identical to previously described antiserum Rα-70, requiring the presence of the C-terminal residue D70 for binding to β-TG Ag.15
Mass spectrometry
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) was performed with a Bruker-ReflexIII (Bruker-Franzen Analytik, Bremen, Germany) in reflector time-of-flight configuration at an acceleration voltage of 20 kV and with delayed ion extraction. The compounds dissolved in 0.1% TFA at a concentration of less than 0.3 μg/μL, were diluted 1:2 with a freshly prepared matrix solution consisting of saturated 3,5-dimethoxy-4-hydroxy cinnamic acid (Sinapic; Aldrich, Steinheim, Germany) in a 2:1 mixture of 0.1% trifluoroacetic acid/acetonitrile. Aliquots of 0.5 μL were deposited on a metallic sample holder and analyzed immediately after drying in a stream of air. Mass scale calibration was performed externally.
N-terminal amino acid sequencing
N-terminal sequence analyses of β-TG Ag isoforms dissolved in 0.1% TFA were performed by Dr A. Petersen (Forschungszentrum Borstel, Borstel, Germany) on a gas-phase sequencer (model 473A; Applied Biosystems, Foster City, CA). When proteins were analyzed directly from the blot, the area containing the unstained protein was excised, washed with bidistilled water for 1 hour, dried, and sequenced.
Statistical analysis
Data were statistically analyzed using Microcal Origin 4.10 software (Microcal Software, Northampton, MA).
Results
PBP in comparison with CTAP-III shows enhanced potency to desensitize chemokine-induced neutrophil degranulation
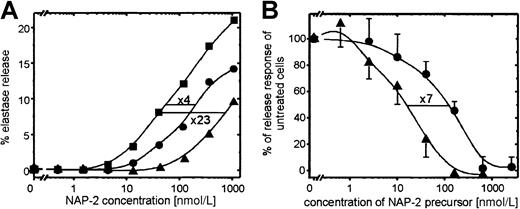
We have previously shown that CTAP-III, the quantitatively prevailing NAP-2 precursor released by platelets, functions as a desensitizing agent for chemokine-induced PMN degranulation. In the current study we were interested in whether PBP—another major platelet-secreted NAP-2 precursor bearing an N-terminal extension by 9 residues longer than that in CTAP-III—would function in a corresponding way. For comparison, we used highly purified PBP and CTAP-III isolated from platelet-release supernatants. As shown in Figure 2, PBP turned out to be considerably more effective than CTAP-III in desensitizing PMN degranulation. On preincubation of PMN with a fixed dosage (50 nmol) of either PBP or CTAP-III for 10 minutes, degranulation in response to increasing concentrations of NAP-2 was significantly reduced (Figure2A). Compared with unexposed control cells, pretreatment with PBP turned the cells approximately 23-fold less sensitive to NAP-2 stimulation, whereas CTAP-III pretreatment reduced their sensitivity by only 4-fold. To determine the relative potencies of the 2 precursors, cells were preincubated with increasing concentrations of PBP or CTAP-III for 10 minutes, and PMN degranulation in response to a fixed dosage of NAP-2 (40 nmol/L) was assessed (Figure 2B). As seen by the respective dose-response curves running parallel to each other, half-maximal down-regulation of the degranulation response required approximately 120 nmol/L CTAP-III and 16 nmol/L PBP, respectively, indicating that PBP is approximately 7-fold more potent a desensitizing agent than CTAP-III. Similarly, the threshold concentration for PBP to induce a measurable degree of desensitization was approximately 6- to 7-fold lower (3 nmol/L) than that for CTAP-III (20 nmol/L).
Desensitizing effect of PBP and CTAP-III on NAP-2–induced PMN degranulation.
(A) Degranulation response of PMN toward increasing concentrations of NAP-2 after pretreatment of cells for 10 minutes with or without 50 nmol/L PBP or CTAP-III. Data from 1 of 3 representative independent experiments with PMN from different donors are shown. (B) Degranulation response of PMN to 40 nmol/L NAP-2 after 10-minute pretreatment with or without increasing concentrations of PBP or CTAP-III. Data are expressed as the percentage of the elastase release response from cells left unexposed to the desensitizing agents, and they represent means ± SD of data obtained in 3 independent experiments. CTAP-III, ●; PBP, ▴; no desensitizing agent, ▪.
Desensitizing effect of PBP and CTAP-III on NAP-2–induced PMN degranulation.
(A) Degranulation response of PMN toward increasing concentrations of NAP-2 after pretreatment of cells for 10 minutes with or without 50 nmol/L PBP or CTAP-III. Data from 1 of 3 representative independent experiments with PMN from different donors are shown. (B) Degranulation response of PMN to 40 nmol/L NAP-2 after 10-minute pretreatment with or without increasing concentrations of PBP or CTAP-III. Data are expressed as the percentage of the elastase release response from cells left unexposed to the desensitizing agents, and they represent means ± SD of data obtained in 3 independent experiments. CTAP-III, ●; PBP, ▴; no desensitizing agent, ▪.
Neutrophils generate NAP-2 more rapidly from PBP than from CTAP-III
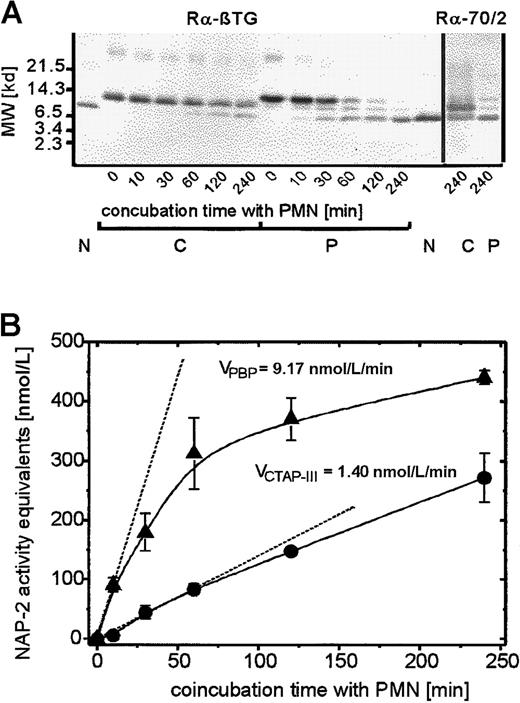
To address the mechanism underlying the enhanced desensitizing effect of PBP on PMN degranulation, we first examined whether this could be caused by the direct interaction of PBP with specific cellular binding sites. In competition binding assays performed at 4°C with up to 10 nmol/L radioiodinated PBP in the absence or presence of a 100-fold molar excess of cold PBP, no specific binding to neutrophils could be detected (data not shown). These results corresponded to those previously obtained with radiolabeled CTAP-III3 and excluded the possibility that PBP-induced desensitization was a directly receptor-mediated event. More likely, the proteolytic generation of an NAP-2–like molecule was involved, similar to the generation of NAP-2 from CTAP-III. To examine this we looked for the presence of NAP-2–like proteolytic truncation products in the supernatants of PMN that had been incubated with 500 nmol/L PBP or, for comparison, with 500 nmol/L CTAP-III. Initially, a relatively long incubation period (240 minutes) was chosen to obtain sufficient amounts of the potential truncation products for biochemical analyses. Separation by SDS-PAGE and subsequent visualization of the blotted proteins by immunostaining with an antiserum to β-TG Ag (Figure3A) revealed that PBP (lane P/0) was almost completely converted to a molecule similar in size to purified NAP-2 (compare lanes P/240 to lane N). In fact, sequencing of this truncation product revealed an N-terminus (A-E-L-R) identical to that of NAP-2. We suspected that this molecule could represent a C-terminally truncated NAP-2 variant. The formation of such a variant from PBP could explain the precursor's enhanced desensitizing effect; we previously found that C-terminally truncated NAP-2 binds to receptors with higher affinity and thereby desensitizes PMN degranulation more potently than full-length NAP-2.16However, as seen on probing with a β-TG Ag antiserum (Rα-70/2) that required the outermost C-terminal amino acid (D70) for binding to NAP-2, the PBP-derived molecule was fairly immunoreactive (Figure 3A, lane P/240, right). This observation indicated that the protein indeed represented full-length NAP-2 and thus excluded that C-terminal truncation was responsible for the enhanced desensitizing capacity of PBP.
Time kinetics of proteolytic processing of PBP and CTAP-III on coincubation with PMN.
Neutrophils were coincubated with 500 nmol/L of either NAP-2 precursor for the time period indicated, and supernatants (SN) were further analyzed. (A) Western blot analysis of SN. 10 μL of each SN was separated by SDS-PAGE, blotted, and immunostained with either antiserum Rα-βTG (binding to all known β-TG Ag isoforms) or Rα-70/2 (detecting the C-terminal residue in β-TG-Ag). 50 ng NAP-2 was run in parallel for comparison. Data from 1 of 3 representative experiments are shown. N indicates NAP-2; P, PBP; C, CTAP-III. (B) Determination of NAP-2 activity equivalents in SN. The potential NAP-2 concentrations in the SN were determined by comparison with purified NAP-2 in the degranulation assay as outlined in “Materials and methods.” Shown are means ± SD of data obtained in 3 independent experiments. The initial velocity, V, was defined as the increase in NAP-2 concentration per minute on the basis of the first time point at which significantly elevated NAP-2 levels could be detected. This time point differed between the precursors (C, 30 minutes; P, 10 minutes). ●, CTAP-III (500 nmol/L); ▴, PBP, (500 nmol/L).
Time kinetics of proteolytic processing of PBP and CTAP-III on coincubation with PMN.
Neutrophils were coincubated with 500 nmol/L of either NAP-2 precursor for the time period indicated, and supernatants (SN) were further analyzed. (A) Western blot analysis of SN. 10 μL of each SN was separated by SDS-PAGE, blotted, and immunostained with either antiserum Rα-βTG (binding to all known β-TG Ag isoforms) or Rα-70/2 (detecting the C-terminal residue in β-TG-Ag). 50 ng NAP-2 was run in parallel for comparison. Data from 1 of 3 representative experiments are shown. N indicates NAP-2; P, PBP; C, CTAP-III. (B) Determination of NAP-2 activity equivalents in SN. The potential NAP-2 concentrations in the SN were determined by comparison with purified NAP-2 in the degranulation assay as outlined in “Materials and methods.” Shown are means ± SD of data obtained in 3 independent experiments. The initial velocity, V, was defined as the increase in NAP-2 concentration per minute on the basis of the first time point at which significantly elevated NAP-2 levels could be detected. This time point differed between the precursors (C, 30 minutes; P, 10 minutes). ●, CTAP-III (500 nmol/L); ▴, PBP, (500 nmol/L).
More detailed analysis of the time course of NAP-2 formation by neutrophils exposed to either PBP or CTAP-III pointed to a different mechanism that might be effective and that appeared to be based on a higher susceptibility of PBP to cleavage by PMN. Although the 240-minute coincubation of PMN with PBP led to practically complete conversion of the precursor to NAP-2, this was not the case with CTAP-III, which became only partially processed, as indicated by a more than 50% proportion of the precursor remaining detectable on immunoblotting (Figure 3A; compare left panels of lanes C/240 and P/240 and right panels of lanes C/240 and P/240). Further comparison of supernatants obtained after various shorter incubation periods indeed showed that with PBP-exposed cells, a detectable NAP-2 band appeared approximately 4- to 6-fold earlier than with cells exposed to CTAP-III (Figure 3A; compare lanes P/10 and C/60). This observation correlated well with the data shown in Figure 3B, in which, using purified NAP-2 as a standard, the same supernatants were analyzed for the presence of NAP-2–like biologic activity in the PMN degranulation assay. Consistently, NAP-2–like biologic activity was generated by neutrophils more quickly from PBP than from CTAP-III, and this difference appeared most drastic at early time points. Thus, the initial velocity of NAP-2 generation from 500 nmol/L PBP (≈9 nmol/L NAP-2/min) was determined to be approximately 6 times higher than that from the same concentration of CTAP-III (≈1.4 nmol/L NAP-2/min). Interestingly, this difference in the velocities of NAP-2 generation from the 2 precursors correlates well with the approximately 7-fold different potencies for the desensitization of PMN degranulation. Hence, the more rapid accumulation of NAP-2 with PBP-exposed cells most likely causes the more potent PMN desensitization by PBP.
Conversion of PBP to NAP-2 is mediated by a single, cathepsin G–like enzyme
To obtain further insight into the mechanism(s) responsible for enhanced NAP-2 generation from PBP, we next focused on the enzyme(s) involved in precursor processing by PMN. Concerning CTAP-III, we showed15 that its conversion to NAP-2 is catalyzed by a single PMN-associated cathepsin G–like enzyme and could be mimicked by purified cathepsin G. Under either condition, cleavage occurred in a single step behind the only tyrosine within CTAP-III, giving rise to biologically active NAP-2 (compare Figure 1). Because PBP bears the same cleavage site, cathepsin G was likely to be involved in the processing of this NAP-2 precursor as well. However, as evident in the Western blot shown in Figure 3, panel A, PMN-mediated processing of PBP, in contrast to that of CTAP-III, not only yielded the one truncation product we identified as NAP-2, it also gave rise to another molecule that became visible as an immunoreactive band (intermediate size, approximately 9 kd) and that comigrated with CTAP-III. Different from NAP-2, the latter molecule exhibited only a transient appearance; it was detectable after 10 minutes of processing and disappeared after 240 minutes. Furthermore, with longer developing times of the blot (leading to overstaining of the major bands), a faint band became detectable that migrated at approximately 2 kd (data not shown). These observations led us to assume that PBP was targeted by several enzymes that cleaved the precursor at different sites and possibly cooperated in its conversion to NAP-2. However, control experiments performed by coincubating PBP with purified cathepsin G (Figure4) surprisingly led to results essentially identical to those obtained with neutrophils. Cathepsin G alone was capable of generating the same 3 fragments from PBP (Figure4A), ie, a protein comigrating with NAP-2 and accumulating over time, a transiently appearing protein of intermediate size similar to CTAP-III, and a faintly visible 2-kd fragment. Moreover, the enzyme similarly catalyzed the generation of NAP-2–like biologic activity approximately 6 times more rapidly from PBP than from CTAP-III, as seen by a comparison of the initial velocities for NAP-2 formation, which were approximately 41 nmol/L per minute and 7 nmol/L per minute, respectively (Figure 4B). Indeed, N-terminal sequencing and detection by the C-terminus–specific antiserum Rα-70/2 identified this NAP-2–like molecule as NAP-2. These data, demonstrating that cathepsin G alone catalyzes the enhanced formation of NAP-2 from PBP in a way comparable to that seen with neutrophils, strongly suggest that cell-mediated PBP conversion depends on a single enzyme, most likely identical to neutrophil cathepsin G.
Time kinetics of proteolytic processing of PBP and CTAP-III on coincubation with purified cathepsin G.
Cathepsin G (1 μg/mL) was coincubated with 500 nmol/L of either NAP-2 precursor for the time periods indicated, and supernatants (SNs) were taken and analyzed exactly as reported in the legend to Figure 3A. (A) Western blot analysis of SN: 10 μL of each SN was separated by SDS-PAGE, blotted, and subsequently immunostained with antiserum Rα-βTG. 50 ng NAP-2 was run in parallel for comparison. Data from 1 of 3 representative experiments are shown. N, NAP-2; P, PBP; C, CTAP-III. (B) Determination of NAP-2 activity equivalents in SN (compare with legend to Figure 3B). Shown are means ± SD of data obtained in 3 independent experiments. ●, CTAP-III (500 nmol/L); ▴, PBP, (500 nmol/L).
Time kinetics of proteolytic processing of PBP and CTAP-III on coincubation with purified cathepsin G.
Cathepsin G (1 μg/mL) was coincubated with 500 nmol/L of either NAP-2 precursor for the time periods indicated, and supernatants (SNs) were taken and analyzed exactly as reported in the legend to Figure 3A. (A) Western blot analysis of SN: 10 μL of each SN was separated by SDS-PAGE, blotted, and subsequently immunostained with antiserum Rα-βTG. 50 ng NAP-2 was run in parallel for comparison. Data from 1 of 3 representative experiments are shown. N, NAP-2; P, PBP; C, CTAP-III. (B) Determination of NAP-2 activity equivalents in SN (compare with legend to Figure 3B). Shown are means ± SD of data obtained in 3 independent experiments. ●, CTAP-III (500 nmol/L); ▴, PBP, (500 nmol/L).
Molecular identity and functional impact of the PBP fragments generated by neutrophils
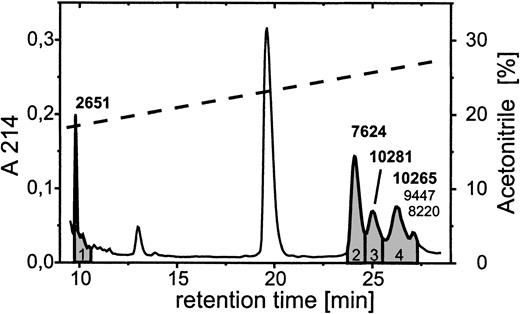
Further analyses revealed the identity and the functional involvement of the other fragments generated from PBP by neutrophils. N-terminal sequence analyses of the ≈2 kd band directly from the blot yielded a sequence reading S-S-T-K, indicating that the fragment represented an N-terminal stretch of PBP. Because of an insufficient amount of material, no result was obtained for the 9-kd truncation product. We therefore chose a different approach by using reverse-phase HPLC on a C2/C18 column for the separation of supernatants obtained after 10 minutes of coincubation of neutrophils with 2 μmol/L PBP (Figure 5). Mass spectrometric analyses of the single-protein peaks yielded molecular masses for the major components contained in peaks 3 and 4 of 10 281 and 10 265, respectively, corresponding to the calculated masses of PBP in its oxidized (10 278) and nonoxidized (10 262) forms. Additionally, peak 4 contained low amounts of 2 components of intermediate size. These had molecular masses of 9447 and 8220, which corresponded most closely to PBP cleavage products of 86 and 75 residues in length, representing CTAP-III extended by one N-terminal residue (R-CTAP-III) and NAP-2 extended by 5 N-terminal residues (DSDLY-NAP-2) (calculated masses, 9446 and 8219, respectively). Peaks 1 and 2 contained only one component each, with masses of 2651 and 7624, respectively. Although the former mass was closest to that of the N-terminal stretch of PBP, encompassing residues −24 through −1 (calculated mass, 2655), the latter was identical to that calculated for NAP-2 (7624). These results indicate that PMN cleave PBP at 3 different sites, first directly behind the Tyr at position −1, giving rise to NAP-2 and the 24-aa residue N-terminal peptide, second at a dibasic cleavage site between Lys-Arg at positions −16 and −17, leading to the formation of the fragment one residue longer than CTAP-III, and third behind the Leu at position −6, giving rise to the molecule 5 residues longer than NAP-2. Most likely, the amounts of the latter truncation product were too low to become detected by Western blotting. Nevertheless, these data demonstrate that formation of the intermediate-size fragments is not a prerequisite for the generation of NAP-2. Although it appears most unlikely that R-CTAP-III and DSDLY-NAP-2 contribute to neutrophil functional desensitization before becoming converted into NAP-2, a potential desensitizing function for the N-terminal fragment PBP (−24 to −1) could not be excluded. We produced a corresponding synthetic peptide and evaluated its impact on NAP-2–induced degranulation. However, in assays using up to 2 μmol/L of the peptide for preincubation of PMN, no change of NAP-2–promoted degranulation was observed (data not shown). Thus our data demonstrate that NAP-2 is the only PBP-derived truncation product responsible for neutrophil desensitization.
Separation of PBP truncation products generated on coincubation with PMN.
Neutrophils (1 × 107/mL) were coincubated with 2 μmol/L PBP for 10 minutes at 37° C in D-PBS and after acidification with TFA 500 μL of the cell-free supernatant (SN) was applied to an analytical C2/C18 column equilibrated in 0.1% TFA. The column was developed with a linear gradient of 17.5% to 37.5% acetonitrile in 0.1% TFA (dashed line). The profile of eluting proteins as detected at λ = 214 nm (solid line) was compared to that of SN from PMN not having received PBP, and only those peaks (1-4, shaded) that newly emerged in the SN of PBP-supplemented cells, compared with cells not having received PBP, were collected and further analyzed by mass spectroscopy. The molecular masses given above peaks 1 to 3 are representative for the respective major component detected, whereas for peak 4 the masses of the major (bold) and 2 minor components are given.
Separation of PBP truncation products generated on coincubation with PMN.
Neutrophils (1 × 107/mL) were coincubated with 2 μmol/L PBP for 10 minutes at 37° C in D-PBS and after acidification with TFA 500 μL of the cell-free supernatant (SN) was applied to an analytical C2/C18 column equilibrated in 0.1% TFA. The column was developed with a linear gradient of 17.5% to 37.5% acetonitrile in 0.1% TFA (dashed line). The profile of eluting proteins as detected at λ = 214 nm (solid line) was compared to that of SN from PMN not having received PBP, and only those peaks (1-4, shaded) that newly emerged in the SN of PBP-supplemented cells, compared with cells not having received PBP, were collected and further analyzed by mass spectroscopy. The molecular masses given above peaks 1 to 3 are representative for the respective major component detected, whereas for peak 4 the masses of the major (bold) and 2 minor components are given.
Preincubation of PMN with PBP leads to enhanced down-regulation of CXCR-1 and CXCR-2 compared to preincubation with CTAP-III
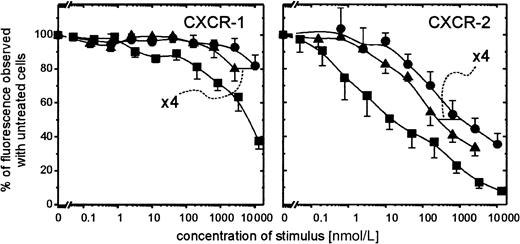
The evident correlation existing between the extremely rapid conversion of PBP to NAP-2 by neutrophils and its similarly enhanced capacity to desensitize the degranulation response of the processing cells led us to analyze how this precursor affected the surface expression of NAP-2 binding sites on PMN. As we have shown,3 preincubation of PMN with CTAP-III leads to compromised binding of radiolabeled NAP-2 to PMN, and, in fact, the same was the case for PBP. Hence, preincubation of PMN for 5 minutes with 400 nmol/L PBP reduced binding of 0.5 nmol/L radioiodinated NAP-2 to background level (data not shown). However, we wanted to know to what degree the expression of the different receptors known for NAP-2, namely CXCR-1 and CXCR-2, was affected. Therefore, by using monoclonal antibodies SE-2 and RII115 specific for CXCR-1 and CXCR-2, respectively, we assessed by FACS analyses the presence of these receptors on the surface of PMN on 10 minutes pretreatment with different concentrations of PBP or CTAP-III. As shown in Figure6, preincubation of PMN with PBP reduced the expression of both receptors approximately 4-fold more potently than did pretreatment with CTAP-III. These data correspond relatively well with the ≈6- to 7-fold enhanced desensitizing capacity of PBP and its similarly more rapid conversion to NAP-2, suggesting that enhanced receptor down-regulation by PBP could indeed be caused by the higher quantity of NAP-2 accumulating around the processing cells. Further support for NAP-2 as the active mediator may be derived from the observation that approximately 250-fold lower precursor concentrations are required to down-regulate the NAP-2 high-affinity receptor CXCR-2 (Kd ≈1 nmol/L9) compared with the low-affinity receptor CXCR-1 (Kd ≈200 nmol/L9) (minimal effective doses of PBP, ≈10 nmol/L and 2.5 μmol/L, respectively). In fact, a comparable (≈400-fold) difference in potency to down-regulate CXCR-2 versus CXCR-1 was observed when NAP-2 itself was tested, which down-regulated 50% of CXCR-2 at ≈15 nmol/L, whereas it took ≈6 μmol/L to see the same effect with CXCR-1 (compare Figure6). Thus, our data suggest that PBP (and CTAP-III) mediates its desensitizing effect predominantly through the preferential interaction of NAP-2 with CXCR-2.
Surface expression of CXCR-1 and CXCR-2 on PMN at 10-minute preincubation with PBP, CTAP-III, or NAP-2.
PMN (1 × 106/mL) were pretreated for 10 minutes at 37°C with the indicated concentrations of the respective stimulus. Surface-expressed receptors were subsequently immunodetected using the monoclonal antibodies versus CXCR-1, termed SE-2, or versus CXCR-2, termed RII-115, as primary and subsequently GαMig-FITC as secondary antibody as outlined in “Materials and methods.” Under every treatment applied the population stained homogeneously. Background stainings of isotype-matched controls were subtracted. The impact of stimulus preincubation on receptor detection was expressed as a percentage of the median fluorescence observed with PMN stained upon preincubation without the stimulus. Shown are means ± SD of data obtained from 3 independent experiments. ▪, NAP-2; ●, CTAP-III; ▴, PBP.
Surface expression of CXCR-1 and CXCR-2 on PMN at 10-minute preincubation with PBP, CTAP-III, or NAP-2.
PMN (1 × 106/mL) were pretreated for 10 minutes at 37°C with the indicated concentrations of the respective stimulus. Surface-expressed receptors were subsequently immunodetected using the monoclonal antibodies versus CXCR-1, termed SE-2, or versus CXCR-2, termed RII-115, as primary and subsequently GαMig-FITC as secondary antibody as outlined in “Materials and methods.” Under every treatment applied the population stained homogeneously. Background stainings of isotype-matched controls were subtracted. The impact of stimulus preincubation on receptor detection was expressed as a percentage of the median fluorescence observed with PMN stained upon preincubation without the stimulus. Shown are means ± SD of data obtained from 3 independent experiments. ▪, NAP-2; ●, CTAP-III; ▴, PBP.
PBP inhibits CXCR-2–mediated neutrophil chemotaxis toward NAP-2 more potently than does CTAP-III
The observation that PBP potently reduced the expression of CXCR-2 led us to look at its effect on PMN chemotaxis. We recently reported12 that the chemotactic response of PMN toward NAP-2 consists of 2 optima (compare Figure7B), one at very low NAP-2 concentrations (≈5 nmol/L) because of the interaction with CXCR-2 and the other at ≈400-fold higher concentrations because of the interaction with CXCR-1. Therefore, CXCR-2 especially appeared to be important for PMN attraction in response to the initially low amounts of NAP-2 arising from platelet-released precursors immediately after platelet degranulation. However, during this early phase, the concentration of precursors would still be high. Hence, we wanted to know whether the presence of PBP or CTAP-III would have an impact on PMN chemotaxis induced by NAP-2 concentrations addressing CXCR-2. Thus, we analyzed the chemotactic migration of PMN toward 5 nmol/L NAP-2 mixed with increasing dosages of either precursor. In contrast to the conditions applied in the degranulation assay, PMN were not pretreated with the respective precursor but were exposed to NAP-2 and precursors simultaneously. Though without such a prior period of desensitization PBP or CTAP-III never affected the degranulation response toward NAP-2 (data not shown), this was different for the chemotactic response (Figure 7A). Most interestingly, both precursors inhibited NAP-2–induced chemotaxis, with CTAP-III reducing the chemotactic response by 50% at ≈1100 nmol/L and PBP exhibiting a corresponding effect at ≈160 nmol/L, the latter being approximately 7 times more potent. Moreover, analyses of the dose-dependent chemotactic response toward NAP-2 in the presence of 1 μmol/L PBP showed that though the first (CXCR-2–associated) optimum was virtually abrogated, the second (CXCR-1–associated) optimum remained unchanged (Figure 7B). These data demonstrate that PBP does not alter the responsiveness of the cells in general but selectively down-regulates CXCR-2–dependent PMN chemotaxis.
Chemotactic migration of PMN toward NAP-2 in the presence of PBP or CTAP-III.
NAP-2 precursors were mixed with NAP-2 at the concentrations indicated and were added to the lower compartment of the chemotaxis chamber. Subsequently, PMN (2 × 105/mL) was added to the upper chamber, incubated for 1 hour at 37°C, and numbers of migrated cells were determined. (A) Impact of increasing concentrations of PBP (▴) or CTAP-III (●) on PMN chemotaxis induced by 5 nmol/L NAP-2. Results are expressed as a percentage of the number of cells that migrated to 5 nmol/L NAP-2 alone. (B) Impact of 1 μmol/L PBP (▴) on the characteristic chemotactic response of PMN toward NAP-2. ▪, buffer. (A, B) Means ± SD of data obtained in 3 independent experiments are shown.
Chemotactic migration of PMN toward NAP-2 in the presence of PBP or CTAP-III.
NAP-2 precursors were mixed with NAP-2 at the concentrations indicated and were added to the lower compartment of the chemotaxis chamber. Subsequently, PMN (2 × 105/mL) was added to the upper chamber, incubated for 1 hour at 37°C, and numbers of migrated cells were determined. (A) Impact of increasing concentrations of PBP (▴) or CTAP-III (●) on PMN chemotaxis induced by 5 nmol/L NAP-2. Results are expressed as a percentage of the number of cells that migrated to 5 nmol/L NAP-2 alone. (B) Impact of 1 μmol/L PBP (▴) on the characteristic chemotactic response of PMN toward NAP-2. ▪, buffer. (A, B) Means ± SD of data obtained in 3 independent experiments are shown.
In summary, the fact that PBP, and to a lesser extent CTAP-III, are not only capable of desensitizing PMN degranulation but also inhibit neutrophil chemotaxis strongly suggests that these molecules play an important role in delimiting neutrophil activation during wound repair.
Discussion
In the current study we have shown that PBP acts as an outstandingly potent agent to down-modulate neutrophil activation. Compared with the NAP-2 precursor CTAP-III, PBP proved to be approximately 6 to 7 times more potent to desensitize NAP-2–induced neutrophil degranulation. By several lines of evidence we have shown that the underlying mechanism for enhanced activity of PBP is the more rapid formation of NAP-2 from this precursor. Our data suggest that the serine protease cathepsin G or a closely related enzyme is responsible for this conversion. This contention is strongly supported by the similar kinetics of NAP-2 generation from PBP by the purified enzyme compared with PMN (compare Figures 2 and 3). Moreover, the pattern of truncation products observed, composed of 3 different molecules detectable by antiserum Rα-βTG, was virtually identical. In fact, the abundance of multiple PBP fragments in PMN supernatants first led us to believe that an enzyme other than cathepsin G was involved in PBP processing. Cathepsin G was originally described as strongly preferring aromatic residues at the P1 position of the substrate cleavage site.17 This suggested that the processing of PBP, which bears only one such residue (the tyrosine at position −1; compare Figure 1), should result in only 2 truncation products, namely NAP-2 and the N-terminal PBP fragment PBP [−24 to −1]. Indeed, mass spectroscopic analysis verified the formation of this fragment, proving that NAP-2 may become generated from PBP through cleavage at a single site. However, we observed 2 additional N-terminal truncation products, an elongated 75-residue variant of NAP-2 (DSDLY-NAP-2), most likely a byproduct generated through cleavage by neutrophil elastase behind Leu-6, and an 86-residue molecule (R-CTAP-III) arising through cleavage at a dibasic site between residues K-17 and R-16 within the PBP N-terminus. Interestingly, the generation of the latter truncation product could be explained by a recent report18 showing that cathepsin G exhibits a dual, so-called Janus-like specificity, accepting not only aromatic residues but also lysines in P1. Compared with NAP-2, R-CTAP-III did not accumulate during the enzymatic reaction, though it became detectable practically simultaneously with NAP-2 at very early time points. This was most likely because R-CTAP-III itself may serve as a substrate for further conversion to NAP-2, as indicated by its complete disappearance after long incubation times. Its turnover, similar to that of CTAP-III, appears to be slower than that of PBP because R-CTAP-III remains detectable after complete degradation of the former has taken place. Irrespective of these considerations, our data show that neither R-CTAP-III nor DSDLY-NAP-2 is a required intermediate for PBP conversion to NAP-2 but that each is a side-product that may give rise to NAP-2 during further processing. This is corroborated by our observation (unpublished, 1997) that chemical modification of lysines in PBP inhibits cathepsin G–mediated formation of the R-CTAP-III fragment but not that of NAP-2 or the N-terminal PBP-fragment (data not shown). Hence, our data concomitantly point at cathepsin G as the enzyme solely responsible for the conversion of PBP to NAP-2 by neutrophils. We can only speculate on why PBP is processed more rapidly than CTAP-III. Although structural analyses of PBP and CTAP-III have shown that backbone-folding is essentially the same in both proteins, the elongated N-terminus in PBP appears to confer additional α-helix formation within this region.19 This may improve the enzyme's accessibility to its cleavage site behind the tyrosine in position −1. Most likely helix formation is not directly involved, but it entails changes in the tertiary PBP structure19 that facilitate the enzyme's access to the cleavage site.
Based on our data, we suggest that NAP-2—on generation from PBP and from CTAP-III—causes functional desensitization of PMN mainly by the down-regulation of the high-affinity receptor for NAP-2, CXCR-2. Although we have shown that the treatment of PMN with CTAP-III leads to a loss of high-affinity binding sites for radioiodinated NAP-2,3 we here present direct evidence for the involvement of CXCR-2. FACS analyses using receptor-specific antibodies revealed that PBP, and less potently CTAP-III, caused a strong reduction of immunoreactive CXCR-2 on the cell surface of PMN (compare Figure 6). Although the expression of CXCR-1 was also compromised, that effect was only observed at approximately 200-fold higher precursor concentrations (more than 1 μmol/L with PBP, more than 10 μmol/L with CTAP-III), which are less likely to occur under physiologic conditions. These findings suggest that functions mediated by CXCR-2 would be attenuated when neutrophils encounter NAP-2 precursors.
Interestingly, this conclusion is supported by our discovery that NAP-2 precursor molecules inhibit NAP-2–induced PMN chemotaxis (compare Figure 7A). Strikingly, as shown for PBP in Figure 7, panel B, this inhibitory effect was only observed for the first of the 2 optima that characterize the chemotactic response of neutrophils toward NAP-2. Because we had previously shown that this first optimum at approximately 5 nmol/L NAP-2 was mediated by CXCR-2,12 PBP appears to specifically inhibit CXCR-2–mediated chemotaxis. Surprisingly, this effect, in contrast to the desensitization of degranulation, does not require the pretreatment of PMN with NAP-2 precursors but appears to depend on a high precursor–NAP-2 ratio in the lower compartment of the chemotaxis chamber. This phenomenon is most easily explained by the assumption that PMN, before they come upon chemotactically active NAP-2 concentrations, first encounter precursor concentrations that after conversion to NAP-2 are sufficient to counteract the original NAP-2 gradient and to down-modulate CXCR-2.
Thus, our data attribute anti-inflammatory properties to PBP and CTAP-III, which are directed against homologous PMN activation by NAP-2 or other CXCR-2 ligands, such as MGSA, but also affect IL-8. In fact, we have previously shown3 that CTAP-III pretreatment of PMN significantly reduces IL-8–induced degranulation. Correspondingly, we have observed the significantly reduced efficiency of IL-8 to induce PMN chemotaxis in the presence of PBP (data not shown). In addition, PMN desensitization may affect heterologous agonists such as fMLP. Cross-desensitizing activity of IL-8 on fMLP-induced PMN calcium flux has already been demonstrated,20 and we are investigating the impact of NAP-2 precursors on fMLP-induced degranulation. Possibly, as PBP proves a more potent anti-inflammatory agent than CTAP-III, the relative proportion of these agents stored within platelets influences their desensitizing capacity. Because the contribution of PBP to the total β-TG Ag content in platelets varies from 11% to 49%,11 undesired PMN activation after wounding might occur more readily in persons with lower percentages of PBP in their platelets.
Despite their anti-inflammatory properties, it has to be kept in mind that, after all, PBP and CTAP-III represent the source for NAP-2 that, by itself, can potently activate PMN. Taking both characteristics into account, we suggest that the NAP-2–precursor system may have a dual function and be effective during different phases of wound repair. Thus, during the initial hemostatic phase on wounding, NAP-2 generated directly after platelet aggregation from PBP and CTAP-III most likely desensitizes degranulation, especially of those PMN entrapped within the hemostatic plug. Such functional attenuation is probably important for protecting the plug and regenerating tissue from deleterious neutrophil enzymes. By the same token, our data suggest that during the initial prevalence of NAP-2 precursors over NAP-2, further PMN will be prevented from migrating to the site of tissue damage. However, because this prevalence will decline within the thrombus over time due to successive PMN-mediated conversion of the precursors to active NAP-2, the inhibitory effect will decline as well. Therefore, NAP-2 in a later, less critical phase of tissue regeneration is probably very important for the attraction and activation of PMN to the site of wound repair to ensure antimicrobial defense.21
Acknowledgments
We thank Dr A. Petersen for N-terminal sequencing of β-TG Ag isoforms. We especially thank C. Pongratz and G. Kornrumpf for perfect technical assistance.
Supported in part by Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 367, Projekt C4 and Graduiertenkolleg 288, Projekt B4). J.E.E. is a recipient of fellowship EH 188/1-1 from the Deutsche Forschungsgemeinschaft.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ernst Brandt, Department of Immunology and Cell Biology, Forschungszentrum Borstel, Parkallee 22, D-23845 Borstel, Germany; e-mail: ebrandt@fz-borstel.de.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal