Abstract
The intron 22 inversion represents the most prevalentfactor VIII gene defect in severe hemophilia A, accounting for about 40% of all mutations. It is hypothesized that the inversion mutations occur almost exclusively in germ cells during meiotic cell division by intrachromosomal recombination between 1 of 2 telomeric copies of the Int22h region and its intragenic homologue. The majority of inversion mutations originate in male germ cells, where the lack of bivalent formation may facilitate flipping of the telomeric end of the single X chromosome. This is the first intron 22 inversion that presents as a somatic mosaicism in a female, affecting only about 50% of lymphocyte and fibroblast cells of the proposita. Supposing a post-zygotic de novo mutation as the usual cause of somatic mosaicism, the finding would imply that the intron 22 inversion mutation is not restricted to meiotic cell divisions but can also occur during mitotic cell divisions, either in germ cell precursors or in somatic cells.
Introduction
Hemophilia A is the most common severe bleeding disorder in humans, affecting 1 in 5000 male births. The disease is caused by a wide range of heterogeneous mutations in thefactor VIII gene and leads to a partial or total deficiency of the factor VIII protein activity.1 In severe hemophilia A the intron 22 inversion is the most prevalent mutation, accounting for about 40%-50% of all mutations.2-4 It is widely accepted that the inversion is caused by an intrachromosomal recombination between a 9.6-kb sequence (Int22h region) within intron 22 of the factor VIII gene and 1 of 2 almost identical copies located about 300 kb distal to the factor VIII gene at the telomeric end of the X chromosome.5 Previous studies have shown that the majority of the inversion mutations were of paternal origin. The male:female ratio has been calculated to be between 15:14 and 300:1.6 The strong bias toward male origin of the inversion mutation can be explained by a model suggesting that during male meiosis, the absence of a second X chromosome facilitates flipping of the telomeric end, which is believed to precede the intrachromosomal recombination. In contrast, during female meiosis the pairing of the 2 homologous X chromosomes would prevent this process. The suggested pathomechanism of the intron 22 inversion and its predominantly paternal origin has led to the general belief that this mutation is almost exclusively of meiotic origin, although there is no direct experimental evidence for this hypothesis. Herein, we report the first intron 22 inversion that presents in a female as a somatic mosaicism, thus pointing to a mitotic origin of this mutation during early embryogenesis.
Study design
Hemophilia A family
The index patient is the only hemophiliac in the family. He is suffering from severe hemophilia A, and he developed an inhibitor at the age of 6 months. Mutation analysis revealed a distal intron 22 inversion. Haplotype analysis showed that the patient's factor VIII gene derived from the maternal grandmother, who herself was not a carrier. The factor VIII gene haplotype of the index patient was not inherited to his 2 sisters and his 2 brothers (data not shown).
Southern blot analysis
DNA from lymphocytes was prepared by a standard procedure yielding high molecular weight DNA. Fibroblast cells were taken from the proposita by skin puncture and cultured until cell growth was sufficient for DNA extraction. Fibroblast RNA was digested with ribonuclease (RNAse). The intron 22 probe (p482.6) was generated by polymerase chain reaction (PCR),4 labeled with phosphorous 32 (32P), and hybridized to a Southern blot analysis of BclI-digested DNA from lymphocyte and fibroblast cells. Densitometric quantification of the Southern blot signals was achieved on a Phosphorimager using the ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Results and discussion
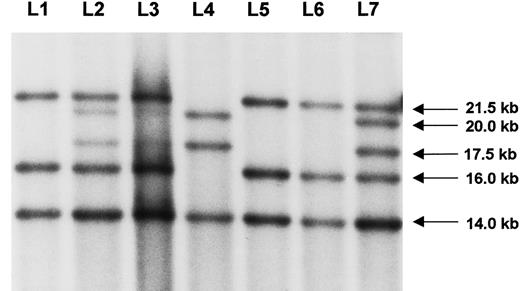
The mutation in this family was identified by Southern blot analysis in the severely affected hemophilia A patient as a typical distal intron 22 inversion (Figure 1, L4). Subsequent testing of lymphocyte DNA from the mother (L2) revealed much fainter signals for the bands that show the distal intron 22 inversion (20 kb and 17.5 kb) as compared to the pattern of a typical heterozygote carrier of this mutation (L7). These results were confirmed on several independent blots from different DNA samples. Densitometric quantification of the signals in lymphocyte cells revealed a reduced density of the inversion-specific bands to about 50% in contrast to the 100% observed in a typical female carrier (Figure 2, L2 and L7). Consequently, only about 50% of the mother's lymphocyte chromosomes are likely to carry the disease allele. Thus, the distal intron 22 inversion presents as a somatic mosaicism in the proposita. Numeric aberrations of the X chromosomes as a cause for the uncommon Southern blot pattern were excluded by the presence of a normal 46, XX karyotype in the proposita.
Southern blot analysis.
The index patient (L4) had a distal intron 22 inversion with the typical signal shift to 20 kb and 17.5 kb, respectively. The patient's mother exhibited disproportionally weaker inversion-specific signals in lymphocytes (L2) and also in fibroblasts (L3), thus representing a somatic mosaic. A typical pattern of a heterozygous female carrier (L7) showed similar densities of the corresponding signals. Healthy controls are given in L1, L5, and L6, respectively. Arrows on the right indicate the size of the bands.
Southern blot analysis.
The index patient (L4) had a distal intron 22 inversion with the typical signal shift to 20 kb and 17.5 kb, respectively. The patient's mother exhibited disproportionally weaker inversion-specific signals in lymphocytes (L2) and also in fibroblasts (L3), thus representing a somatic mosaic. A typical pattern of a heterozygous female carrier (L7) showed similar densities of the corresponding signals. Healthy controls are given in L1, L5, and L6, respectively. Arrows on the right indicate the size of the bands.
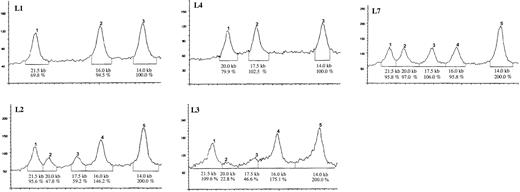
Densitometric analysis of the Southern blot analysis shown in Figure 1.
Below each curve the size of the corresponding Southern blot band is given in kb. The signal density is given in percent of area under the curve where the 14-kb signal, which is not involved in the distal intron 22 inversion, was set to 100% (corresponding to one X chromosome in L1 and L2) and 200% (representing the 2 X chromosomes in L2, L3, and L7). L1: Normal pattern with bands at 21.5 kb, 16 kb, and 14 kb. L4: Distal intron 22 inversion in the index patient with bands at 20 kb, 17.5 kb, and 14 kb. L7: Heterozygous female carrier with equal 50% densities of the bands at 21.5 kb, 20 kb, 17.5 kb, and 16 kb. The band at 14 kb shows double density because it represents the proximal telomeric copy of the F8A gene that is not involved in the inversion process. L2 and L3: Somatic mosaicism in lymphocytes (L2) and fibroblasts (L3) of the patient's mother, with reduced density of the signals showing the inversion at 20 kb and 17.5 kb and corresponding increase in density of the bands at 21.5 kb and 16 kb, respectively.
Densitometric analysis of the Southern blot analysis shown in Figure 1.
Below each curve the size of the corresponding Southern blot band is given in kb. The signal density is given in percent of area under the curve where the 14-kb signal, which is not involved in the distal intron 22 inversion, was set to 100% (corresponding to one X chromosome in L1 and L2) and 200% (representing the 2 X chromosomes in L2, L3, and L7). L1: Normal pattern with bands at 21.5 kb, 16 kb, and 14 kb. L4: Distal intron 22 inversion in the index patient with bands at 20 kb, 17.5 kb, and 14 kb. L7: Heterozygous female carrier with equal 50% densities of the bands at 21.5 kb, 20 kb, 17.5 kb, and 16 kb. The band at 14 kb shows double density because it represents the proximal telomeric copy of the F8A gene that is not involved in the inversion process. L2 and L3: Somatic mosaicism in lymphocytes (L2) and fibroblasts (L3) of the patient's mother, with reduced density of the signals showing the inversion at 20 kb and 17.5 kb and corresponding increase in density of the bands at 21.5 kb and 16 kb, respectively.
Because the proportion of mosaic cells may vary in different tissues, fibroblasts were taken from the proposita by skin biopsy and cultured until the cell growth was sufficient for DNA extraction. The autoradiogram was of lower quality, probably due to some residual RNA content. Nevertheless, faint bands deriving from the inversion clearly indicated the presence of a mosaicism in fibroblasts (Figure 1, L3). The higher density of the normal signals, compared to the mosaic lymphocytes (Figure 1, L2), suggested that fewer fibroblasts than lymphocytes carry the distal intron 22 inversion. Densitometric quantification confirmed this interpretation (Figure2, L3).
Because a considerable proportion of cells carry the mutation in lymphocytes and additionally in fibroblasts, it must be assumed that the somatic mosaicism developed within the very first days of embryogenesis. It is therefore likely that tissues deriving from other cell lineages may also carry the mutation. For germ cells the presence of the inversion mutation is demonstrated by the hemophilic son. For liver cells the normal factor VIII levels of greater than 100% may argue for the presence of a mosaicism, although different degrees of lyonization of the 2 X chromosomes should also be considered.
Somatic mosaicisms usually are caused by de novo mutations in early embryogenesis. However, the finding in the proposita may also be the result of a human chimerism caused by the fusion of dizygotic twins, of which one is a carrier of the inversion and the other is not.7 8
Somatic mosaicism of the intron 22 inversion caused by a post-zygotic de novo mutation would imply that this mutation is not, as suggested before, exclusively restricted to meiotic cell divisions, but it may also occur during mitotic cell divisions either in germ cells or in somatic cells. Because the Southern blot technique is not very sensitive for the detection of mosaic mutations, with a presumptive detection limit of 10%-20% mutated cells, mosaicisms involving the inversion mutation may occur with unknown frequency. Therefore, genetic counseling of apparent de novo cases should take into account the possibility of somatic and germline mosaicism of the intron 22 inversion mutation.
Supported by a grant (J.O.) from the Stiftung Hämotherapie-Forschung, Königsberg, Germany, and grants Ol 21/18-1 and Ol 100/1-2 from the Deutsche Forschungsgemeinschaft, Bonn, Germany.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Johannes Oldenburg, Institute of Human Genetics, Biozentrum, Am Hubland, 97074 Würzburg, Germany; e-mail:j.oldenburg@biozentrum.uni-wuerzburg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal