Abstract
Primary diffuse large B-cell lymphomas (DLBCLs) are aggressive tumors accounting for approximately 40% of B-cell malignancies. The immunoglobulin (Ig) variable region genes have undergone rearrangement and are commonly somatically mutated. The majority show intraclonal variation which indicates that somatic mutation has continued after transformation. Typically, cells of DLBCLs express Ig of a single isotype, but there may be accompanying cells that express alternative isotypes. To probe the status of the isotype switch process in DLBCL, 4 cases of tumor-derived constant region transcripts of all isotypes were investigated. Following the identification of the VDJ sequences, the presence of the major isotype expected from immunohistochemical analysis was confirmed at the RNA level. Another 3-4 alternative isotypes were revealed in all cases, some of which could also be detected by immunohistochemistry. All cases were somatically mutated with an intraclonal variation. In 2 cases there were clearly distinct patterns of somatic mutation between isotypes, which was consistent with independent evolution of the tumor subpopulations. There was apparent clustering of mutational patterns into either an IgMD/IgG3/IgA set or an IgG1/IgA set, indicating that the switch to IgA can occur by different routes. Alternative isotype expression is evident in DLBCL at both the RNA and protein levels. The pattern of mutation indicates that switching is occurring in subpopulations of the tumor after malignant transformation. The findings support the concept that isotype switch events may be a feature of DLBCL.
Introduction
Primary diffuse large B-cell lymphomas (DLBCLs) are clinically aggressive lymphomas that account for approximately 40% of all B-cell malignancies. They were previously thought to represent malignant counterparts of germinal center B cells.1However, recent analysis using microarrays suggests that there are at least 2 subsets, with gene expression patterns similar either to germinal center B cells or to activated B cells.2 DLBCLs are composed of sheets of blasts, which typically express the B-cell markers CD19, CD20, and CD22 and the surface immunoglobulin (Ig). The Ig variable region genes in DLBCL are commonly somatically mutated,3-7 and this applies also to morphologically distinct subsets of DLBCL that are sited in the central nervous system (CNS) or skin and to T-cell–rich B-cell lymphoma.6,8-10Somatic mutation can continue in B-cell lymphomas located in a germinal center environment, thereby leading to intraclonal sequence variation.3,11-14 In DLBCL, this feature has been observed in some studies3,15,16 but not in all studies.5 These differences may reflect the recently observed heterogeneity of DLBCLs2 or the narrowing of intraclonal heterogeneity, which can occur after treatment.3 12
Isotype switch events in IgM+/IgD+ (IgM/D) B cells generally occur by deletional recombination,17 with sequences between the μ-switch region (S-μ) and the switch regions upstream of the C-γ, C-α, or C-ε constant regions removed. Apart from IgD, which does not have an upstream switch region and is commonly coexpressed with IgM, a B cell should therefore be able to express only a single isotype. However, several B-cell tumors that apparently express several clonally related isotypes at the transcript level have been described. Tumor-derived VHDJHsequences combined with C-γ and C-α have been detected in IgM+ chronic lymphocytic leukemia (CLL).18 In the rare IgM+ multiple myeloma (MM), combined C-γ transcripts were identified.19Conversely, C-μ transcripts have been detected in IgG+MM.20-22 In these cases, somatic mutational patterns were closely similar between the various isotypes, which leaves open the question of whether there was heterogeneity within the population or whether one cell could produce more than one isotype.
This point was investigated in a study of a subset of lymphoplasmacytoid lymphoma, where tumor cells expressing IgM and IgG protein were evident.23 Transcript analysis of 5 cases showed that both isotypes were clonally related. Interestingly, in 2 of 5 cases, where cells were apparently expressing both IgM and IgG protein, somatic mutational patterns were divergent, which is consistent with 2 separate clonally related tumor populations. This demonstrated the danger of drawing conclusions from Ig phenotypic profiles. It also indicated that some tumor cells can undergo isotype switch and can continue to coexist with the nonswitched population.
In GC-derived lymphomas, such as follicular lymphomas (FLs) or DLBCLs, there are cases in which the tumor cells express isotype proteins other than IgM.24-26 We previously investigated 1 FL case and 2 cases of DLBCL at the transcript level3 and could detect clonally related alternative isotype transcripts. The evidence points to the use of deletional switch mechanisms for the production of these transcripts. Using fiber-FISH (fluorescence in situ hybridization) on a group of FL patients, Vaandrager et al27 observed that there was no mechanism for class switching other than deletion, in spite of detecting complex constant region rearrangements downstream of the functional constant region on the productive allele. Tumors of GC origin, such as Burkitt's lymphoma, can be induced to switch in vitro by a deletional mechanism.28 However, there are also models in which one cell can produce more than one isotype simultaneously, perhaps indicating RNA processing or trans-splicing.29
In this study we investigated the nature of isotype switch variants in primary DLBCL. We found evidence for class switching to a range of isotypes with distinct somatic mutational patterns, which is consistent with the existence of isotype switch variant subpopulations within the tumor.
Materials and methods
Clinical information and immunochemistry
Four cases of nodal primary DLBCL were randomly chosen for this study based on the availability of frozen material from the time of diagnosis. All cases showed typical morphology, with sheets of blasts replacing the nodal architecture. The tumor cells were CD20+, CD79a+, CD3−, and CD45RO−.30
The patients were staged according to the Ann Arbor criteria31 (Table 1). Routine flow cytometric analyses were available for 3 cases. Additionally, sections were cut from paraffin-embedded material and stained using an established indirect immunoperoxidase method with appropriate controls.30 32 Primary monoclonal antibodies (mAbs) were specific for IgD (Dako, High Wycombe, England) and IgM, IgG, and IgA (M. Glennie, Tenovus, Southampton, England).
Summary of clinical data and VH region gene analysis of four cases of DLBCL
| Case no. . | Patient age (y)/gender . | Tumor stage . | Primary site . | VH germline donor . | Tumor-related sequences/ total sequences, no. . | Homology to germline, % . | JH germline donor . |
|---|---|---|---|---|---|---|---|
| 5049 | 74/M | IA | cervical node | V4-34 | 10/10 | 94.6 | JH4a |
| 5126 | 50/F | IVB | supraclavicular node | V3-11 | 18/21 | 93.8 | JH4a |
| 5184 | 37/M | IA | submandibular node | V3-21 | 12/27 | 90.4 | JH4b |
| 5334 | 77/M | IIIA | inguinal node | V1-02 | 12/17 | 89.2 | JH5b |
| Case no. . | Patient age (y)/gender . | Tumor stage . | Primary site . | VH germline donor . | Tumor-related sequences/ total sequences, no. . | Homology to germline, % . | JH germline donor . |
|---|---|---|---|---|---|---|---|
| 5049 | 74/M | IA | cervical node | V4-34 | 10/10 | 94.6 | JH4a |
| 5126 | 50/F | IVB | supraclavicular node | V3-11 | 18/21 | 93.8 | JH4a |
| 5184 | 37/M | IA | submandibular node | V3-21 | 12/27 | 90.4 | JH4b |
| 5334 | 77/M | IIIA | inguinal node | V1-02 | 12/17 | 89.2 | JH5b |
The percent of homology to germline is calculated from mutations shared by all clonally related sequences of a biopsy. M indicates male; F, female.
Identification of the tumor-related variable heavy chain region genes
Double sets of 5 μm frozen tissue were cut in a cryostat. RNA was extracted with RNAzol (Cinna Biotecx Laboratories, Houston, TX). Reverse transcription was carried out with oligo-d(T)18primer and Superscript reverse transcriptase (RT) (Gibco Life Technologies, Oxford, England) in a final volume of 20 μL. Both procedures followed the manufacturers' protocols. ForVH gene analysis, 50 μL polymerase chain reactions (PCRs) were performed using 2 μL complementary DNA (cDNA),Taq, and buffer solutions from Qiagen (Qiagen, Crawley, England). Mixed 5′-oligonucleotide primers (5′-primers) specific for the VH leader or VHframework region 1 (FWR1) sequences33,34 were used together with a consensus 3′-JH primer (3′-primer).3 35 At least 2 separate PCRs were performed for each sample.
The products were analyzed on agarose gels and purified with the Gene Clean kit (Bio 101, Vista, CA). DNA was ligated into a pGEM-T vector (Promega, Southampton, England), cloned into JM109 (Promega), and sequenced with Big Dye, a GeneAmp 9600 PCR system, and an ABI Prism 377 DNA sequencer (PerkinElmer Biosystems, Warrington, England). M13 forward and reverse primers were used to sequence in both directions. The Taq error rate was determined using a known plasmid template from the leader to C-μ in a nested PCR. We sequenced 20 clones with an error rate of 0.2 of 1000 nucelotides. Sahota et al36 additionally assessed the Taqerror rate starting from a DNA template (β-globin) in tumor samples with a similar rate of error at 0.26 of 1000 nucleotides. The sequences were analyzed with Mac Vector 4.5.3 software (Oxford Molecular, England), and aligned to Entrez and V-BASE.37
Identification of isotype transcripts
For the identification of the tumor-derived isotype profiles, nested PCRs were performed. A touchdown protocol was used, starting with 65°C annealing and then reducing the annealing temperature by 1 degree per cycle to a final temperature of 59°C. This annealing temperature was maintained for a total of 20 cycles (first round) or 25 cycles (second round). Initial amplification from cDNA was carried out with the family-specific VH leader or FRW1 5′-primers together with an outer constant region 3′-primer.38 39 In case 5049, where the germline was identified as V4-34, analysis of isotype variants was carried out with a V4-34–specific FRW1 primer. Sequences for the external IgM primer, the primer pairs for IgG and IgA, and the external IgE primer are shown in Table 2. The IgG primers allow distinction between the IgG subclasses. For the secondary reaction, internal FRW1, FRW2, or FRW3 primers were used with an internal constant region 3′-primer. Where the secondary reactions failed with the consensus primers, patient-specific primers, based on the 3′-end of the VH gene segment and the first half of CDR3 (Table 2), were used together with 20 pmol/50 μL inner constant region 3′-primer. At least 2 separate PCR amplifications were performed for each constant region and biopsy. Denaturation, annealing, and extension were for 5, 5, and 15 seconds, respectively. Cloning and sequencing were performed as for the VHsequences. Products with a length of less than 150 nucleotides were ligated directly without purification to improve the efficiency of the ligation.
Constant region primers and case-specific primers
| Primer . | Sequence . |
|---|---|
| 3′-primers constant region primer name | |
| Outer IgM | GGA ATT CTC ACA GGA GAC GAG G |
| Outer IgG | CTG AGT TCC ACG ACA CCG TCA |
| Inner IgG | CTG AGT TCC ACG ACACCG TCA |
| Outer IgA | TTC GCT CCA GGT CAC ACT GAG T |
| Inner IgA | ATC TGG CTG GGT GCT GCA GAG GCT |
| Outer IgE | TGT CCC GTT GAG GGA GCC TGT |
| 5′-primers (CDR3 case-specific) | |
| 5049 | T ATG TAT TAC TGT GCG AGG GAA AAC GAC |
| 5126 | C GTC TAT TTC TGT GCG AGA CAA TCC ACG CGA |
| 5184 | GGT CTC TAT TAT TGT GCG AGA GAG CCC TAT |
| 5334 | TAC TGT GCR AAG GAT CGT GGC CTG CAA GCC |
| Primer . | Sequence . |
|---|---|
| 3′-primers constant region primer name | |
| Outer IgM | GGA ATT CTC ACA GGA GAC GAG G |
| Outer IgG | CTG AGT TCC ACG ACA CCG TCA |
| Inner IgG | CTG AGT TCC ACG ACACCG TCA |
| Outer IgA | TTC GCT CCA GGT CAC ACT GAG T |
| Inner IgA | ATC TGG CTG GGT GCT GCA GAG GCT |
| Outer IgE | TGT CCC GTT GAG GGA GCC TGT |
| 5′-primers (CDR3 case-specific) | |
| 5049 | T ATG TAT TAC TGT GCG AGG GAA AAC GAC |
| 5126 | C GTC TAT TTC TGT GCG AGA CAA TCC ACG CGA |
| 5184 | GGT CTC TAT TAT TGT GCG AGA GAG CCC TAT |
| 5334 | TAC TGT GCR AAG GAT CGT GGC CTG CAA GCC |
Results
Histology and VHgene analysis
All 4 cases were classified as primary DLBCL and had extensive replacement of the primary site by tumor cells. Predominant repeatedVHDJH sequences with a CDR3 “clonal signature” were identified in all cases (Table 1), which indicated derivation from the tumor cells.40 Other sequences detected were individually distinct and likely to be derived from normal B cells. All tumor-related VH genes revealed a significant level of somatic mutation, with 89.2%-94.6% homology to the closest germline VH segment. The deduced amino acid sequences and the replacement/silent mutations of the dominant subclone of each case are summarized in Figure1. No unequivocal D segment alignments could be made.41
Alignment of the deduced amino acid sequences of the dominant tumor clone to the closest germline VH and JH segments for 4 cases of DLBCL.
Dashes indicate homology to germline. Capital or lower-case mismatch letters indicate replacement or silent mutations, respectively. Full JH alignment was made by taking into account the result of sequencing of the constant region transcripts.
Alignment of the deduced amino acid sequences of the dominant tumor clone to the closest germline VH and JH segments for 4 cases of DLBCL.
Dashes indicate homology to germline. Capital or lower-case mismatch letters indicate replacement or silent mutations, respectively. Full JH alignment was made by taking into account the result of sequencing of the constant region transcripts.
Patterns of somatic mutation
To investigate intraclonal variation in the tumor-derived sequences, multiple clones were analyzed. Detailed results for the alignment to VH and JHgermlines for case 5334 are shown in Figure2. The majority of somatic mutations were present in all clones. Additionally there were mutations found in only some subclones. When the same mutation was detected in more than one subclone (eg, in codon 8 or 28), it was unlikely to be related toTaq errors and was consistent with ongoing mutational activity. Some mutations were found in only one sequence. While for each individual mutation the possibility remains that the mutation might be due to Taq error, the overall level of mutations was 5.6, 14.7, and 11.8 mutations of 1000 nucleotides sequenced for cases 5126, 5184, and 5334, respectively, which was a 28-fold to 73-fold excess of the Taq error rate (see “Materials and methods”). Therefore, the large majority of the observed nucleotide changes are likely to represent intraclonal sequence variation. Case 5049 showed no evidence for ongoing mutation in 10 of 10 identical sequences from leader to JH, thereby initially implying that this case was clonally homogeneous. However, further amplification of 49 clonally related sequences from leader and FWR into the constant regions using nonnested and seminested PCR revealed the presence of intraclonal heterogeneity at a low level of 2.4 of 1000 nucleotides for all 59 sequences, approximately a 10-fold excess of the Taqerror (Figure 3).
Intraclonal variation of the clonally related VHDJH sequences obtained from case 5334.
Individual clonally-related sequences are compared to the closest germline VH gene segments V1-02 andJH5a. Sequence identity is shown by dashes, and mutations are shown by mismatched characters. Two sequences were repeated, and all others were unique. The rate of intraclonal heterogeneity was calculated at 11.8 of 1000 nucleotides.
Intraclonal variation of the clonally related VHDJH sequences obtained from case 5334.
Individual clonally-related sequences are compared to the closest germline VH gene segments V1-02 andJH5a. Sequence identity is shown by dashes, and mutations are shown by mismatched characters. Two sequences were repeated, and all others were unique. The rate of intraclonal heterogeneity was calculated at 11.8 of 1000 nucleotides.
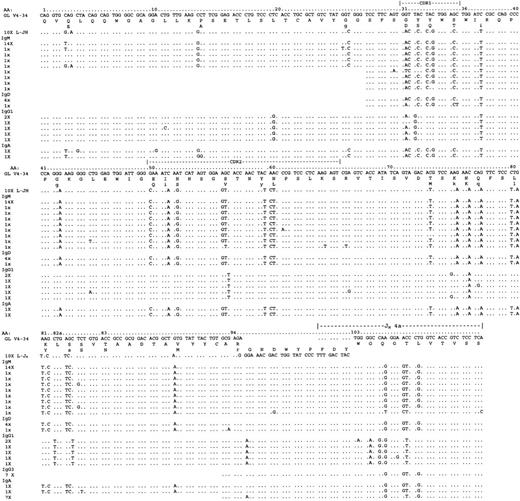
Analysis of VH constant region transcripts from case 5049.
Alignment is made to the closest germline gene segments V4-34 and JH4a and to the LHJH sequences. Using VH primers together with constant region primers specific for IgM, IgD, IgG, and IgA, transcripts for IgM, IgD, IgG1, IgG3, and IgA were identified. Mutational patterns indicated 2 distinct clusters of transcripts: IgM/D, IgG3, and IgA on one hand and IgG1 and IgA on the other. Only nonidentical sequences are shown; the numbers at the left of each line indicate the frequency of the repeated sequences. Constant region sequences are not shown. In total, 59 tumor-related sequences were analyzed.
Analysis of VH constant region transcripts from case 5049.
Alignment is made to the closest germline gene segments V4-34 and JH4a and to the LHJH sequences. Using VH primers together with constant region primers specific for IgM, IgD, IgG, and IgA, transcripts for IgM, IgD, IgG1, IgG3, and IgA were identified. Mutational patterns indicated 2 distinct clusters of transcripts: IgM/D, IgG3, and IgA on one hand and IgG1 and IgA on the other. Only nonidentical sequences are shown; the numbers at the left of each line indicate the frequency of the repeated sequences. Constant region sequences are not shown. In total, 59 tumor-related sequences were analyzed.
Transcripts for alternative isotypes
Immunophenotypically, all cases were reported as IgM+. To assess the presence of isotype transcript tumor-derivedVHDJH, clonal sequences linked to C-μ, C-δ, C-γ, C-α, or C-ε were sought using frozen tissue. For all 4 cases we were able to demonstrate the presence of transcripts for IgM, IgD, IgG, and IgA (Table 3). Tumor-related IgE sequences were not detected, even though unrelated IgE sequences could be amplified. In case 5049, transcript analysis was facilitated because the V4-34 gene allows the use of a specific FWR1 primer. This was used in single-round PCRs together with outer constant region primers. To further probe the presence of transcripts for the IgG subclasses and to expand on the number of IgA transcripts detected by the initial reactions, nested PCRs were obtained with a CDR3-specific 5′-primer and inner constant region primers. A total of 59 clonally related sequences were found. Figure 3shows all subclones, with their observed frequency indicated to the left of a particular sequence. We detected the μ-, δ-, γ-1-, γ-3-, and α-transcripts. Intraclonal heterogeneity was clearly documented in these sequences, both within and between isotypes.
Detected constant region transcripts in frozen biopsies
| Case no. . | IgM . | IgD . | IgG . | IgA . | IgE . |
|---|---|---|---|---|---|
| 5049 | + | + | γ1+ | + | − |
| γ3+ | |||||
| 5126 | + | + | γ3+ | + | − |
| 5184 | + | + | γ1+ | + | − |
| 5334 | + | + | γ1+ | + | − |
| Case no. . | IgM . | IgD . | IgG . | IgA . | IgE . |
|---|---|---|---|---|---|
| 5049 | + | + | γ1+ | + | − |
| γ3+ | |||||
| 5126 | + | + | γ3+ | + | − |
| 5184 | + | + | γ1+ | + | − |
| 5334 | + | + | γ1+ | + | − |
+ indicates the presence of the transcripts; −, the absence of the transcripts. Detected IgG subclasses (γ1 and γ3) are shown.
Assessment of the mutational patterns of different isotypes revealed that the μ- and δ-transcripts were closely related. The IgG transcripts segregated into 2 groups: IgG3 and IgG1 transcripts. The IgG3 transcripts, detected only by nested PCR, were closely related to the IgM/D transcripts in that they shared their FWR4 mutations in codons 105, 107, and 108. (The codon numbering of Chothia et al42 is used, in which the heavy chain FWR1-4 comprises codons 1-30, 36-49, 66-94, and 103-113, respectively.) IgG1 transcripts were readily detectable in single-round PCRs. While some mutations were shared with the IgM/D sequences (codons 37, 76, 105, and 107), the IgG1 sequences showed marked deviation from the IgM/D mutational pattern. Transcripts lacked multiple mutations observed in the μ/δ-set and had acquired their own characteristic set (eg, codons 20, 56, 82a, and 82b, and in FWR4). These findings suggest a class switch event from IgM to IgG1 at an earlier time point in a subset of the tumor cells, which continued to undergo further rounds of somatic mutation separately from the IgM/D population.
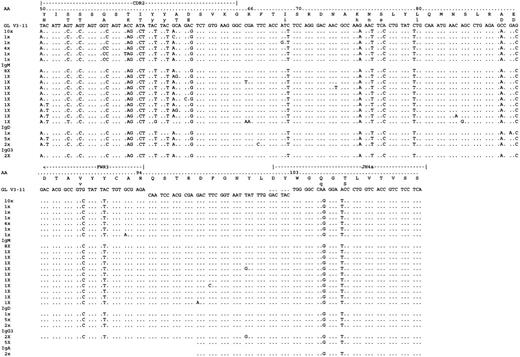
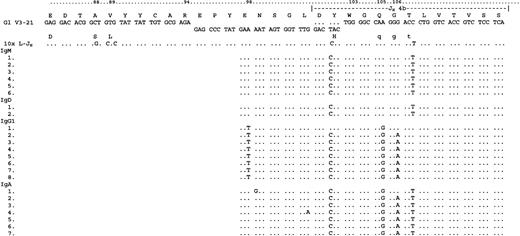
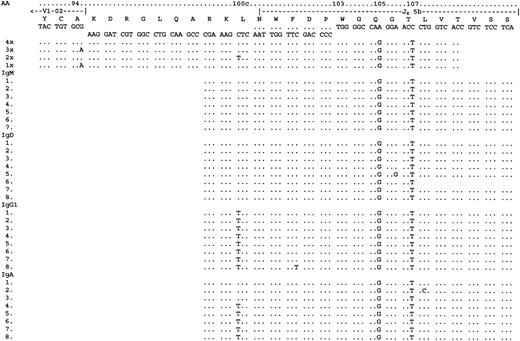
The α-transcripts showed mutations suggestive of 2 separate switch events. One group (detected in the PCR from FWR1 to C-α) was closely related to the surviving IgM/D sequences. Further subclones were obtained in nested PCRs. While these sequences were short, they suggest that these transcripts were derived from a precursor cell and shared with the IgG1 sequences (codon 95). In case 5126, constant region transcripts beginning at CDR2 were obtained in nested PCRs. The sequences were detected for IgM, IgD, IgG3, and IgA (Figure4). The μ-, δ-, α-, and γ-3-sequences were again closely related and showed multiple common mutations. Intraclonal heterogeneity was evident in the IgM/D/G classes. No IgG1 was found. For 5184 transcripts were obtained downstream from CDR3, and there was some evidence of intraclonal heterogeneity in these short sequences (Figure5). However IgM and IgD transcripts showed sequence identity, and γ-3 transcripts were not detected. As for case 5049, IgG1 sequences diverged from the IgM/D population, thereby showing 3 mutations in codons 98, 105, and 106, and no mutations in IgM/D. Those in codons 105 and 106 were shared with the α-transcripts, which may indicate a second switch event in a cell population that had switched from IgM/D to IgG1. In case 5334, transcripts were detected for IgM, IgD, IgA, and IgG1 by nested PCR from CDR3. A low degree of intraclonal heterogeneity was visible in the short sequences (Figure 6). IgM and IgD sequences were virtually identical. IgG1 transcripts again differed by a further mutation in codon 100c. As in the previous cases, some IgA transcripts appeared to be related to the IgM/D group, whereas others had acquired the additional mutation in codon 100c, which was observed in the IgG1 group. These sequences may again provide circumstantial evidence for switching from IgM-> IgG1-> IgA.
Analysis of VH constant region transcripts from case 5126.
Alignment is made to the germline segments V3-21 and JH4b and shown from CDR2. Transcripts for IgM, IgD, IgG3, and IgA were detected with highly similar sequences. Only nonidentical clones are shown. In total 52 tumor-related sequences were analyzed. Constant region sequences are not shown.
Analysis of VH constant region transcripts from case 5126.
Alignment is made to the germline segments V3-21 and JH4b and shown from CDR2. Transcripts for IgM, IgD, IgG3, and IgA were detected with highly similar sequences. Only nonidentical clones are shown. In total 52 tumor-related sequences were analyzed. Constant region sequences are not shown.
Analysis of VH constant region transcripts from case 5184.
LHJH sequences are shown from FWR3 for reference. Transcripts for IgM, IgD, IgG1, and IgA were detected using a CDR3-specific primer together with constant region primers. As for case 5049, IgG1 sequences diverged from the IgM/D population. Mutations in codons 105 and 106 were shared with the α-transcripts, perhaps indicating a second switch event in a cell population that had switched from IgM/D to IgG1. The figure presents the 33 sequences that were analyzed. Constant region sequences are not shown.
Analysis of VH constant region transcripts from case 5184.
LHJH sequences are shown from FWR3 for reference. Transcripts for IgM, IgD, IgG1, and IgA were detected using a CDR3-specific primer together with constant region primers. As for case 5049, IgG1 sequences diverged from the IgM/D population. Mutations in codons 105 and 106 were shared with the α-transcripts, perhaps indicating a second switch event in a cell population that had switched from IgM/D to IgG1. The figure presents the 33 sequences that were analyzed. Constant region sequences are not shown.
Analysis of VH constant region transcripts from case 5334.
LHJH sequences are shown for reference from FWR3. IgM, IgD, IgG1, and IgA transcripts were detected using a CDR3-specific primer together with constant region primers. IgM and IgD sequences were virtually identical. IgG1 transcripts differed by a further mutation in codon 100c. Some IgA transcripts appeared to be related to the IgM/D group, whereas others had acquired the additional mutation in codon 100c, which was observed in the IgG1 group. All sequences are shown for the constant region transcripts. In total, 43 tumor-related sequences were analyzed.
Analysis of VH constant region transcripts from case 5334.
LHJH sequences are shown for reference from FWR3. IgM, IgD, IgG1, and IgA transcripts were detected using a CDR3-specific primer together with constant region primers. IgM and IgD sequences were virtually identical. IgG1 transcripts differed by a further mutation in codon 100c. Some IgA transcripts appeared to be related to the IgM/D group, whereas others had acquired the additional mutation in codon 100c, which was observed in the IgG1 group. All sequences are shown for the constant region transcripts. In total, 43 tumor-related sequences were analyzed.
Analysis of Ig isotype protein expression
Table 4 shows the immunophenotypic analyses performed. Flow cytometry at diagnosis identified only the dominant isotype, and minor populations of other isotypes were not sought at this stage. For the immunohistochemical analyses, the difficulties of using anti-Ig mAbs in paraffin-embedded material are well known, and the staining for alternative isotypes was only taken as positive when accompanied by a characteristic perinuclear pattern.30 Case 5049 expressed IgM/D/κ by flow cytometry, and immunohistochemistry also revealed a subpopulation of IgG+ tumor cells, but no clear evidence for IgA. However, transcripts for IgM, IgD, IgG, and IgA were detectable (Table 3). Morphologically, about one-third of the cells expressed IgG protein, implying that a subgroup of cells were double-positives for IgM/G. For case 5126, tumor cells were λ+, and it was possible to use anti-κ as a background control. In addition to the major IgM+ population, a few IgG+ and IgA+ cells with characteristic staining patterns were observed. Transcripts for IgM, IgD, IgG, and IgA, the latter only by nested PCR, were detected. There were no flow cytometric data available for case 5184. Immunohistochemistry showed that all cells were κ–light chain restricted and expressed IgM. Additionally 30% coexpressed IgD. Although protein expression was not detectable, we were able to additionally detect IgG and IgA transcripts. In case 5334, initial phenotyping by flow cytometry had shown staining for IgM/D/κ. In the histochemical profile, only plasma cells showed convincing staining for IgM, IgG, and IgA, which is probably due to poor conservation of the specimen. At an RNA level we could detect transcripts for IgM, D, G, and A, only with a nested PCR approach, which possibly reflected this reduced quality.
Immunoglobulin isotype protein expression
| Case no. . | Flow cytometry . | Immunohistochemistry, % of positive tumor cells . | ||||
|---|---|---|---|---|---|---|
| IgM . | IgD . | IgG . | IgA . | Light chain . | ||
| 5049 | sIgM/D/κ | 100 | − | 30 | − | 100 κ |
| 5126 | SigM/λ | 100 | − | 5-10 | single cells4-150 | 100 λ |
| 5184 | na | 100 | 30 | − | − | 100 κ |
| 5334 | sIgM/D/κ | na4-151 | na4-151 | na4-151 | na4-151 | na4-151 |
| Case no. . | Flow cytometry . | Immunohistochemistry, % of positive tumor cells . | ||||
|---|---|---|---|---|---|---|
| IgM . | IgD . | IgG . | IgA . | Light chain . | ||
| 5049 | sIgM/D/κ | 100 | − | 30 | − | 100 κ |
| 5126 | SigM/λ | 100 | − | 5-10 | single cells4-150 | 100 λ |
| 5184 | na | 100 | 30 | − | − | 100 κ |
| 5334 | sIgM/D/κ | na4-151 | na4-151 | na4-151 | na4-151 | na4-151 |
na indicates not available; − indicates negative.
Indicates that single IgA-bearing cells were detected. Because there were no positive confluent groups of cells, however, it was not possible to definitely attribute these cells to the tumor cell population on morphological grounds.
The morphology in case 5334 was poorly maintained in the paraffin-embedded material.
Discussion
A consensus is beginning to emerge on the nature ofVH genes used by tumor cells in DLBCL. While there has been some controversy concerning a possible bias inVH gene usage and in the level of ongoing somatic mutation,5 there now seems to be agreement that there is no obvious bias and that most cases show evidence for intraclonal variation.3,4,6,10,16 However, it is clear from clinical course, morphology, and more recently from microarray analysis that DLBCL is a heterogeneous disease.2Expression of Ig is almost always retained by neoplastic B cells, but some cases of DLBCL appear Ig−,1 and some have been reported to express more than one isotype.43These data might suggest dysregulation of the Ig gene expression, and this has been indicated for some cases of FL.27 For DLBCL, the limits of immunohistochemical analysis of Ig have hampered the investigation of possible isotype variants at the protein level. However, analysis of RNA transcripts allows deductions to be made about the status of the transcribedVHDJH constant region gene sequence. This then provides insight into the functional IgM and potential isotype switch events and accompanying somatic mutation, which may be occurring in the germinal center site.
In a previous investigation of 2 cases of IgM+ DLBCL, we found the expected CDR3 C-μ transcripts and also detected CDR3 C-γ-3 transcripts.3 In one of the cases, transcripts from IgA and IgE were additionally obtained. Because only limited sequence was available, it was not possible then to view the pattern of somatic mutation in the full VH sequences of the alternative isotype transcripts. In each of the 4 cases presented here, we have been able to confirm the presence of multiple isotypes and to use the mutational patterns in the full sequences to probe the relationship between them. Identification of theVHDJH constant region transcripts indicates functional switch recombination within the tumor clone. Matching to Ig protein expression runs into the problems of immunophenotypic definition, but there was evidence for translation of at least some of the alternative transcripts.
IgD transcripts were present in all cases, presumably arising from RNA processing.44 More surprisingly, IgG and IgA transcripts were also detected in all cases. For IgG, it was possible from the sequences to assign the subclass: 2 cases were IgG1 only; 1 case, IgG3 only; and 1 case, IgG1 and IgG3. There was no other IgG subclass detected, and IgE was not found in these cases. Dominance of a switch to IgG1 and IgG3 indicates a possible influence of IL-10, a cytokine known to stimulate switching of CD40-activated naive B cells to these isotypes in vitro.45,46 A similar parallel switching to IgG1 and IgG3 has been observed in vivo in Lyme borreliosis, where interferon (IFN)-γ is a predominant cytokine.47
The finding of clonally related IgMD, IgG1, IgG3, and IgA transcripts indicates that the tumor cells are heterogeneous in their response to switch stimuli and that only some cells have undergone the presumed deletional recombination events.
In all cases the level of somatic mutation was quite high in all isotypes, including IgMD, and although ongoing somatic mutational activity tends to blur mutational differences, there were clearly distinct patterns evident in the switch variants. The close similarity between IgMD and IgG3, especially present in case 5126 and confirmed in case 5049, suggests direct switching, with little or no further accumulation of mutations. In contrast, there were markedly different mutational patterns between IgMD and IgG1, evident in case 5049 and confirmed in case 5184, which suggests that the switch to IgG1 had occurred prior to the acquisition of the majority of the mutations seen in the major IgMD population. On the route to IgG1, mutational activity continued, either in an unidentified IgMD sister cell or in the IgG1-switched cells. The history of IgA transcripts is more complex because it is most similar to the IgMD and IgG3 sequences (cases 5049 and 5126). However, there are common mutations between IgA and IgG1 in cases 5334 and 5184 and in one set of sequences in case 5049. Although the data are limited, there is a suggestion that IgA might be generated by 2 routes, one via IgG3 and the other via IgG1. By either route, few further mutations appear to accumulate. Control of the switch to IgA is not completely understood and may vary with the location of the B cell. Knockout mouse models have shown that switching in the mucosal site can occur in the absence of T cells.48 Experiments in vitro have indicated that the CD40 engagement of naive B cells is sufficient to induce switching to IgA by the release of endogenous transforming growth factor (TGF)–β.49
In summary, events in DLBCL indicate that the tumor cell population is arrested at a stage where both somatic mutation and isotype switch events can continue after transformation. Most of the mutational activity appears to be over by the IgMD+ stage, although there may be a further low level following the isotype switch. The IgG isotype variants probably reflect an influence of TH1 cells. The variants appear functional at the RNA level, and protein expression is seen at least for some of the transcripts. Mutational patterns are consistent with the existence of subpopulations within the tumor and with parallel switch events to either IgG3 or IgG1, which can then switch to IgA. The findings do not necessarily reflect dysregulation, but rather that the tumor cells are responding to normal signals without being able to achieve a fully differentiated state.
Acknowledgments
The authors are grateful to Emeritus Professor Dennis Wright and Dr Bridget Wilkins, the Department of Histopathology, Southampton University, Southampton, England, for expert help in evaluating the tumor morphology and immunocytochemistry.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Christian H. Ottensmeier, Molecular Immunology Group, Tenovus Laboratory, Cancer Sciences Division, University of Southampton, Tremona Road, Southampton SO16 6YD, England; e-mail: cho@soton.ac.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal