Abstract
Granzyme B (GrB) and perforin (PFN) are the major components of cytoplasmic granules contained in immune cellular effectors. The granule secretory pathway is one of the mechanisms by which these cells exert their cellular cytotoxicity. Recently, it has been reported that GrB and PFN are also present in circulating hemopoietic CD34+ progenitor cells mobilized by chemotherapy and granulocyte-colony stimulating factor, whereas these proteins are undetected in steady-state peripheral CD34+ cells. In this study, we hypothesized that anticancer agents may increase GrB and PFN expression in immature myeloid leukemic cells and that these treated leukemic cells become cellular effectors through a granule-dependent mechanism. Our results show that KG1a, HEL, and TF-1 CD34+acute myeloblastic leukemia cells expressed both GrB and PFN. Moreover, ionizing radiation, aracytine, and etoposide not only increase GrB and PFN expression but also conferred potent cellular cytotoxicity to these cells toward various cellular targets. Cellular cytotoxicity required cell-cell contact, was not influenced by anti-tumor necrosis factor α or anti-Fas blocking antibodies, and was abrogated by GrB inhibitors or antisense. These results suggest that, when exposed to genotoxic agents, immature leukemic cells acquire potent GrB- and PFN-dependent cellular cytotoxicity that can be potentially directed against normal residual myeloid progenitors or immune effectors.
Introduction
The granule secretory pathway is one of the mechanisms by which cytotoxic lymphocytes (CTLs) and natural killer (NK) cells exert their cytotoxicity against virus-infected, alloreactive, or transformed cells. The contact between effector cells and aberrant target cells induces exocytosis from the CTL or the NK cells, of granules that contain the potential cytolytic effector molecules. The most prominent components of cytotoxic granules are perforin (PFN) and a family of proteases, most importantly granzyme A (GrA) and granzyme B (GrB) in humans.1 Although it has been documented that PFN alone may induce, at least in some circumstances, cell lysis,2 it is generally believed that the main function of PFN is to create pores in the target cell membrane, which may disrupt osmoregulation and facilitate transfer of the granzymes.3,4 Although exogenous granzymes can also enter target cells by themselves, via a classical endocytic pathway, they are not cytotoxic unless liberated from endosomes into the cytosol by PFN or another endosomolytic agent, such as adenovirus.5 Previous studies have established that the GrB/PFN system plays a central role in cellular immunity. Indeed, targeted mutation of the PFN gene in mice results in profound immunosuppression and altered skin and tumor allograft rejections,6 whereas GrB homozygous null mutant mice have a severely depressed ability to cause target cell lysis.7Moreover, it has been previously shown that interleukin-2-induced NK/lymphokine activated killer (LAK) cell cytotoxic activity measured in a 51Cr-release short-term assay can be mainly attributed to PFN.8
More recently, GrB was detected in normal and tumor nonlymphoid cells, such as keratinocytes,9 Kupffer cells,10 and Reed-Sternberg cells.11 Albeit controversial,12 this protein was also found in some myeloid cell lines, including FDCP-Mix, a murine growth factor-dependent stem cell line,13 as well as KG1a, a human acute myeloid leukemia (AML) cell line derived from an early stage of hemopoiesis as attested by its CD34+CD38− immature differentiation phenotype.14Moreover, it has been reported that messenger RNAs and proteins of GrB and PFN were strongly expressed in normal circulating hemopoietic progenitor CD34+ cells mobilized by chemotherapy and granulocyte colony-stimulating factor (G-CSF), whereas steady-state bone marrow CD34+ or CD34+ cells mobilized by G-CSF alone were negative.14 Thus, the latter study suggests that anticancer agents may enhance both GrB and PFN expression in immature myeloid cells. On the basis of these observations, we hypothesized that genotoxic agents may enhance GrB and PFN expression in CD34+ AML cells and that increased GrB/PFN expression may confer to leukemic cells a potent cellular cytotoxicity directed against cells of various origin.
In this study, we showed that genotoxic stress increased the expression of both GrB and PFN and influenced their cellular distribution in KG1a, HEL, and TF-1 CD34+ AML cells. Moreover, we have found that when treated with ionizing radiation (IR), etoposide (VP-16), and aracytine (Ara-C), these cells became capable of inducing rapid cell lysis of both lymphoid and myeloid cellular targets through a GrB-dependent mechanism.
Materials and methods
Cell cultures
HEL, TF-1 (erythromyeloblastic), KG1a (promyeloblastic), U937 (monocytic), K562 (erythroblastic), Jurkat (T-lymphoid), MCF-7 (breast carcinoma), HeLa (neck carcinoma), and NIHOVCARIII (ovary cancer) human cell lines were obtained from the American Type Culture Collection (ATCC; Rockville, MD). NK cell line (NKL) was a generous gift from Pr E. Vivier (INSERM U.136, CIML, Marseille, France), and peripheral blood lymphocytes were obtained from a healthy donor. Cells were maintained in IMDM medium (HEL, TF-1, and KG1a) or RPMI-1640 medium (U937, K562, Jurkat, MCF-7, HeLa, NIHOVCARIII, NKL, and PBL). These media were supplemented with 20% fetal calf serum (FCS) for IMDM, 10% FCS for RPMI-1640, or AB human serum (NKL), 2 mmol/L L-glutamine (Life Technologie, Cergy Pontoise, France), 200 U/mL penicillin and 100 μg/mL streptomycin (Life Technologie), 25 μmol/L sodium pyruvate (Life Technologie) (NKL), and 100 U/mL recombinant human interleukin-2 (rh-IL-2) (NKL and PBL). All cells were maintained at 37°C in a fully humidified 5% CO2 incubator.
Drugs and reagents
3,4-Dichloroisocoumarin (DCIC) was purchased from ICN Biomedical (Aurora, OH) and dissolved in dimethyl sulfoxide (DMSO). VP-16 (Sandoz, Rueil-Malmaison, France), Ara-C (UpJohn, Paris, France), and daunorubicin (DNR) (Cerubidine, Roger Bellon, Neuilly/Seine, France) were used at appropriated concentrations. Irradiation was realized using Theratron 60Co-gamma radiation (Department of Radiation Oncology, Institut Claudius Regaud, Toulouse, France). Glycosylated rh-IL-2 was a kind gift from Synthelabo-Sanofi (Toulouse, France). Antisense oligonucleotides and controls directed against GrB have been designed and manufactured by Biognostik (Göttingen, Germany). Other products were purchased from Sigma (Saint Quentin-Fallavier, France).
Flow cytometry analysis of GrB and PFN expression
Cells were fixed with 4% (w/v) paraformaldehyde for 10 minutes at room temperature (RT), then permeabilized by 0.3% saponin-PBS (10 minutes at RT), and resuspended in 20% FCS-PBS for 20 minutes at RT. Cells were then incubated for 25 minutes at RT with anti-human GrB monoclonal antibody (MoAb; clone GrB-7; Euromedex, Souffelweyersheim, France). This MoAb is an immunoglobulin G2a (IgG2a) that reacts specifically with human serine protease GrB and does not cross-react with human GrA.11,15-17 Alternatively, cells were incubated with an anti-PFN MoAb (clone KM585; Kamiya, Seattle, WA). This MoAb is a rat IgG2a that recognizes murine PFN but cross-reacts with human PFN.18 Nonimmune mouse or rat IgG2a was used as control (Beckman-Coulters, Roissy, France). Each antibody was used at 1:20 in PBS containing 1% FCS. After two washes in PBS, cells were incubated for 30 minutes at RT with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse (GrB) or anti-rat (PFN) IgG (Beckman-Coulters, Roissy, France) diluted at 1:50 in 1% FCS-PBS. Before analysis on a flow cytometer (FACScan Becton Dickinson Co, San Jose, CA), cells were resuspended in 0.5% formaldehyde-PBS. rh-IL-2 treated peripheral blood lymphocytes (PBL-IL2) were used as positive controls for GrB and PFN expression.
Confocal laser scanning microscopy (CLSM) analysis
Aliquots of 1-2 × 105 cells were cytocentrifuged and fixed with 4% (w/v) paraformaldehyde (10 minutes at RT), washed twice with PBS, and permeabilized using 100% methanol (2 minutes at RT). Coverslips were washed with PBS, saturated for 20 minutes at RT with 20% FCS-PBS and incubated with anti-human GrB MoAb or mouse isotype-matched control, both at a 1:20 dilution in 1% FCS-PBS, followed by FITC-conjugated goat anti-mouse IgG (Beckman-Coulters) diluted at 1:100 in 1% FCS-PBS for 25 minutes at RT. After washing, the coverslips were sealed and examined with confocal imaging system that was a Zeiss (Oberkochen, Germany) scanning assembly incorporating argon and helium/neon lasers coupled to a Zeiss Axiovert 100 fluorescence microscope.
Western blot analysis
Ten million cells were washed twice in serum-free medium and lysed by resuspension in lysis buffer containing 10 mmol/L Hepes (pH 7.8), 100 mmol/L EDTA, 100 mmol/L EGTA, 1 mmol/L DTT, 1 mmol/L PMSF, 2 μmol/L pepstatin A, 0.6 μmol/L leupeptin, and 1 μg/mL aprotinin on ice for 15 minutes. Nonidet P-40 was then added at 0.6% final, and the nuclear pellet was recovered after centrifugation at 1200g for 30 minutes at 4°C. Cytoplasmic lysate (postnuclear supernatant) was collected and stored at 20°C. The nuclear pellet was resuspended in 20 mmol/L Hepes (pH 7.8), 400 mmol/L NaCl, 1 mmol/L EDTA, and 1 mmol/L EGTA. Aliquots were then incubated at 4°C for 30 minutes, centrifuged at 21 000g for 30 minutes at 4°C, and supernatants containing nuclear protein were collected. Nuclear and cytoplasmic fractions were boiled in sample buffer containing 0.4% β-mercaptoethanol, separated on 12.5% (w/v) SDS-PAGE, and transferred electrophoretically onto nylon membranes (Hybond-C extra; Amersham Life Science, Cergy-Pontoise, France). Nonspecific binding sites were blocked in 10 mmol/L Tris buffered saline (TBS) containing 0.1% Tween-20, and 10% nonfat milk. Membranes were incubated overnight at 4°C with mouse anti-GrB or rat anti-PFN MoAb both diluted at 1:200 in 10 mmol/L TBS containing 0.1% Tween-20 and 1% nonfat milk. Membranes were then washed five times at RT, and bound Ig was detected with anti-mouse IgG or anti-rat IgG coupled to horseradish peroxidase (Beckman-Coulters). The signal was visualized by enhanced chemiluminescence (Amersham, Buckinghamshire, UK) and autoradiography.
Cytotoxicity assays
The CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega, Madison, WI) was used to evaluate stress-induced cytotoxicity.19,20 This assay is a colorimetric alternative to 51Cr-release cytotoxicity assay based on the measurement of lactate dehydrogenase (LDH), a stable cytosolic enzyme that is released on cell lysis, in much the same way as51Cr is released in radioactive assay and that allows discrimination between effectors and targets LDH release.21 Variations on this technology have been reported for measuring natural cytotoxicity and have been demonstrated to be identical (within experimental error) to values determined in parallel 51Cr-release assays and was performed according to manufacturer recommendations.
Briefly, effector cells (KG1a, HEL, TF-1, PBL-IL2, and NKL: 8 × 106 or 4 × 106 cells/mL) were pretreated with cytotoxic agents, washed, then resuspended in RPMI-1640 supplemented with 5% FCS, and mixed with target cells (U937, Jurkat, and K562: 1 × 105 cells/mL) in U-bottom 96-microwell plates (Nunc, Roskilde, Denmark) at various effector-to-target (E:T) ratio in triplicates. Microplates were spun for 3 minutes at 200g and incubated for 4 hours at 37°C, 5% CO2. Supernatant (50 μL) was collected from each well and added to 50 μL reconstituted Substrate Mix for 30 minutes in the dark at RT. Enzymatic reaction was stopped by adding Stop Solution. Counting was realized by recording absorbance at 490 nm. Maximum release (TM) was determined by lysing target cells with 20 μL of Lysis Solution. Spontaneous release (TS) was determined by incubation of target cells in medium in the absence of effector cells. Effector spontaneous release (ES) was done with effector cells alone at the same E:T ratio. Results are expressed as a percentage of cytotoxicity, using the following formula: % cytotoxicity = [(experimental − ES − TS) / (TM − TS)] × 100.
Blocking experiments were performed with DCIC (50 μmol/L), a broadly reactive serine esterase inhibitor, that has been shown to neutralize granzyme enzymatic activity,22,23 MgCl2/EGTA (1 mmol/L) that blocks degranulation and prevents PFN polymerization,24 or phosphorothioate oligodeoxynucleotides (2 μmol/L) antisense or sense directed against GrB.
Statistics
Quantitative experiments were analyzed, using the Studentt test. All P values resulted from the use of 2-sided tests.
Results
GrB expression in leukemic cell lines
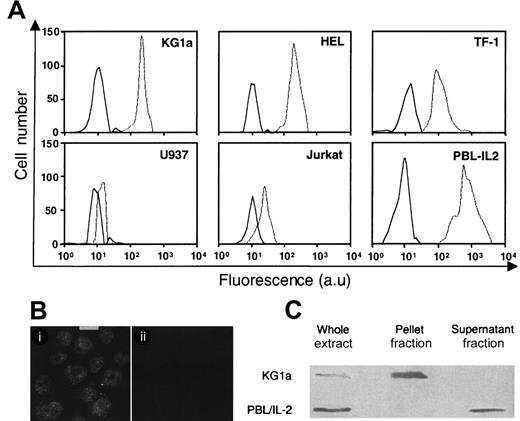
The steady-state levels and cellular distribution of GrB in leukemic cells were investigated respectively by flow cytometry, confocal microscopy, and Western blot analysis. As shown in Figure1A, GrB was detected in CD34+KG1a, HEL, and TF-1 cells, whereas CD34− U937 cells and Jurkat cells lacked significant GrB expression. However, CD34+ AML cell GrB expression was lower than that measured in PBL-IL2. Remarkably, confocal microscopy showed that GrB was mainly localized in the nucleus of KG1a cells (Figure 1B), whereas very low level, if any, was detected in the cytoplasm. Similar results were obtained in HEL and TF-1 cells (data not shown). Fractionation studies showed that GrB was almost exclusively contained in the nuclear pellet as shown for KG1a in Figure 1C. These results established that, in KG1a cells, GrB is predominantly located into the nucleus. In contrast, in PBL-IL2, GrB was exclusively found in the postnuclear fraction, suggesting that it is predominantly located in the cytoplasm (Figure 1C).
GrB expression in leukemic cell lines.
(A) Flow cytometry analysis of GrB expression as determined on KG1a, HEL, TF-1, U937, Jurkat, and activated peripheral blood lymphocytes (PBL-IL2). GrB expression in PBL-IL2 was assessed as positive control. Experiments were performed by using flow cytometry with specific anti-GrB (dotted line). Each histogram is representative of 4 to 6 different experiments. Irrelevant isotopic murine monoclonal antibody (MoAb) (black line) was used as internal control (au: arbitrary units). (B) Subcellular distribution of GrB in KG1a cells. Localization of GrB in KG1a cells was performed by confocal analysis using specific anti-GrB (i) or irrelevant isotopic murine MoAb (ii) (bar: 5 μm). (C) GrB in nuclear (pellet fraction) and cytoplasmic (supernatant fraction) lysates in KG1a cells. Western blot analysis of nuclear and cytoplasmic fractions from KG1a was performed by using MoAb-detecting GrB (32 kDa; diluted 1/200). For each lane, the nuclear and cytoplasmic lysates were loaded with the same number of cell equivalents.
GrB expression in leukemic cell lines.
(A) Flow cytometry analysis of GrB expression as determined on KG1a, HEL, TF-1, U937, Jurkat, and activated peripheral blood lymphocytes (PBL-IL2). GrB expression in PBL-IL2 was assessed as positive control. Experiments were performed by using flow cytometry with specific anti-GrB (dotted line). Each histogram is representative of 4 to 6 different experiments. Irrelevant isotopic murine monoclonal antibody (MoAb) (black line) was used as internal control (au: arbitrary units). (B) Subcellular distribution of GrB in KG1a cells. Localization of GrB in KG1a cells was performed by confocal analysis using specific anti-GrB (i) or irrelevant isotopic murine MoAb (ii) (bar: 5 μm). (C) GrB in nuclear (pellet fraction) and cytoplasmic (supernatant fraction) lysates in KG1a cells. Western blot analysis of nuclear and cytoplasmic fractions from KG1a was performed by using MoAb-detecting GrB (32 kDa; diluted 1/200). For each lane, the nuclear and cytoplasmic lysates were loaded with the same number of cell equivalents.
Effects of IR on GrB expression
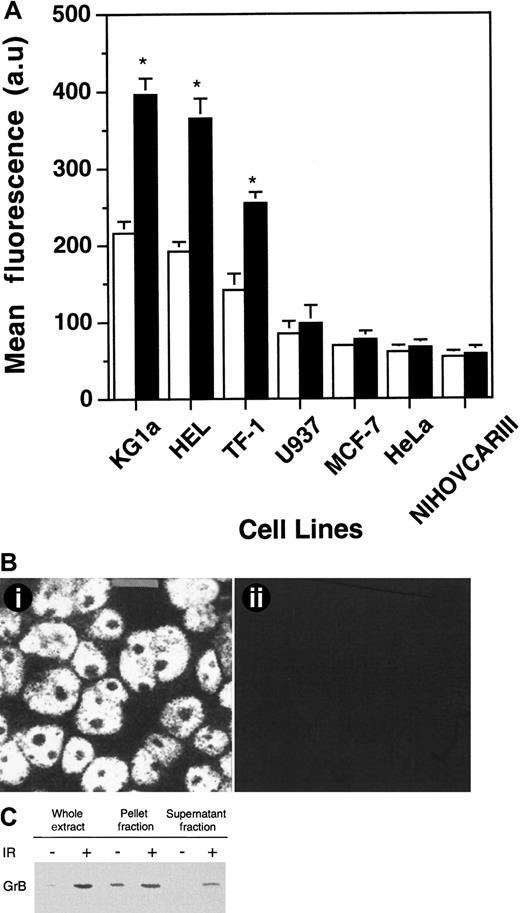
We next investigated the effects of IR on expression and cellular distribution of GrB in CD34+ leukemic cells. In repeated experiments (n = 5), cytometric analysis showed that γ-irradiation induced a modest but reproducible increase in GrB expression in KG1a, HEL, and TF-1 cells (Figure2A). Indeed, γ-radiation induced a dose- and time-dependent increase of GrB expression (data not shown) with a maximum amplification at 72 hours in cells treated with 4 Gy exposure. In these conditions, CD34+ cells retained viability as assessed by trypan blue exclusion (data not shown). However, γ-irradiation, used at subtoxic doses, induced no GrB expression in U937, Jurkat, and all epithelial cell lines tested, including MCF-7, HeLa, and NIHOVCARIII (Figure 2A). Confocal analysis showed that nuclear GrB-associated fluorescence was increased in γ-irradiated KG1a cells, compared with untreated cells. However, the most prominent finding was that GrB became easily detectable in the cytoplasm with a punctate distribution (Figure 2B). Western blot analysis performed on whole extracts of KG1a cells showed a significant increase in GrB expression, thus confirming the above findings. Moreover, fractionation studies revealed that IR-induced GrB expression increase was still more significant in the cytoplasm compared with the nuclear fraction (Figure 2C).
IR-induced GrB expression.
(A) Flow cytometry analysis of IR-induced GrB expression as determined on KG1a, HEL, TF-1, U937, MCF-7, HeLa, and NIHOVCARIII cell lines. Experiments were performed by using flow cytometry with specific anti-GrB MoAb on cells irradiated (black histograms) or not (white histograms). Each histogram is the mean of five different experiments and is expressed relative to nonirradiated cells.*P < .05 (au: arbitrary units). (B) IR-induced subcellular redistribution of GrB in KG1a cells. Localization of GrB after irradiation in KG1a cells was performed by confocal analysis using specific anti-GrB (i) or irrelevant isotopic murine MoAb (ii) (bar: 5 μm). (C) IR-induced GrB expression in nuclear (pellet fraction) and cytoplasmic (supernatant fraction) lysates in KG1a cells. Western blot analysis of nuclear and cytoplasmic fractions from KG1a was performed using MoAb-detecting GrB (32 kDa; diluted 1/200). For each lane, the nuclear and cytoplasmic lysates were loaded with the same number of cell equivalents.
IR-induced GrB expression.
(A) Flow cytometry analysis of IR-induced GrB expression as determined on KG1a, HEL, TF-1, U937, MCF-7, HeLa, and NIHOVCARIII cell lines. Experiments were performed by using flow cytometry with specific anti-GrB MoAb on cells irradiated (black histograms) or not (white histograms). Each histogram is the mean of five different experiments and is expressed relative to nonirradiated cells.*P < .05 (au: arbitrary units). (B) IR-induced subcellular redistribution of GrB in KG1a cells. Localization of GrB after irradiation in KG1a cells was performed by confocal analysis using specific anti-GrB (i) or irrelevant isotopic murine MoAb (ii) (bar: 5 μm). (C) IR-induced GrB expression in nuclear (pellet fraction) and cytoplasmic (supernatant fraction) lysates in KG1a cells. Western blot analysis of nuclear and cytoplasmic fractions from KG1a was performed using MoAb-detecting GrB (32 kDa; diluted 1/200). For each lane, the nuclear and cytoplasmic lysates were loaded with the same number of cell equivalents.
Effects of IR on PFN expression
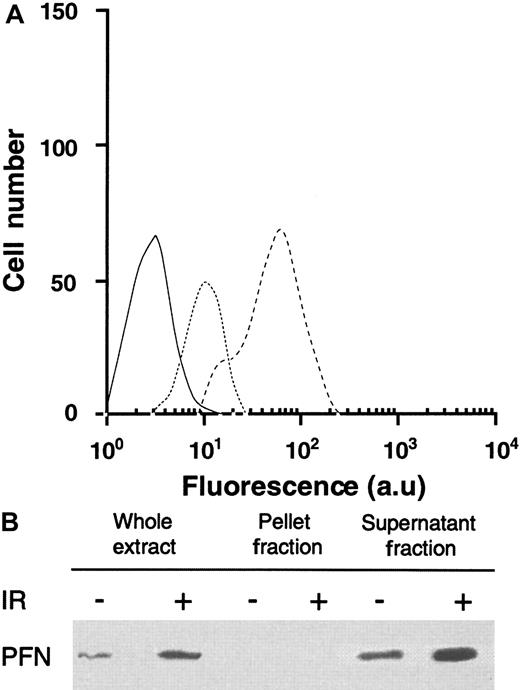
Flow cytometry and Western blot analysis showed that KG1a cells expressed a modest level of PFN expression that was significantly increased by γ-irradiation (Figure 3A). Moreover, fractionation experiments revealed that KG1a cell nuclei were devoid of PFN and that IR increased cytoplasmic PFN expression (Figure 3B).
IR-induced PFN expression in KG1a cells.
(A) Flow cytometry analysis of IR-induced PFN expression as determined on KG1a cells. Experiments were performed by flow cytometry with specific anti-PFN on KG1a irradiated (dashed line), or not (dotted line). Each histogram is representative of four different experiments. Irrelevant isotopic murine MoAb (black line) was used as internal control (au: arbitrary units). (B) IR-induced PFN expression in nuclear (pellet fraction) and cytoplasmic (supernatant fraction) lysates in KG1a cells. Western blot analysis of nuclear and cytoplasmic fractions from KG1a was performed using MoAb-detecting PFN (65 kDa; diluted 1/200). For each lane, the nuclear and cytoplasmic lysates were loaded with the same number of cell equivalents.
IR-induced PFN expression in KG1a cells.
(A) Flow cytometry analysis of IR-induced PFN expression as determined on KG1a cells. Experiments were performed by flow cytometry with specific anti-PFN on KG1a irradiated (dashed line), or not (dotted line). Each histogram is representative of four different experiments. Irrelevant isotopic murine MoAb (black line) was used as internal control (au: arbitrary units). (B) IR-induced PFN expression in nuclear (pellet fraction) and cytoplasmic (supernatant fraction) lysates in KG1a cells. Western blot analysis of nuclear and cytoplasmic fractions from KG1a was performed using MoAb-detecting PFN (65 kDa; diluted 1/200). For each lane, the nuclear and cytoplasmic lysates were loaded with the same number of cell equivalents.
Effects of genotoxic drugs on GrB and PFN expression
The effect of other anticancer agents on GrB and PFN expression was also investigated in KG1a cells. As depicted in Figure4, treatment with clinically relevant doses of Ara-C (40 μmol/L) or VP-16 (20 μmol/L), but not DNR (1 μmol/L), increased both GrB and PFN expression at 24 hours, compared with untreated cells (Figure 4). Similar results were obtained in drug-treated TF-1 and HEL cell lines (data not shown). At these doses, cell viability remained intact.
Effects of genotoxic drugs on GrB and PFN expression.
Flow cytometry analysis of cytotoxic drug-induced GrB and PFN expression as determined on KG1a cells. Cells were treated or not with 40 μmol/L AraC, 20 μmol/L VP-16, or 1 μmol/L DNR. Experiments were performed by flow cytometry with specific anti-GrB (white histograms) or anti-PFN (black histograms) MoAbs. Each histogram is the mean of five different experiments and is expressed relative to controls (au: arbitrary units). *P < .05.
Effects of genotoxic drugs on GrB and PFN expression.
Flow cytometry analysis of cytotoxic drug-induced GrB and PFN expression as determined on KG1a cells. Cells were treated or not with 40 μmol/L AraC, 20 μmol/L VP-16, or 1 μmol/L DNR. Experiments were performed by flow cytometry with specific anti-GrB (white histograms) or anti-PFN (black histograms) MoAbs. Each histogram is the mean of five different experiments and is expressed relative to controls (au: arbitrary units). *P < .05.
Cellular cytotoxicity of irradiated KG1a cells
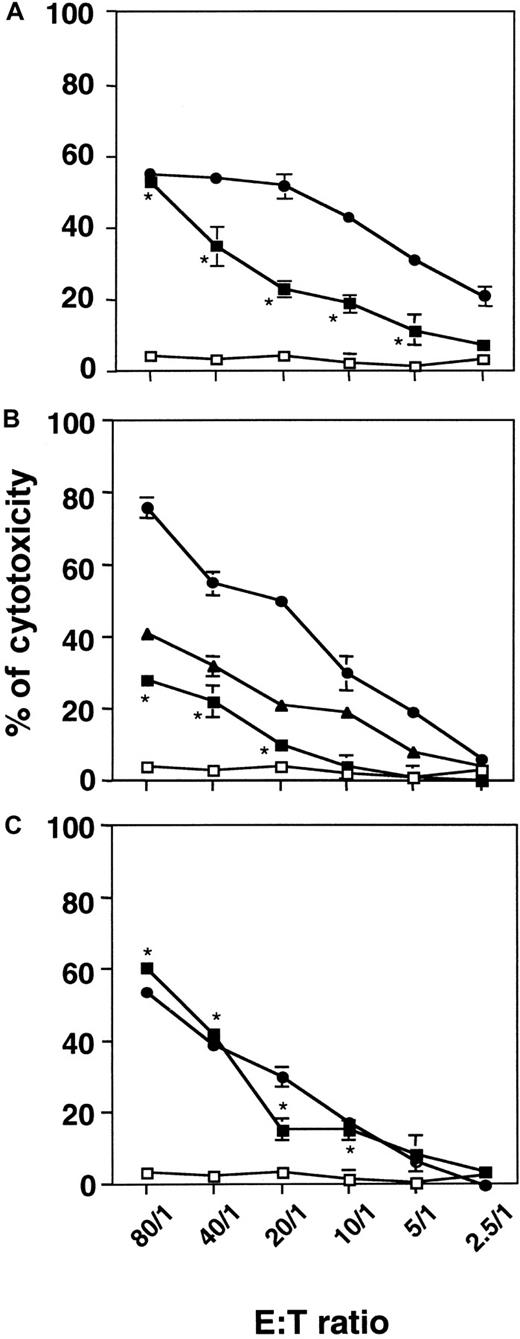
On the basis of the observation that, in the immune system, CTL and NK/LAK cells can kill their cellular targets through the GrB/PFN system, we hypothesized that stressed CD34+ AML cells could acquire cytotoxic potential because of GrB/PFN overexpression and redistribution. γ-Irradiated or untreated KG1a cells were co-cultured with various cellular targets at an E:T ratio between 2.5:1 and 80:1. Untreated KG1a cells displayed no cytotoxicity toward target cells (Figure 5). When irradiated at a dose of 4 Gy and then incubated for 72 hours, KG1a cells exhibited significant cytotoxicity toward U937 cells (Figure 5A), K562 cells (Figure 5B), and Jurkat cells (Figure 5C). Interestingly, irradiated KG1a cells exerted a cytotoxic effect comparable to that observed with PBL-IL2 toward Jurkat target cells. In addition, in the 4-hour assay, we were unable to detect any apoptotic features in target cells as evaluated by DAPI staining and DNA fragmentation measurement (data not shown). Moreover, K562, U937, and Jurkat cells displayed no cytotoxicity toward KG1a cells (data not shown).
Cellular cytotoxicity of irradiated KG1a cells toward myeloid (U937, K562) or lymphoid (Jurkat) cell lines.
Irradiated KG1a cells were cultured for 72 hours. Cytotoxicity was measured by using a 4-hour nonradioactive LDH-release assay at the indicated E:T ratio. Untreated KG1a (■), irradiated KG1a (▪), NKL (▴), or PBL-IL2 (●) cells (used as an internal control) were co-incubated with U937 (A), K562 (B), or Jurkat (C) cells as targets. Results are the mean ± standard deviation of three independent experiments performed in triplicate. *P = 0.02 for E:T ratio of 80:1 to 10:1.
Cellular cytotoxicity of irradiated KG1a cells toward myeloid (U937, K562) or lymphoid (Jurkat) cell lines.
Irradiated KG1a cells were cultured for 72 hours. Cytotoxicity was measured by using a 4-hour nonradioactive LDH-release assay at the indicated E:T ratio. Untreated KG1a (■), irradiated KG1a (▪), NKL (▴), or PBL-IL2 (●) cells (used as an internal control) were co-incubated with U937 (A), K562 (B), or Jurkat (C) cells as targets. Results are the mean ± standard deviation of three independent experiments performed in triplicate. *P = 0.02 for E:T ratio of 80:1 to 10:1.
Cellular cytotoxicity of drug-treated KG1a cells
We also investigated the influence of drugs on KG1a cellular cytotoxicity. We found that when treated with Ara-C (40 μmol/L) or with VP-16 (20 μmol/L) for 24 hours, but not with DNR, KG1a, HEL, and TF-1 cells displayed significant cytotoxicity toward K562 or Jurkat cells (Table 1).
Cytotoxicity of drug-treated CD34+ cells toward K562 or Jurkat cells at an effector-to-target ratio of 40:1
| Effector cells and treatment . | Target cells (% of cytotoxicity) . | |
|---|---|---|
| K562 . | Jurkat . | |
| KG1a | 1.7 (±0.7) | 3 (±0.9) |
| KG1a + Ara-C | 24.5 (±4.3)* | 39 (±7.1)* |
| KG1a + VP-16 | 14.7 (±2.2)* | 16 (±2.8)* |
| KG1a + DNR | 0.9 (±1.2) | 0.4 (±0.9) |
| TF1 | ND | 2.6 (±1.1) |
| TF1 + Ara-C | ND | 23.5 (±4.3)* |
| TF1 + VP-16 | ND | ND |
| TF1 + DNR | ND | 1.8 (±2.2) |
| HEL | ND | 0.5 (±0.1) |
| HEL + Ara-C | ND | 23.4 (±3.5)* |
| HEL + VP-16 | ND | 21.4 (±1.7)* |
| HEL + DNR | ND | 1.1 (±0.9) |
| Effector cells and treatment . | Target cells (% of cytotoxicity) . | |
|---|---|---|
| K562 . | Jurkat . | |
| KG1a | 1.7 (±0.7) | 3 (±0.9) |
| KG1a + Ara-C | 24.5 (±4.3)* | 39 (±7.1)* |
| KG1a + VP-16 | 14.7 (±2.2)* | 16 (±2.8)* |
| KG1a + DNR | 0.9 (±1.2) | 0.4 (±0.9) |
| TF1 | ND | 2.6 (±1.1) |
| TF1 + Ara-C | ND | 23.5 (±4.3)* |
| TF1 + VP-16 | ND | ND |
| TF1 + DNR | ND | 1.8 (±2.2) |
| HEL | ND | 0.5 (±0.1) |
| HEL + Ara-C | ND | 23.4 (±3.5)* |
| HEL + VP-16 | ND | 21.4 (±1.7)* |
| HEL + DNR | ND | 1.1 (±0.9) |
KG1a, TF-1, and HEL were treated with aracytine (Ara-C; 40 μmol/L), etoposide (VP-16; 20 μmol/L), or daunorubicin (DNR; 1 μmol/L) for 24 hours. Cytotoxicity was measured by using a 4-hour nonradioactive lactate dehydrogenase-release assay. Results are the mean ± standard deviation of four independent experiments performed in triplicate. ND: not determined.
P < .05.
Effects of GrB and PFN inhibitors
To investigate the role of the GrB/PFN system in the mechanism by which irradiated KG1a cells exerted their lytic ability, we used pharmacological agents known to modulate GrB/PFN cytotoxic function. These experiments were performed with K562 and Jurkat cells as targets at various E:T ratios. Therefore, irradiated KG1a cells were pre-incubated with DCIC, a GrB inhibitor, washed, and allowed to react with target cells. As depicted in Table2, at an E:T ratio of 40:1, pretreatment with DCIC resulted in total abrogation of irradiated-KG1a cell cytotoxicity. Similar results were obtained at the other E:T ratios (data not shown). As expected, in the same conditions, DCIC also significantly reduced PBL-IL2 cytotoxicity. Moreover, when treated KG1a cells or PBL-IL2 were co-incubated with MgCl2/EDTA, a PFN inhibitor, cytotoxicity was no longer observed. To investigate other mechanisms implicated in cellular-mediated cytotoxicity, irradiated KG1a cells and target cells were co-cultured in the presence of anti-tumor necrosis factor α (TNF-α) or anti-CD95 (Fas/APO-1) blocking antibodies. As shown in Table 2, these antibodies had no influence on irradiated-KG1a cellular cytotoxicity.
Effects of specific GrB and PFN inhibitors
| Effector cells and treatment . | Target cells (% of cytotoxicity) . | |
|---|---|---|
| K562 . | Jurkat . | |
| KG1a | 2.8 (±0.4) | 2 (±0.5) |
| KG1a + IR | 22 (±4.2) | 28.5 (±2.4) |
| KG1a + IR + DCIC | 0.8 (±0.2)* | 2.7 (±1.4)* |
| KG1a + IR + MgCl2/EGTA | 0.6 (±0.3)* | 1 (±2.4)* |
| KG1a + IR + anti-TNF-α (blocking) | 19.8 (±1.2) | 25.5 (±1.8) |
| KG1a + IR + anti-CD95 (blocking) | ND | 22 (±3.4) |
| PBL/IL-2 | 50 (±5.5) | 55 (±4.3) |
| PBL/IL-2 + DCIC | 11.7 (±2.1)* | 8 (±1.4)* |
| PBL/IL-2 + MgCl2/EGTA | 3 (±1.3)* | 4.3 (±2.7)* |
| Effector cells and treatment . | Target cells (% of cytotoxicity) . | |
|---|---|---|
| K562 . | Jurkat . | |
| KG1a | 2.8 (±0.4) | 2 (±0.5) |
| KG1a + IR | 22 (±4.2) | 28.5 (±2.4) |
| KG1a + IR + DCIC | 0.8 (±0.2)* | 2.7 (±1.4)* |
| KG1a + IR + MgCl2/EGTA | 0.6 (±0.3)* | 1 (±2.4)* |
| KG1a + IR + anti-TNF-α (blocking) | 19.8 (±1.2) | 25.5 (±1.8) |
| KG1a + IR + anti-CD95 (blocking) | ND | 22 (±3.4) |
| PBL/IL-2 | 50 (±5.5) | 55 (±4.3) |
| PBL/IL-2 + DCIC | 11.7 (±2.1)* | 8 (±1.4)* |
| PBL/IL-2 + MgCl2/EGTA | 3 (±1.3)* | 4.3 (±2.7)* |
KG1a cells were treated with either DCIC (20 μmol/L; GrB inhibitor) or MgCl2/EGTA (1.5 mmol/L; PFN inhibitor) after irradiation (4 Gy) and incubation for 72 hours and then were allowed to react with target cells (K562 or Jurkat) for 4 hours. Effector and target cells were co-incubated in the presence of anti-TNF-α or anti-CD95 blocking monoclonal antibodies for 4 hours. Cytotoxicity was measured by a 4-hour nonradioactive lactate dehydrogenase-release assay. Results are the mean ± standard deviation of three independent experiments performed in triplicate.
GrB indicates granzyme B; PFN, perforin; IR, ionizing radiation; DCIC, 3,4-dichloroisocoumarin; TNF-α, tumor necrosis factor α.
P = .02.
Effects of GrB antisense oligonucleotides
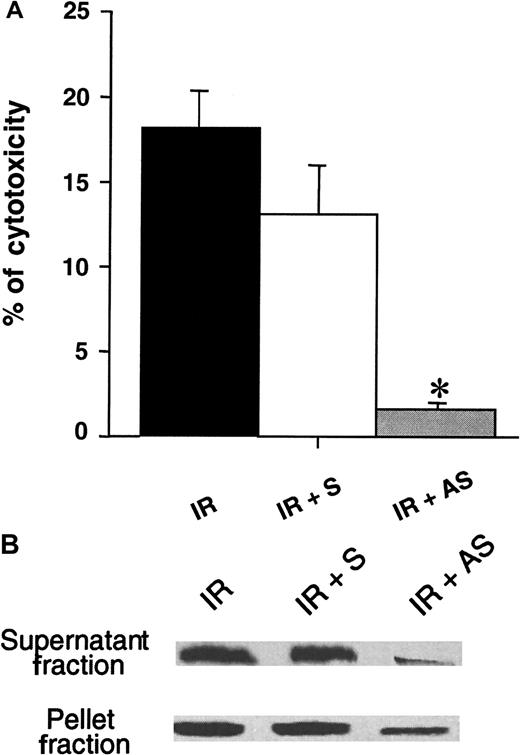
To ascertain the involvement of the lytic protein GrB in cell-mediated cytotoxicity, we first exposed KG1a cells to the action of antisense oligonucleotides directed against GrB. Cells were then irradiated at a dose of 4 Gy, incubated for 72 hours, and then allowed to react with K562 used as target cells at various E:T ratios for 4 hours. In fact, GrB antisense, but not sense, oligonucleotides strongly diminished irradiated-KG1a cell cytotoxicity as depicted in Figure6a for an E:T ratio of 40:1. In parallel, exposure to antisense oligonucleotide resulted in a decrease of GrB expression, more significantly in the cytoplasm than in the nucleus, whereas exposition to control oligonucleotide did not affect significantly GrB expression (Figure 6B).
Effect of antisense oligonucleotides directed toward GrB.
(A) KG1a cells were incubated with antisense GrB or sense (control) oligonucleotides for 14 hours, irradiated, cultured for 72 hours, and then co-incubated with K562 cells as targets. Cytotoxicity was measured by using a 4-hour nonradioactive LDH-release assay at 40:1 E:T ratio. Results are the mean ± standard deviation of three independent experiments performed in triplicate. *P = .02. (B) Western blot analysis of antisense or sense oligonucleotides on GrB expression after irradiation and cellular fractionation. IR: irradiation, S: sense, AS: antisense.
Effect of antisense oligonucleotides directed toward GrB.
(A) KG1a cells were incubated with antisense GrB or sense (control) oligonucleotides for 14 hours, irradiated, cultured for 72 hours, and then co-incubated with K562 cells as targets. Cytotoxicity was measured by using a 4-hour nonradioactive LDH-release assay at 40:1 E:T ratio. Results are the mean ± standard deviation of three independent experiments performed in triplicate. *P = .02. (B) Western blot analysis of antisense or sense oligonucleotides on GrB expression after irradiation and cellular fractionation. IR: irradiation, S: sense, AS: antisense.
Discussion
In this report, we show that KG1a, TF-1, and HEL cells express both GrB and PFN as assessed by flow cytometry, confocal microscopy, and Western blot analysis. For GrB detection, we have used GrB-7 MoAb that has been characterized elsewhere.15Briefly, GrB-7 has been raised against recombinant GrB and reacts with isolated GrB from activated cytotoxic lymphocytes, but it does not recognize GrA.15 In fact, this antibody has been used in a large number of immunohistochemical studies.11,15-17,25,26Moreover, we have used in parallel B18.1 MoAb that also reacts specifically against human GrB.9,14,27 B18.1 MoAb provided similar results with GrB-7 in flow cytometry and confocal microscopy studies (data not shown) but, at least in our hands, it was found to be less reliable than GrB-7 for Western blot analysis. For this reason, the study was conducted with GrB-7 MoAb. For PFN detection, we have used KM585 MoAb,18 a rat MoAb raised against mouse recombinant PFN that cross-reacts with human PFN.18 This antibody was also used in immunohistochemical studies and Western blot analysis.18,28-31 Moreover, we have used in parallel δG9 MoAb, a purified mouse anti-human PFN.32-34 This antibody provided similar results with KM585 in flow cytometry and confocal microscopy studies (data not shown) but was nonapplicable for Western blot. For this reason, the study was conducted with KM585.
The fact that KG1a cells express both GrB and PFN has been previously reported.14 However, intracellular distribution of these lytic proteins have not been documented. In our study, cellular fractionation followed by Western blot analysis showed that PFN was exclusively distributed in the cytoplasm of KG1a cells. In contrast, we found that GrB was mainly contained in the nucleus. This result contrasts with that generally observed in activated T or NK/LAK cells in which GrB and PFN are internalized into cytoplasmic granules.35 Although nuclear localization of GrB has been previously described,36 the fact that KG1a cells express cytoplasmic PFN and nuclear GrB while retaining viability is somewhat intriguing. Indeed, recent work37 supports the idea that translocation of GrB to the nucleus plays a central role in effecting the nuclear changes associated with cell death. One can hypothesize that GrB is complexed to other molecule(s) in the nucleus and stored in an inactive form as it has been previously proposed.36
Our study shows that IR and some chemotherapeutic compounds increase PFN and GrB expression in KG1a, HEL, and TF-1 cells. The reason why DNR did not induce GrB and PFN expression is still unclear but could be due to vesicular drug sequestration as we have previously described.38 Fractionation studies combined with Western blot analysis showed that PFN remained localized in the cytoplasm, whereas GrB expression increased in both nuclear and cytoplasmic compartments. This finding suggests that GrB is not translocated from the nucleus to the cytoplasm and that GrB redistribution is due to de novo synthesis activated by genotoxic stress. Indeed, we observed that actinomycin D as well as cycloheximide treatments dramatically decreased stress-induced GrB expression (data not shown). The mechanism by which anticancer agents enhanced GrB and PFN expression remains to be determined. However, it should be noted that, on one hand, GrB promoter contains AP-1 binding sites, and that, on the other hand, γ-irradiation, Ara-C, and VP-16 activate AP-1.39-41Whether or not AP-1 is involved in GrB up-regulation should be examined.
In this study, we show for the first time that leukemic cells treated with genotoxic agents acquire major histocompatibility complex (MHC)-unrestricted potent lytic ability toward targets from various origins. This finding may have important implications in AML therapy. Indeed, it is conceivable that Ara-C, which is used as part of front-line therapy, renders blast cells cytotoxic for nonleukemic bone marrow cells, including residual normal myeloid progenitors and immune cellular effectors. Moreover, blast lytic activity should be facilitated by a high E:T ratio in the overt phase of the disease.
The mechanism by which stress agents render leukemic cells cytotoxic has been examined. Irradiated or drug-treated KG1a cells displayed no lytic ability when separated from target cells by nylon membrane (data not shown). The fact that cell-cell contact is required for cellular cytotoxicity precludes the role of soluble cytotoxic molecules, including those which have been found to be produced under stress conditions, such as TNF-α,42,43 tumor-growth factor β,44 and Fas-ligand.45 Moreover, anti-TNF-α and anti-Fas blocking antibodies had no influence on target cell lysis. In addition, stress-induced KG1a cells were highly efficient against K562 that are known to be resistant to TNF-α and Fas agonist.46,47 Altogether, these results strongly argue against the role of these soluble mediators or even other yet unidentified soluble molecules that would have been produced by leukemic cells under stress conditions. It has been reported recently that γ-irradiation induces, at least in peripheral blood mononuclear cells, the production of membrane-bound TNF-α (mTNF-α), and that mTNF-α mediates cell-cell contact- dependent cellular cytotoxicity toward endothelial cells that could be blocked by anti-TNF-α antibody.44 The role of mTNF-α seems also unlikely because anti-TNF-α had no influence on target cell lysis induced by irradiated KG1a cells.
On the basis of these findings and considerations, we hypothesized that GrB and PFN could be involved in the cellular cytotoxicity of stressed leukemic cells. Several lines of evidence support this hypothesis: (1) the requirement of cell-cell contact for target lysis, (2) the blocking effects of serine esterase inhibitor and MgCl2/EGTA, and (3) the blocking effect of antisense oligonucleotides directed against GrB.
To conclude, we propose a model in which some leukemic cells constitutively express cytoplasmic PFN and nuclear GrB. Under normal conditions, these proteins are sequestered in distinct intracellular compartments and confer no lytic activity to leukemia cells. However, under genotoxic stress, both PFN and GrB are newly synthesized, stored in cytoplasmic cytotoxic granules, and confer potent MHC-unrestricted cellular cytotoxicity to leukemia cells.
Supported by the Association pour la Recherche contre le Cancer (ARC) (grant 9296) and la Faculté de Médecine Toulouse-Rangueil. A.P.B. is the recipient of a grant from the Ministère de l'Education Nationale, de l'Enseignement Supérieur, et de la Recherche (MENESR).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anne Quillet-Mary, INSERM E9910, Institut Claudius Régaud, 20 rue du Pont St Pierre, 31052 Toulouse, France; e-mail:quillet_mary@icr.fnclcc.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal