Abstract
PBX1 is a proto-oncogene that plays important roles in pattern formation during development. It was discovered as a fusion with the E2A gene after chromosomal translocations in a subset of acute leukemias. The resulting E2a-Pbx1 chimeric proteins display potent oncogenic properties that appear to require dimerization with Hox DNA binding partners. To define molecular pathways that may be impacted by E2a-Pbx1, a genetic screen consisting of neonatal retroviral infection was used to identify genes that accelerate development of T-cell tumors in E2A-PBX1 transgenic mice. Retroviral insertions in the Notch1 gene were observed in 88% of tumors arising with a shortened latency. Among these, approximately half created a NotchIC allele, encoding the intracellular, signaling portion of Notch1, suggesting a synergistic interaction between the Notch and E2a-Pbx1 pathways in oncogenesis. The remaining proviral insertions involvingNotch1 occurred in a more 3′ exon, resulting in truncating mutations that deleted the carboxy-terminal region ofNotch1 containing negative regulatory sequences (Notch1ΔC). In contrast toNotchIC, forced expression ofNotch1ΔC in transgenic mice did not perturb thymocyte growth or differentiation. However, mice transgenic for both the E2A-PBX1 and Notch1ΔC genes displayed a substantially shortened latency for tumor development compared with E2A-PBX1 single transgenic mice. These studies reveal a novel mechanism for oncogenic activation ofNotch1 and demonstrate a collaborative relationship between 2 cellular oncogenes that also contribute to cell fate determination during embryonic development.
Introduction
A number of genes whose products regulate animal development are also targets for acquired mutations in human cancers.1 The characterization of these so-called developmental oncogenes has contributed to our current understanding that neoplastic transformation can result from the misregulation of molecular mechanisms governing cell fate determination and cellular identity. However, the pathogenetic connections between disrupted development and cancer are poorly understood at a molecular level.
PBX1 is a proto-oncogene that plays an important role in pattern formation during development. It codes for a homeodomain protein that is the mammalian homolog ofDrosophila extradenticle (exd).2 Pbx and exd proteins serve as dimerization partners for a wide variety of Hox proteins3-7 and make critical contributions to the segment-specific execution of Hox programs in arthropods and vertebrates.8-10 In a subset of pediatric acute lymphoblastic leukemias, chromosomal translocations fuse thePBX1 and E2A genes, resulting in the production of chimeric E2a-Pbx1 proteins.11,12 E2a proteins, members of the bHLH family of master developmental regulators, contribute to tissue-specific regulation of the IG genes and are essential for B-lymphoid lineage development.13 Fusion of the strong transcriptional activation domains of E2a onto the DNA-binding domain of Pbx1 results in a profound disruption of Pbx1 transcriptional regulatory properties as evaluated using in vitro assays.14-16 Furthermore, E2a-Pbx1 displays potent oncogenic properties, in a variety of cell types, that appear to require dimerization with Hox DNA-binding partners.17 18Taken together, the available data strongly suggest that E2a-Pbx1–mediated oncogenesis results from perturbations of Hox-dependent transcriptional pathways that normally orchestrate the differentiation programs of lymphoid progenitors. However, the subordinate genes and transcriptional pathways that are subject to Hox-Pbx regulation in normal and neoplastic lymphocytes remain undefined.
Notch1, like Pbx1, plays important roles in cell fate determination and lymphoid oncogenesis. It codes for a highly conserved transmembrane protein whose extracellular portion contains a ligand-binding domain, composed of 36 EGF repeats, as well as 3 lin12/Notch repeats of uncertain function.19-21The ligands for Notch1 are also cell surface proteins whose interactions induce the release and nuclear migration of the intracellular portion (NotchIC) of Notch1.22-24 Forced expression ofNotchIC, which lacks the extracellular ligand-binding domain, circumvents the need for ligand to activate theNotch1 pathway thereby creating a constitutiveNotch1 signal. This has been shown to suppress myogenesis and neurogenesis and results in profound alterations on the fate decisions of maturing thymocytes.25-28,NotchIC was originally isolated as a lymphoid oncogene (TAN1) in a subset of T-cell acute lymphoblastic leukemias that harbor chromosomal translocations that truncate and ectopically express the signaling form of Notch1.29Neoplastic transformation of various cell types by NotchIChas subsequently been reported.30-32
In the studies reported here, we used a genetic screen to identify genes that accelerate development of T-cell tumors inE2A-PBX1 transgenic mice. Mutations of the Notch1gene were observed in 88% of tumors arising with a shortened latency. Although a significant number induced the intracellular signaling form of Notch1, more than 50% of the cases harbored activating mutations that preserved the ligand-binding domain of Notch1. These observations indicate that Notch1 collaborates with E2a-Pbx1 in lymphoid oncogenesis and reveal a novel mechanism for oncogenic activation of Notch1.
Materials and methods
Transgenic mice and retroviral infections
E2A-PBX1 transgenic mice (line Tg.E2a-Pbx1a as reported previously33) were maintained on an FVB-inbred genetic background. Neonates (1-2 days of age) resulting from backcross matings of E2A-PBX1 mice were injected intraperitoneally with Moloney murine leukemia virus (MMuLV) (105 pfu in 50 μL RPMI) obtained from the American Type Culture Collection (strain E-286). Animals were killed when they became moribund and tumors were removed for histologic, molecular, and FACS analyses. LCK-Notch1ΔC andLCK-Notch1 transgenic mice were constructed by microinjection of FVB pronuclei with purified transgene insert DNA using standard techniques.34 The transgene construct consisted of an EcoRI-HindIII fragment of the ratNotch1 complementary DNA (cDNA) (a kind gift from G. Weinmaster) cloned downstream of an LCK promoter.35 For the LCK-Notch1ΔC construct, Notch1coding sequence was prematurely truncated in its 3′ portion (nucleotide 7117 of the Notch1 cDNA) by insertion of a synthetic oligonucleotide encoding an in-frame FLAG epitope sequence and translation stop codon using standard cloning techniques. Downstream of the stop codon, the transgene contained transcriptional termination and polyadenylation signals encoded by a fragment of the human growth hormone gene. Two founder LCK-Notch1 lines were obtained and displayed similar properties. Four founderLCK-Notch1ΔC mice were obtained, 3 of which passed the transgene to their progeny. Two lines were selected for further study, each of which gave essentially identical results. Transgenic mice were maintained on an FVB-inbred genetic background by backcross matings and intercrossed with Tg.E2a-Pbx1a mice to obtain double-transgenic progeny. Transgene expression was evaluated by quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) using primers specific for human growth hormone sequences encoded by the transgene construct and commercially prepared reagents (Preamplification System) under conditions recommended by the supplier (Gibco BRL, Rockville, MD).
Southern and Western blotting
DNA isolated from tumors was subjected to Southern blot analyses using standard procedures. For detection of MMuLV proviral integrations, a 560-base pair (bp) fragment containing the U3 portion of the MMuLV long terminal repeat (LTR) was used as a probe onEcoRI- and NheI-digested DNA. Rearrangements of the Notch1 gene induced by proviral insertions were detected using 2 different Notch1 cDNA probes. A probe specific for the midportion (cluster region I) of the Notch1 gene consisted of a cDNA fragment spanning nucleotides 3851-4861 and was used on EcoRV-digested DNA for detection of proviral insertions. Rearrangements of the Notch1 gene due to proviral insertions at its 3′ end (cluster region II) were detected with a 0.9-kilobase (kb) fragment of Notch1 cDNA (nucleotides 7053-7957) on DNA digested with KpnI. T-cell receptor gene rearrangements were evaluated using a Jβ2-specific probe on EcoRI-digested DNA.
Fresh or snap-frozen tissues were lysed in SDS gel-loading buffer lacking bromophenol blue.36 Proteins (50 μg) were fractionated through either 6% or 10% SDS-polyacrylamide denaturing gels, transferred to nitrocellulose by electroblotting, and subjected to Western blot analysis.36 Primary antibodies were reactive with the FLAG epitope and secondary antibodies consisted of peroxidase-conjugated polyclonal goat antirabbit IgG. All antibodies were used at a dilution of 1:5000, and immune complexes were visualized by chemiluminescence (Amersham) performed according to the manufacturer's recommendations.
Characterization of proviral integration sites
Proviral insertion sites were characterized in tumors from MMuLV-injected E2A-PBX1 mice at 2 months of age before outward signs of disease to favor isolation of proviral integrations that occurred early in the tumor process. Seminested PCR was used in combination with Southern blot enrichment to isolate sites of MMuLV proviral insertions. DNA was purified from tumors, digested with the restriction enzymes EcoRI and NheI, and then subjected to electrophoresis through a 0.8% agarose gel. Portions of the agarose gel containing DNA fragments in the size range corresponding to the integration of interest were excised and DNA was purified from the agarose matrix using glass bead absorption (Gene Clean, Bio 101). The purified DNA fragments were then ligated to pUC19 DNA that had been predigested with EcoRI andXbaI. Seminested PCR was performed directly on 10% of the ligation reaction mix using Expand High Fidelity polymerases (Boehringer Mannheim, Mannheim, Germany) and primers designed by Natarajan et al37 in the reverse orientation. The identity of mouse genomic DNA flanking the proviral LTR was determined by nucleotide sequencing of the resulting PCR products using an automated sequencer (model 373; Applied Biosystems, Foster City, CA) and the manufacturer's recommended protocols. The resulting nucleotide sequences of flanking Notch1 exonic DNA were used to design oligonucleotide primers for use in PCR to screen for proviral integrations into the Notch1 gene in additional tumors. PCR was performed on tumor DNA using primers homologous to the MMuLV LTR (5′-CCCGTGTATCCAATAAACCCTCTTG-3′) and Notch1(5′-GCCGTAGTGGGTTGTACTGGC-3′). Amplified DNA fragments were gel purified and used as templates for automated sequence analyses.
Notch1-MLV fusion RNA transcripts were detected and analyzed by RT-PCR. Polyadenylated RNA was isolated from snap frozen tumor tissues using Fastrack 2.0 (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. Complementary DNA was synthesized using oligo (dT)12-18 and Superscript II reverse transcriptase (Gibco BRL) according to the manufacturer's protocol. PCR was performed using primers homologous to the U3 portion of the MMuLV LTR (5′-CCATCTGTTCCTGACCTTGATCTG-3′ or 5′-CAAGAGGGTTTATTGGATACACGGG-3′) and Notch1(5′-CACCTGCCTGGTATGCCTGACA-3′ or 5′-CCTGGGCATCAGCCACTTGAATG-3′) with Expand High Fidelity polymerases. The resulting DNA fragments were subjected to automated nucleotide sequencing.
Tumor analyses
Freshly dissected tissues were fixed overnight in buffered formalin and then embedded in paraffin using standard techniques. Paraffin-embedded tissues were sectioned at 5 to 6 μm thickness, mounted on slides, deparaffinized, and stained with hematoxylin and eosin. For FACS analyses, cells freshly isolated from the thymus were stained, and the fluorescence was analyzed using a dual-laser FACS Vantage (Becton Dickinson Immunocytometry Systems, Mountain View, CA) with 4 decade logarithmic amplifier. Dead cells were detected by propidium iodide staining (1 μg/mL) and gated out electronically. Residual red blood cells were also gated out electronically. All antibodies were purchased from Pharmingen Research Products (San Diego, CA). Specificities of antibodies were as follows: FITC-conjugated 145-2C11 (anti-CD3ε); phycoerythrin (PE)-conjugated RM4-5 (anti-CD4); and biotinylated 53-6.7 (anti-CD8α).
Results
Neonatal retroviral infection accelerates development of thymic lymphomas in E2A-PBX1 transgenic mice
Transgenic mice expressing the E2A-PBX1 chimeric oncogene in the T-lymphoid compartment are highly susceptible to development of malignant lymphomas that arise with an average latency of more than 150 days.33 The long latency and clonal composition of the lymphomas indicated that forced expression ofE2A-PBX1 alone was not sufficient for induction of the malignant phenotype, which likely required one or more secondary events in addition to the E2A-PBX1 transgene. We used retroviral mutagenesis38,39 in a genetic screen to identify genes that cooperate with E2A-PBX1 to accelerate the development of tumors and shorten the survival of E2A-PBX1 mice. Newborn mice resulting from backcrosses of E2A-PBX1 transgenic mice were injected with MMuLV. Malignant lymphomas developed with a mean latency of 75 days in transgenic mice that were infected neonatally with virus (Figure 1A), which was significantly shorter than the time required for lymphoma development in transgenic littermates that had not been injected with virus (mean = 180 days). In addition, it was significantly different from that displayed by virus-injected, nontransgenic littermates that exhibited a reduced survival compared with nontransgenic, noninjected mice, consistent with previous observations.40
Neonatal MMuLV infection accelerates development of thymic lymphomas in
E2A-PBX1 transgenic mice. (A) The survival for a cohort (n = 10) of E2A-PBX1 mice is compared with that for transgenic (n = 9) and nontransgenic (n = 10) littermates injected neonatally with MMuLV. Deaths were the result of malignant lymphoma confirmed by histologic examination. (B) DNA isolated from lymphomas was analyzed by Southern blot analysis using a probe specific for the MMuLV LTR. DNA was digested with NheI (N), which cuts in the U3 region of the proviral LTR, and EcoRI, which bisects the provirus (N+E). Using these conditions, proviral DNAs were detected as 2 bands, a common one of 2 kb containing a portion of the MMuLV genome and 5′ LTR, as well as a second band of unique size containing the 3′ LTR and flanking mouse genomic DNA. Clonal LTR integrations are indicated by arrows. En, endogenous cross-hybridizing retroviral genome. Ex, fragment of exogenous viral genomes containing 5′ LTR released by double enzyme digestion.
Neonatal MMuLV infection accelerates development of thymic lymphomas in
E2A-PBX1 transgenic mice. (A) The survival for a cohort (n = 10) of E2A-PBX1 mice is compared with that for transgenic (n = 9) and nontransgenic (n = 10) littermates injected neonatally with MMuLV. Deaths were the result of malignant lymphoma confirmed by histologic examination. (B) DNA isolated from lymphomas was analyzed by Southern blot analysis using a probe specific for the MMuLV LTR. DNA was digested with NheI (N), which cuts in the U3 region of the proviral LTR, and EcoRI, which bisects the provirus (N+E). Using these conditions, proviral DNAs were detected as 2 bands, a common one of 2 kb containing a portion of the MMuLV genome and 5′ LTR, as well as a second band of unique size containing the 3′ LTR and flanking mouse genomic DNA. Clonal LTR integrations are indicated by arrows. En, endogenous cross-hybridizing retroviral genome. Ex, fragment of exogenous viral genomes containing 5′ LTR released by double enzyme digestion.
Tumors that arose in MMuLV-infected E2A-PBX1 mice primarily involved the thymus and were essentially indistinguishable from those in E2A-PBX1 mice (data not shown). They comprised clonal populations of T-lineage progenitors expressing surface antigens CD3, 4, and 8 characteristic of mid-thymocytes. Histologic features were those of diffuse high-grade lymphomas that were usually metastatic to the spleen, lymph nodes, liver, and lungs.
Proviral integration patterns were evaluated by Southern blot analysis using a probe specific for the U3 region of the viral LTR. In addition to endogenous proviral genomes detected with the U3 probe, tumors characteristically displayed 1 to 5 unique bands corresponding to acquired proviral integrations (Figure 1B). Therefore, neonatal MMuLV injection of E2A-PBX1 mice resulted in a substantially accelerated development of thymic lymphomas that harbored clonal integrations of proviral DNA.
Notch1 is frequently targeted by MMuLV proviral insertions in E2A-PBX1 mice
To characterize the sites of MMuLV insertion, genomic DNA fragments that hybridized with the U3 probe were cloned and sequenced. Tumors with the least number of proviral insertions were used in an effort to isolate those that may be specifically cooperating with the transgene. Hybridizing DNA fragments of interest were purified by agarose gel electrophoresis, ligated into pUC19, and used as templates for seminested PCR. Resulting DNA fragments were subjected to nucleotide sequence analyses. Using this approach, one of several proviral insertions analyzed was found to involve theNotch1 gene.
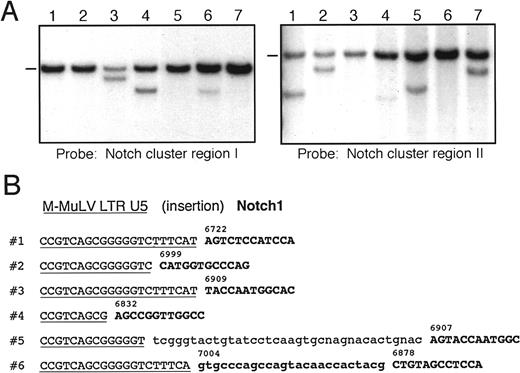
To establish the prevalence of proviral insertions inNotch1, DNA from accelerated tumors was examined by Southern blot analysis using probes specific for Notch1. A frequent site (cluster region I) for proviral insertions into Notch1was identified by previous studies of malignant lymphomas induced by MMuLV neonatal infections in MMTVD/myctransgenic mice.41 Using a probe for this central region of the Notch1 gene that spans exons encoding the LNR and a portion of the EGFR domains, approximately 50% of accelerated tumors from E2A-PBX1 mice displayed Notch1 gene rearrangements (Figure 2A). Thus, a substantial proportion of the accelerated tumors contained proviral-induced mutations of the Notch1 gene, thereby implicating it as a frequent collaborating gene inE2A-PBX1–initiated lymphomagenesis.
Proviral insertions in accelerated tumors of
E2A-PBX1 mice frequently target theNotch1 gene with a predilection for its 3′ exonic sequences. (A) Southern blot analyses of tumor DNAs using probes specific for cluster regions I (left panel) or II (right panel) in theNotch1 gene. Analysis for cluster region I used aNotch1 cDNA fragment spanning nucleotides 3851-4861 onEcoRV-digested DNA. Rearrangements affecting cluster region II were detected with a 0.9-kb fragment of Notch1 cDNA (nucleotides 7053-7957) on DNA digested with KpnI. Dashes indicate migrations of germline Notch1 DNA fragments. All other bands correspond to clonally rearranged Notch1 genes. (B) Nucleotide sequences are shown for junctions of LTR and genomic DNA at proviral integration sites in tumors. Sequences homologous to the U5 portion of the 3′ LTR are underlined; Notch1 exonic sequences are in bold and numbers indicate corresponding nucleotide position of the Notch1 open reading frame. Nucleotide insertions observed in 2 of the cases are shown in lower case.
Proviral insertions in accelerated tumors of
E2A-PBX1 mice frequently target theNotch1 gene with a predilection for its 3′ exonic sequences. (A) Southern blot analyses of tumor DNAs using probes specific for cluster regions I (left panel) or II (right panel) in theNotch1 gene. Analysis for cluster region I used aNotch1 cDNA fragment spanning nucleotides 3851-4861 onEcoRV-digested DNA. Rearrangements affecting cluster region II were detected with a 0.9-kb fragment of Notch1 cDNA (nucleotides 7053-7957) on DNA digested with KpnI. Dashes indicate migrations of germline Notch1 DNA fragments. All other bands correspond to clonally rearranged Notch1 genes. (B) Nucleotide sequences are shown for junctions of LTR and genomic DNA at proviral integration sites in tumors. Sequences homologous to the U5 portion of the 3′ LTR are underlined; Notch1 exonic sequences are in bold and numbers indicate corresponding nucleotide position of the Notch1 open reading frame. Nucleotide insertions observed in 2 of the cases are shown in lower case.
However, a Notch1 DNA rearrangement was not detected in cluster region I for the tumor from which we originally cloned aNotch1 proviral insertion. Therefore, Southern blot analyses were also conducted using an additional Notch1 probe flanking the site of this proviral insertion. Notch1 gene rearrangements were detected in more than 50% of cases with this second probe. Most of the cases detected with this probe lacked rearrangements in cluster region I (Figure 2A). This probe therefore defined a second cluster region for proviral insertions that appeared to be distant from cluster region I by mapping and sequencing analyses (see below). Rare tumors contained DNA rearrangements in both regions of Notch1 (Figure 2A, lane 4). Different hybridization intensities of rearranged bands for cluster region I versus II in cases with double insertions suggested that they were present in different subpopulations within the same tumor mass. Overall, 88% of the accelerated tumors that arose in E2A-PBX1 mice contained DNA rearrangements of the Notch1 gene, indicating that it was targeted at high frequency by proviral insertions, which clustered in 2 different regions of the gene.
A second cluster site for proviral insertions in E2A-PBX1 mice targets Notch1 3′ exonic sequences
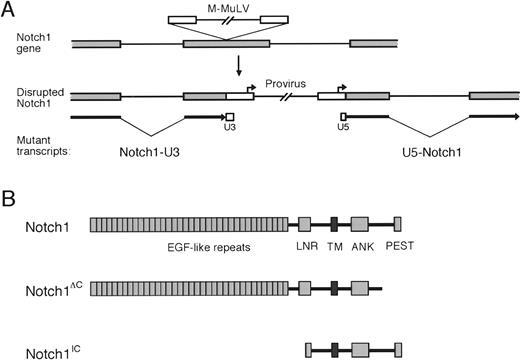
To further characterize the proviral insertions in cluster region II of the Notch1 gene, PCR was performed on tumor DNAs using primers homologous to the proviral LTR andNotch1 exonic sequences from cluster region II. Amplification products were observed in several tumors fromE2A-PBX1 transgenic mice injected with MMuLV but not in tumors from noninjected mice. Nucleotide sequence analyses of the amplification products showed similar configurations of U5 LTR sequences fused to Notch1 (Figure 2B). The precise point of fusion with Notch1 differed in each of the analyzed tumors but occurred in a relatively short segment of the gene-encoding nucleotides 6832-7225 of the Notch1 cDNA and contained in a single exon. In 2 of the tumors, nucleotide insertions were observed between LTR and Notch1 sequences. In one case, the inserted nucleotides were of unknown origin (case 5), in the other they originated from further 3′ in Notch1 (case 6). The fusion of LTR and Notch1 coding sequences indicated that proviral integrations had occurred into Notch1 exonic sequences (schematically shown in Figure 3A). The exon that was targeted by these events constituted a 3′ portion of theNotch1 gene coding for amino acids between the ankyrin repeats and the C-terminal PEST domain.
Effects of MMuLV insertions on the
Notch1 gene, transcripts, and protein products.(A) Disruption of Notch1 exonic sequences by the MMuLV proviral genome is shown schematically. Structures of mutatedNotch1 transcripts that could potentially result from integration into Notch1 exons are shown below. (B) Predicted Notch1 protein products resulting from 2 types of retroviral insertions in thymic lymphomas are compared with wild-type Notch1. Notch 3′ deletions remove all of PEST domain and varying portions of the transactivation domain reported by Kurooka et al.42
Effects of MMuLV insertions on the
Notch1 gene, transcripts, and protein products.(A) Disruption of Notch1 exonic sequences by the MMuLV proviral genome is shown schematically. Structures of mutatedNotch1 transcripts that could potentially result from integration into Notch1 exons are shown below. (B) Predicted Notch1 protein products resulting from 2 types of retroviral insertions in thymic lymphomas are compared with wild-type Notch1. Notch 3′ deletions remove all of PEST domain and varying portions of the transactivation domain reported by Kurooka et al.42
Lymphomas arising with shortened latencies express mutated Notch1 gene products
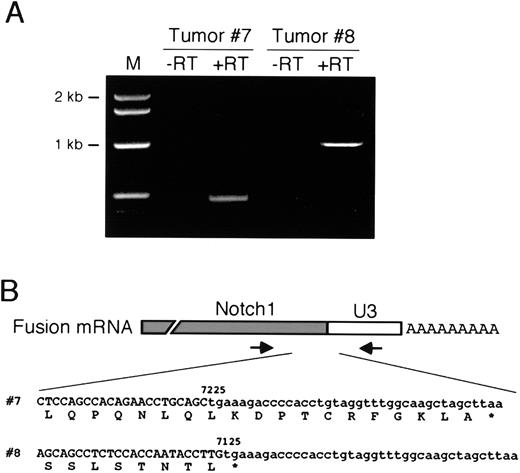
Proviral insertions into exonic regions ofNotch1 were predicted to disrupt the production of appropriately processed Notch1 transcripts. Because proviral LTRs contain transcriptional initiation and termination signals, they can induce production of prematurely truncated 5′ and/or ectopically initiated 3′ transcripts fused at one end to retroviral LTR sequences (shown schematically in Figure 3A). Potential synthesis ofNotch1-LTR fusion transcripts was evaluated by RT-PCR analysis of RNA using primers homologous to Notch1 and the U3 segment of the LTR. Under these conditions, specific amplification products were detected in accelerated tumors harboring 3′Notch1 proviral insertions (Figure4A). Nucleotide sequence analyses of the PCR products showed that 5′ Notch1 sequences were fused directly to U3 sequences of the MMuLV LTR (Figure 4B). Translation of the sequences demonstrated that the Notch1 open-reading frame prematurely terminated downstream of the fusion site because of in-frame stop codons in the U3 portion of the chimeric transcripts. As a result, the transcripts were predicted to encode mutated Notch1 proteins lacking C-terminal amino acids that encode the PEST domain (hereafter referred to as Notch1ΔC and schematically illustrated in Figure 3B) and a portion of the transactivation domain.42
Expression of mutated
Notch1 gene products. (A) RT-PCR was performed on RNA isolated from 2 tumors harboring 3′ proviral insertions in theNotch1 gene. Amplification products were only obtained if reverse transcriptase was present in the cDNA synthesis reactions (+RT). (B) Partial nucleotide sequences obtained on the RT-PCR products in panel A span the junctions of Notch1 and LTR U3 segments of the fusion transcripts as shown schematically above. Numbers indicate the Notch1 nucleotide at the site of fusion and are based on the sequence of the wild-type Notch1cDNA.
Expression of mutated
Notch1 gene products. (A) RT-PCR was performed on RNA isolated from 2 tumors harboring 3′ proviral insertions in theNotch1 gene. Amplification products were only obtained if reverse transcriptase was present in the cDNA synthesis reactions (+RT). (B) Partial nucleotide sequences obtained on the RT-PCR products in panel A span the junctions of Notch1 and LTR U3 segments of the fusion transcripts as shown schematically above. Numbers indicate the Notch1 nucleotide at the site of fusion and are based on the sequence of the wild-type Notch1cDNA.
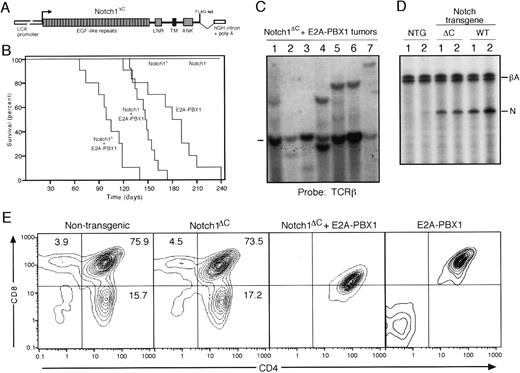
Mice transgenic for both Notch1ΔC and E2A-PBX1 manifest accelerated development of thymic lymphomas
To evaluate the oncogenic properties of Notch1ΔC, we constructed transgenic mice expressing this protein in the thymic compartment. A Notch1 cDNA lacking 3′ sequences encoding the 238 C-terminal amino acids (all of PEST domain and most of the transactivation domain) was expressed under control of the LCK proximal promoter that provides high-level expression in the thymic compartment (Figure5A). Two independent lines ofNotch1ΔC mice were evaluated and found to have no detectable alterations in their thymocyte subset ratios compared with normal mice by FACS analysis at 12 weeks of age (Figure 5E). This contrasts with the substantial elevations in CD8+ T cells observed in Notch1ICΔC transgenic mice28 and with the disorganized thymocyte differentiation observed in E2A-PBX1 mice33(Figure 5E).
Accelerated tumorigenesis in
Notch1ΔC/E2A-PBX1double transgenic mice. (A) Schematic illustration of the transgene construct consisting of aNotch1ΔC cDNA under control of theLCK proximal promoter. (B) The survival for cohorts ofNotch1ΔC/E2A-PBX1 (n = 10) orNotch1/E2A-PBX1 (n = 12) double transgenic mice is compared with that for littermates that are singly transgenic for either E2A-PBX1 (n = 10), Notch1 (n = 10), orNotch1ΔC (n = 10). Deaths were the result of malignant lymphoma confirmed by histologic examination. (C) Southern blot analysis of T-cell receptor gene rearrangements in lymphomas. Dash indicates migration of germline bands. (D) Quantitative RT-PCR analysis of transgene RNA expressed in thymi from wild-type and transgenic mice identified at the top of the gel lanes. Products derived from β-actin control and Notch transgenes are indicated to the right. (E) FACS analysis of CD4 and CD8 expression in thymocytes of 12-week-old wild-type, Notch1ΔC or E2A-PBX1single transgenic and Notch1ΔC/E2A-PBX1 double transgenic mice.
Accelerated tumorigenesis in
Notch1ΔC/E2A-PBX1double transgenic mice. (A) Schematic illustration of the transgene construct consisting of aNotch1ΔC cDNA under control of theLCK proximal promoter. (B) The survival for cohorts ofNotch1ΔC/E2A-PBX1 (n = 10) orNotch1/E2A-PBX1 (n = 12) double transgenic mice is compared with that for littermates that are singly transgenic for either E2A-PBX1 (n = 10), Notch1 (n = 10), orNotch1ΔC (n = 10). Deaths were the result of malignant lymphoma confirmed by histologic examination. (C) Southern blot analysis of T-cell receptor gene rearrangements in lymphomas. Dash indicates migration of germline bands. (D) Quantitative RT-PCR analysis of transgene RNA expressed in thymi from wild-type and transgenic mice identified at the top of the gel lanes. Products derived from β-actin control and Notch transgenes are indicated to the right. (E) FACS analysis of CD4 and CD8 expression in thymocytes of 12-week-old wild-type, Notch1ΔC or E2A-PBX1single transgenic and Notch1ΔC/E2A-PBX1 double transgenic mice.
Notch1ΔC transgenic mice were intercrossed with E2A-PBX1 mice to obtain progeny that were transgenic for both genes. Of the 64 total intercross offspring, a normal male:female ratio was obtained and the expected Mendelian ratios of the various cohorts were observed confirming that theNotch1ΔC/E2A-PBX1 transgene combination was not lethal in utero. Animals from all cohorts displayed normal postnatal development until about 3 to 5 months of age at which point double transgenic mice developed aggressive lymphomas that caused the demise of all animals in this cohort (mean survival = 102 days) (Figure 5B). Single transgenic E2A-PBX1 mice also succumbed to malignant lymphomas but the latency was longer (mean = 180 days) consistent with previous observations.33 The difference in survival was statistically significant (P < .0001). In contrast, none of the single transgenicNotch1ΔC mice developed malignancies over an observation period of 12 months. For comparison, we also created transgenic mice that expressed the wild-type Notch1 gene under control of the LCK promoter. The wild-type and ΔCNotch1 transgenes were expressed at comparable levels in the thymi of transgenic mice as determined by quantitative RT-PCR (Figure5D). Double transgenic Notch1/E2A-PBX1 mice displayed a reduction in tumor latency (mean 148 days) but not as substantial as Notch1ΔC/E2A-PBX1mice (Figure 5B) providing further evidence for the activating effects of the ΔC mutation.
All Notch1ΔC/E2A-PBX1 double transgenic mice succumbed to lymphomas that primarily involved the thymus and spleen with spread to lymph nodes, bone marrow, and liver. The lymphomas were indistinguishable histologically from those arising in E2A-PBX1 mice (Figure 6). FACS analyses demonstrated surface expression of CD3, 4, and 8 consistent with a mid-thymocyte stage of maturation (Figure 5E), a phenotype identical to E2A-PBX1 single transgenic33 and MMuLV accelerated tumors. Analyses ofTCR gene configurations by Southern blotting showed clonally rearranged bands consistent with monoclonal origins for the tumors (Figure 5C). The clonal compositions suggested that the 2 transgenes were not sufficient for lymphomagenesis. This is consistent with the observed latency for tumor development and the absence of neonatal lymphomas. Therefore, as a single transgene,Notch1ΔC did not induce thymic lymphomas or alterations in thymocyte differentiation. However, the C-terminally truncated allele of Notch1 displayed a potent ability to collaborate with E2A-PBX1 and induce a substantial reduction in tumor latency.
Histologic features of tumors arising in transgenic mice.
Tissue sections were stained with hematoxylin and eosin for thymic tumors from E2A-PBX1 single (A) orNotch1ΔC/E2A-PBX1 compound (B) transgenic mice and liver infiltrated by lymphoma (C and D). Magnifications: A, B, and D, 200 ×; C, 40 ×.
Histologic features of tumors arising in transgenic mice.
Tissue sections were stained with hematoxylin and eosin for thymic tumors from E2A-PBX1 single (A) orNotch1ΔC/E2A-PBX1 compound (B) transgenic mice and liver infiltrated by lymphoma (C and D). Magnifications: A, B, and D, 200 ×; C, 40 ×.
Discussion
These studies demonstrate that E2A-PBX1 andNotch1 collaborate in lymphoid oncogenesis and suggest a possible mechanistic link in their mode of collaboration. When forcibly expressed in the lymphoid compartment of transgenic mice,E2A-PBX1 results in the development of T-lineage lymphoblastic lymphomas with a mean latency of 5 to 6 months.33 However, the latency for tumor induction byE2A-PBX1 is substantially shortened by neonatal MMuLV infection (Feldman et al43; this study). As demonstrated here, this acceleration in tumor development is associated with a high frequency (88%) of proviral insertions in the Notch1 gene. Two types of insertions were observed and resulted in either ectopic expression of NotchIC or deletion of the carboxy-terminal portion of Notch1 containing its PEST domain (Notch1ΔC). The latter has not been previously observed as a mechanism forNotch1 oncogenic activation. Unlike NotchIC,Notch1ΔC retains its ligand-binding domain and is not itself oncogenic as a transgene expressed in the thymus. Yet, Notch1ΔC displays a potent ability to collaborate withE2A-PBX1 in transgenic mice. These studies reveal a novel mechanism for oncogenic activation of Notch1 and raise the possibility of a ligand-dependent collaborative relationship between 2 cellular oncogenes that also contribute to cell fate determination during development.
Half of the Notch1 insertions we observed created a mutantNotchIC gene, which is known to contribute to thymocyte transformation in several different settings. It was originally discovered as an oncogene (TAN1) at the sites of chromosomal translocations in a subset of human lymphoblastic lymphomas.29 These translocations bisect theNotch1 gene, resulting in constitutive expression ofNotchIC, which lacks the extracellular ligand-binding domain. A pathogenetic role forNotchIC was conclusively demonstrated experimentally by retroviral gene transfer into primary mouse hematopoietic cells, which resulted in the preferential induction of acute T-lineage lymphomas.32 In studies similar to ours, NotchIC was found to be ectopically expressed after proviral insertions in a subset of thymic lymphomas arising inMMTVD/myc transgenic mice infected neonatally with MMuLV.41 Therefore, in both humans and mice, inappropriate signaling through the Notch1 pathway renders developing thymocytes highly susceptible to oncogenic conversion. Frequent ectopic NotchIC expression in accelerated tumors from E2A-PBX1 mice establishes that events downstream in the Notch1 signaling pathway synergize with E2A-PBX1 in thymocyte oncogenesis.
Although Notch1 signaling pathways have not been fully characterized, some of their downstream effects appear to abrogate bHLH protein expression and/or function through at least 2 distinct pathways. One pathway involves CBF-1/Su(H)44; whereas a second appears to be CBF-1 independent45,46 and impinges on E47 through Ras and Deltex.47 Perturbations of bHLH protein function through ectopic expression48-52 and/or loss of function53-57 may be a final common pathway in thymocyte neoplasia. Notch1 expression is modulated during thymocyte development58 and forced expression of NotchICperturbs the fate decisions of differentiating thymocytes.28,59 Notch1 is also capable of protecting T cells from apoptosis by a pathway that involves interactions with Nur77.60 Thus, it is possible that Notch1 collaborates in thymic lymphomagenesis by suppressing the apoptosis associated with forced expression of E2A-PBX133 61 in addition to perturbing bHLH-mediated developmental programs.
Many of the Notch1 retroviral insertions in accelerated tumors of E2A-PBX1 mice, however, were further 3′ in the gene and resulted in carboxy-terminal Notch1 truncations. Carboxy-terminal deletions mediated by retroviral insertions constitute a potent mechanism for activation of cellular oncogenes as reported for c-myb62 and Tpl-2.63 The C-termini of these proteins negatively regulate their respective functions in transcription and signal transduction.63,64In regards to Notch1, the deleted C-terminal region contains a PEST domain involved in protein turnover by targeting of proteins to the ubiquitin-proteosome complex for subsequent degradation.65Loss of the PEST domain would be expected to render Notch1ΔC more stable than its wild-type counterpart20 possibly enhancing signal transduction after ligand engagement and subsequent proteolytic release22,23of a more stable signaling form of NotchIC. C-terminal deletions comparable to Notch1ΔC of DrosophilaNotch and the C elegans homolog glp-1 produce dominant gain-of-function phenotypes.66,67 These are consistent with a negative regulatory function for sequences C-terminal to the ankyrin repeats of Notch. Interestingly, this region ofDrosophila Notch has been implicated in mediating heterologous interactions with Dishevelled and Numb proteins.68-70 Dishevelled functions genetically on both the Wingless and Notch signaling pathways68 and appears to mediate cross talk between these pathways in part by suppression of Notch. Direct interaction of Dishevelled with the C-terminal region of Notch is proposed to inhibit signal transduction. Thus, the available data are consistent with a negative regulatory role for the Notch C-terminus and its loss would be expected to enhance Notch signaling.
Unlike the more conventional NotchIC, which is a ligand independent oncogenic mutant of the Notch receptor, Notch1ΔC retains its ligand binding domain. Its ability to synergize with E2A-PBX1 in this capacity would therefore appear to require engagement of extracellular ligands to initiate Notch receptor signaling. This is consistent with comparable mutants inDrosophila and C elegans whose gain-of-function properties do not circumvent the need for ligand.67,71 Notch ligands are expressed in the normal thymus,72 although it is not yet clear if they are derived from lymphoid elements or, as in bone marrow, from stromal cells.73 Despite their expression in normal thymus, endogenous levels of Notch ligands did not enhance Notch pathway signaling sufficiently to perturb normal fate decisions of thymocytes in LCK-Notch1ΔC mice. This contrasts with the profound alterations in thymocyte subset ratios in mice transgenic for a mutant Notch1 gene (Notch1ICΔC) containing deletions of both the C-terminal and extracellular domains.28 Furthermore, Notch1ΔCalone was not oncogenic, which also contrasts withNotch1ICΔC, yet displayed strong synergy withE2A-PBX1. This raises the possibility that E2a-Pbx1 stimulates the expression of one or more Notch ligands on thymic stromal cells or neoplastic thymocytes themselves inE2A-PBX1 mice. If E2a-Pbx1 functions upstream of Notch to regulate ligand expression or activity, a possible consequence would be that Notch pathway signaling in thymocytes expressing E2a-Pbx1 may be limited by receptor throughput, not ligand availability. Mutations, such as Notch1ΔC, that enhance Notch receptor-generated signals, may provide a selective advantage toE2A-PBX1–expressing cells leading to accelerated development of lymphomas in Notch1ΔC/E2A-PBX1mice. Recent studies have shown that Rel/NF-κB triggers Notch pathway signaling by inducing expression of the Notch ligand Jagged1.74 This provides important precedent for upstream regulation of Notch ligands by signaling pathways involved in lymphocyte proliferation and function. Additional studies are required to ascertain if any of the several known Notch ligands or modifiers of Notch signaling are affected by E2a-Pbx1.
Acknowledgments
We thank Gerry Weinmaster for Notch cDNAs and helpful discussions. We thank David Feldman, Cathy Schnabel, and Jorge DiMartino for comments on the manuscript. We acknowledge Yelena Marchuk and Yanru Chen for microinjections, Kathie Jones for histotechnology support, and Phil Verzola for photography.
Supported by grants CA34233, AI-07290, and CA42971 from the National Institutes of Health. B.J.F. was supported by funds from the Lucille P. Markey Charitable Trust.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michael L. Cleary, Department of Pathology, Stanford University Medical Center, Stanford, CA 94305; e-mail:michael.cleary@stanford.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal