Abstract
Multiple myeloma (MM) is a hypoproliferative malignancy yielding informative karyotypes in no more than 30% of newly diagnosed cases. Although cytogenetic and molecular deletion of chromosome 13 is associated with poor prognosis, a MM tumor suppressor gene (TSG) has not been identified. To localize a minimal deleted region of chromosome 13, clonotypic plasma cells from 50 consecutive patients with MM were subjected to interphase fluorescence in situ hybridization (FISH) analysis using a panel of 11 probes spanning the entire long arm of chromosome 13. Whereas chromosome 13 abnormalities were absent in plasma cells from 25 normal donors, 86% of patients with MM demonstrated such aberrations. Heterogeneity, both in deletion frequency and extent, was confirmed by simultaneous FISH with 2 chromosome 13 probes. Deletion hot spots were noted at D13S272 (70%) and D13S31 (64%), 2 unlinked loci at 13q14. Homozygous deletions at these loci occurred in 12% (simultaneously in 8%) of the cases. Molecular deletions were found in all 14 patients with morphologic deletions, in 21 of 24 with uninformative karyotypes, and 8 of 12 patients with karyotype abnormalities lacking chromosome 13 deletion. Homozygous deletion of any marker was noted in 4% with low and in 36% with higher plasma cell labeling index greater than 0.4% (P = .01). The absence of increasing deletion incidence and extent with therapy duration suggests that the observed lesions are not induced by treatment. The high incidence and extent of chromosome 13 deletions require the correlation of specific deletion(s) with poor prognosis. These analyses will provide valuable guidance toward cloning of an MM-TSG.
Introduction
Multiple myeloma (MM) is a clonal B-cell malignancy, putatively originating in germinal centers of lymph nodes, that homes to and expands in the bone marrow.1 As a tumor of terminally differentiated plasma cells, its proliferative activity is usually low so that metaphase karyotyping is difficult and informative in only one third of newly diagnosed patients.2,3 However, DNA content abnormalities, as determined by fluorescence in situ hybridization (FISH) and flow cytometry, are almost universal.4-6 In contrast to other hematologic malignancies, karyotypes in MM are typically complex, almost representing a hallmark of this disease.7-9 Despite the genomic chaos, several recurring chromosomal abnormalities have been identified including structural rearrangements (eg, translocations of 14q32), trisomies (eg, chromosome 15), and monosomies (eg, chromosome 13).9 Although recurrent translocations of MYC, BCL1, and BCL2into the immunoglobulin heavy chain locus on chromosome 14q32 are fundamental to the development of many B-cell leukemias and lymphomas,10,11 14q32 translocations in MM involve a great diversity of partner chromosomes,12-16 none of which, however, define a distinct clinical entity.
Recent cytogenetic studies, performed in the context of both standard and high-dose therapy, have shown that partial or complete deletion of chromosome 13 present in 15% to 20% of newly diagnosed MM imparts a poor prognosis.17-20 Similar observations have also been reported for molecular deletions of RB1 andD13S319 in conventional therapy settings.21 22Collectively, these data strongly suggest the presence of MM tumor suppressor gene(s) on chromosome 13. In an effort to identify and eventually clone these gene(s), interphase FISH with 11 probes spanning the long arm of chromosome 13 was used to establish a first-generation molecular deletion map of this chromosome in myeloma.
Materials and methods
Cytogeneticdeletion map
A cytogenetic database of 1000 consecutive patients with MM treated at the University of Arkansas for Medical Sciences was scrutinized for chromosome 13 deletions and a cytogenetic deletion map established.
Probe isolation andlabeling
Eleven oligonucleotide primer pairs, specific for 9 sequence-tagged sites or microsatellite markers and 2 genes covering the q arm of human chromosome 13 and one pair for chromosome 10q were synthesized (Table 1). Chromosome 10 was chosen as a reference probe because structural or numeric abnormalities rarely affect this chromosome in myeloma.9,23 The band positions of probes were determined from the Genome DataBase (http://www.gdb.org) or from previous publications24(Figure 1). Human BAC clones were isolated by hybridization screening of the RPCI-11 BAC library (http://bacpac.med.buffalo.edu) with probes generated by polymerase chain reaction (PCR). High molecular weight BAC DNA was isolated using the KB-100 kit according to manufacturer's instructions (Genome Systems, St. Louis, MO). Spectrum-Green and/or Spectrum-Red-dUTP (Vysis, Downers Grove, IL) were incorporated into BAC DNA by nick translation according to the manufacturer's protocol (Vysis). Briefly, 1.0 μg of BAC DNA was mixed with 0.4 mmol/L Spectrum Green-dUTP, 0.2 mmol/L dTTP, and 0.6 mmol/L of each dATP, dCTP, and dGTP, 5 μL of 10 × reaction buffer and 10 μL of polymerase mix in 50 μL and incubated at 15°C for 3 hours. The reaction was terminated by incubation at 70°C for 10 minutes. The probe cocktail was mixed with 10 μg of human Cot-1 DNA (Life Technologies, Gaithersburg, MD) brought to a volume of 90 μL and 10 μL of 3 mol/L sodium acetate added. Cold ethanol (200 μL) was added and the mixture incubated at −20°C overnight. The sample was centrifuged for 30 minutes, the pellet washed with 70% ethanol and air-dried. The pellet was resuspended in Hybrisol VII (Oncor, Gaithersburg, MD) at a final concentration of 5 ng/μL. The solution was incubated at 75°C for 10 minutes, then 37°C for 2 hours. All probes were mapped to chromosome 13 by simultaneous hybridization of each Spectrum Red-dUTP labeled BAC clone with a Spectrum Green-dUTP labeled whole chromosome painting probe (Vysis) to metaphase chromosomes prepared from stimulated peripheral blood lymphocytes.
List of primers used to synthesize PCR probes for BAC library screening
| Marker . | Band . | Primer sequence . |
|---|---|---|
| 13-07-5′/3′ | 13q11 | AAGGTGCAGCGGGCT |
| CTTCACCTTCAGCCTCG | ||
| D13S221 | 13q12 | TAGCCATGATAGGAAATCAACC |
| GAGATCGTGCAGCACTTGT | ||
| BRCA2 | 13q12 | TAGGGAAAGCTTCATAAGTCAGTCTC |
| TGTCTAGACGTAGGTGAATAGTGAAGACT | ||
| RB | 13q14 | TTTTCCTCACTCATTTCCCACATC |
| ATCTGACACTATCTCCAGGTAAGC | ||
| D13S1150 | 13q14 | CTCTTGAGGGAAAAAAAAAATCA |
| CCAGGCAACCAACCAGTC | ||
| D13S272 | 13q14 | ATACAGACTTCCCAGTGGCT |
| AGCTATTAAAGTTCCCTGGATAAAT | ||
| AFMa301wB5 | 13q14 | GAATGCAGGTGTACCTATCAAC |
| ACTGAGTGACTGCTACCGAG | ||
| D13S25 | 13q14 | AAGCTTCTGTTTGTTCAAGA |
| GATTCTAATCAGATAGACTG | ||
| D13S31 | 13q21 | CAGATCGAGACAATAAGTGC |
| ATCTCTTAATTCCAGCATCC | ||
| D13S1619 | 13q22 | TGTGGTCTCCCAGTCACCTC |
| CCGTTCTTCCAGTGCTTGTC | ||
| D13S285 | 13q34 | ATATATGCACATCCATCCATG |
| GGCCAAAGATAGATAGCAAGGTA | ||
| D10S2142 | 10q | ACACAGTTACACTTGTGCACCC |
| AACAGAGGGAAGTGTCCCCT |
| Marker . | Band . | Primer sequence . |
|---|---|---|
| 13-07-5′/3′ | 13q11 | AAGGTGCAGCGGGCT |
| CTTCACCTTCAGCCTCG | ||
| D13S221 | 13q12 | TAGCCATGATAGGAAATCAACC |
| GAGATCGTGCAGCACTTGT | ||
| BRCA2 | 13q12 | TAGGGAAAGCTTCATAAGTCAGTCTC |
| TGTCTAGACGTAGGTGAATAGTGAAGACT | ||
| RB | 13q14 | TTTTCCTCACTCATTTCCCACATC |
| ATCTGACACTATCTCCAGGTAAGC | ||
| D13S1150 | 13q14 | CTCTTGAGGGAAAAAAAAAATCA |
| CCAGGCAACCAACCAGTC | ||
| D13S272 | 13q14 | ATACAGACTTCCCAGTGGCT |
| AGCTATTAAAGTTCCCTGGATAAAT | ||
| AFMa301wB5 | 13q14 | GAATGCAGGTGTACCTATCAAC |
| ACTGAGTGACTGCTACCGAG | ||
| D13S25 | 13q14 | AAGCTTCTGTTTGTTCAAGA |
| GATTCTAATCAGATAGACTG | ||
| D13S31 | 13q21 | CAGATCGAGACAATAAGTGC |
| ATCTCTTAATTCCAGCATCC | ||
| D13S1619 | 13q22 | TGTGGTCTCCCAGTCACCTC |
| CCGTTCTTCCAGTGCTTGTC | ||
| D13S285 | 13q34 | ATATATGCACATCCATCCATG |
| GGCCAAAGATAGATAGCAAGGTA | ||
| D10S2142 | 10q | ACACAGTTACACTTGTGCACCC |
| AACAGAGGGAAGTGTCCCCT |
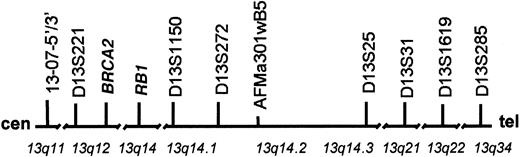
Chromosome 13 map.
This map of chromosome 13 with approximate band positions of TRI-FISH probes is oriented from centromere to telomere (left to right). The band designations are indicated below the line. The genes or sequence tagged site (STS) markers used to clone the BAC probes are depicted above the line. The chromosome length is not to scale and gaps have been placed in the chromosome to indicate large regions of nonlinkage.
Chromosome 13 map.
This map of chromosome 13 with approximate band positions of TRI-FISH probes is oriented from centromere to telomere (left to right). The band designations are indicated below the line. The genes or sequence tagged site (STS) markers used to clone the BAC probes are depicted above the line. The chromosome length is not to scale and gaps have been placed in the chromosome to indicate large regions of nonlinkage.
Triple color interphase FISH (TRI-FISH)
The TRI-FISH protocol was essentially as described by Ahmann and colleagues.27 Briefly, slides prepared above were treated as follows. The cellular DNA was denatured in 90% formamide/2 × standard sodium citrate (SSC) for 5 minutes at 37°C in a coplin jar. The slides were dehydrated in an ice-cold ethanol series (70% for 2 minutes, 85% for 2 minutes, and 100% for 2 minutes) then air-dried. Ten microliters of probe cocktail (1:1 mixture of spectrum-red labeled chromosome 13 probe and chromosome 10 reference probe) was added to each slide, coverslips added, and sealed with rubber cement. The slide was placed in a Hybrite (Vysis) humidified chamber and incubated at 75°C for 10 minutes, then 42°C for 16 hours. The coverslip was removed, and the slides were washed in 65% Formamide/2 × SSC at 45°C for 15 minutes, then once in 2 × SSC at 37°C for 8 minutes, followed by 2 washes in phosphate-buffered detergent (1 × PBD). The slides were then stained with 100 μL of a 1:20 dilution of either goat antihuman-κ or -λ light chain antibody conjugated with 7-amino-4-methylcourmarin-3-acitic acid (AMCA) (Vector Laboratories, Burlingame, CA) for 30 minutes in a humidified chamber. After incubation, the slides were washed 2 times in PBD. To enhance the signal, the slides was incubated with 100 μL of a 1:40 dilution of AMCA labeled rabbit-antigoat IgG antibody and incubated for 30 minutes at room temperature in a humidified chamber. Slides were washed 2 times in 1 × PBD, and Vectashield (Vector Laboratories) antifade and coverslips added. At least 50 AMCA-positive plasma cells were scored for the presence of 2 green and 2 red signals using an Olympus BX60 epifluorescence microscope equipped with appropriate filters for detecting fluoroisothiocyanate (FITC), Texas red, and DAPI. The triple color clone-specific images were captured using a Quips XL genetic workstation (Vysis).
Myeloma bone marrow
Bone marrow aspirates were collected in heparinized containers from patients with active myeloma during scheduled clinic visits. Signed informed consents for use of samples in research studies were obtained and kept on record. Bone marrow cells were separated by density gradient centrifugation over Ficoll-Hypaque (Pharmacia, Piscataway, NJ). The cells in the light density fraction (specific gravity ≤ 1.077) were resuspended in cell culture media plus 10% fetal bovine serum. Approximately 40 000 mononuclear cells were cytocentrifuged at 1000g for 5 minutes at room temperature. Twenty-four slides from each sample were prepared. The slides were air dried overnight, then fixed in 100% ethanol for 5 minutes at room temperature and baked in a dry 37°C incubator for 6 hours. Slides were either used immediately in TRI-FISH experiments or stored at −20°C then used later without loss of signal.
TRI-FISH analysis of normal bone marrow
Twenty-five bone marrow aspirates from normal donors were analyzed to determine the random loss or gain of signals in plasma cells and nonplasma cells for each of the 11 chromosome 13 probes used in this study. The age of the normal allogeneic bone marrow donors ranged from 1 to 40 years with a mean of 30 years, representing 10 female and 15 male donors. An average of 85 plasma cells (range, 30-129) and 104 nonplasma cells (range, 55-128) were examined with each of the 11 probes for chromosome 13. This analysis revealed a deletion or duplication incidence of less than or equal to 6% (SD, 4%) for all probes. The incidence of homozygous deletion for any probe was exceedingly rare (1%). A threshold of 20% for loss or gain in chromosome 13-probe signal in plasma cells was considered evidence that trisomy or monosomy had occurred. The 20% cut-off was also chosen for homozygous deletions to facilitate the localization of potentially important regions of chromosome loss.
Spectral karyotyping (SKY) and metaphase FISH
The SKY procedure has been previously described.34 Metaphase chromosomes were pretreated for FISH by incubation in prewarmed 2 × SSC (pH 7.0, 37°C) for 15 minutes, followed by 0.1 N HCl/0.05% Triton X-100 for 15 minutes, then in 2 × SSC, and finally washed in 1 × PBS. Slides were then immersed in 1% formaldehyde for 10 minutes, washed in 1 × PBS twice and 2 × SSC once, then an ethanol series (70%, 85%, 100%, 2 minutes each), and air dried at room temperature. The BAC probes for D13S221 and D13S285 were directly labeled with Spectrum-Green-dUTP and Spectrum-Red-dUTP, respectively. The chromosome 8 probe, directly labeled with Spectrum Orange-dUTP was obtained from Vysis. The chromosome probes were mixed with Hybrisol VII (Oncor) at a concentration of 5 ng/μL and 10 μL was added to areas containing abundant metapahases. Slides were brought to 75°C for 10 minutes, then to 37°C for more than 16 hours in a humidified chamber. Slides were immersed in prewarmed 65% Formamide/2 × SSC (pH 7.0) for 15 minutes, at 43°C followed by a wash in 2 × SSC for 8 minutes at 37°C, then washed in 1 × PBD (Oncor) twice at room temperature, and finally washed in 1 × PBS. Ten microliters of DAPI/antifade (1:20) was added to each area and the slides viewed.
Results
Conventional cytogenetics of 13q arm deletions in 1000 myeloma patients
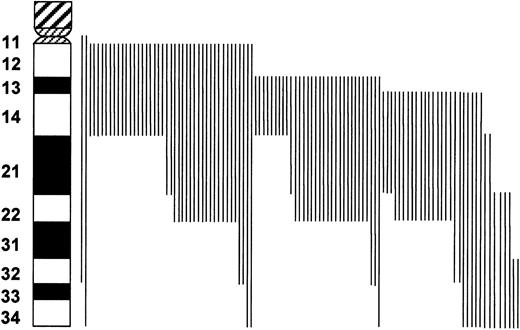
The cytogenetic deletion map of chromosome 13 is depicted in Figure 2. This map shows 13q arm deletions in 106 patients ordered with respect to the centromeric and telomeric deletion boundaries. Ninety-eight (90%) involved 13q14 and 73 (68%) involved the 13q21 region. Except for 8 cases with deletions distal to q14, the 13q14 band was deleted in each case and represented the minimal region of deletion overlap.
Schematic view of chromosome 13 deletions in 106 patients with MM as observed with G banding.
Each vertical line to the right of the chromosome indicates the part of chromosome 13 that has been deleted in one patient. The minimal region of deletion overlap appears to be in the 13q14 region.
Schematic view of chromosome 13 deletions in 106 patients with MM as observed with G banding.
Each vertical line to the right of the chromosome indicates the part of chromosome 13 that has been deleted in one patient. The minimal region of deletion overlap appears to be in the 13q14 region.
Chromosome 13 deletion analysis of monoclonal plasma cells from MM
A panel of 11 probes spanning the long arm of chromosome 13 (Figure 1) was used to analyze 50 patients with MM. TRI-FISH analysis (Figure 3A) showed molecular abnormalities in 86% of patients (Table2). Deletion frequency for any given probe varied and ranged from 20% (case 30) to more than 90% (case 9). Variation in the frequency of heterozygous and especially homozygous deletions of probes along the chromosome was observed within a given patient. Loss of all 11 probes, suggesting monosomy 13, was observed in 15 patients and of at least 8 probes in an additional 5 so that 40% had major molecular deletions. The most commonly deleted markers were D13S272 at 13q14 (76%), representing the sole anomaly in 2 cases (Figure 3B), and D13S31 at the 13q14-q21 band (68%), which were concurrently deleted in 59%. Homozygous deletions (complete loss of signal), present in 9 patients altogether, involved these 2 probes in 6 cases each, of which 4 revealed concurrent deletions of both probes. Homozygous deletions were also observed for the BRCA2locus at 13q12 and the marker D13S1619 at 13q22, but not forRB1. Homozygous deletions were never detected in the absence of heterozygous deletions of other probes. A normal diploid region flanked by trisomic markers was evidence of interstitial deletions in cases 13, 14, 15, 40, and 41. Trisomy restricted to telomeric markers, suggesting translocations of duplicated chromosomal material, was observed in cases 3, 27, 28, 32, and 48.
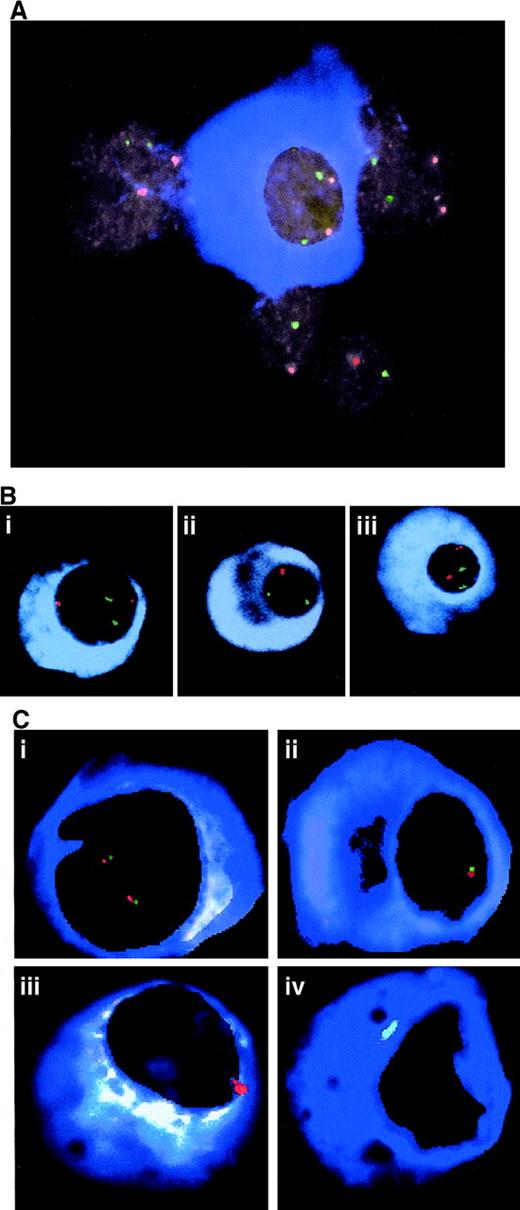
Analysis of bone marrow cells.
(A) Representative cells from a myeloma bone marrow as analyzed by TRI-FISH. A normal plasma cell (blue cytoplasm) is surrounded by several nonplasma cells. Note that all nuclei contain 2 red and 2 green signals. The green signal is derived from the chromosome 10 reference probe and the red signal originates from the chromosome 13 probe D13S272. (B) TRI-FISH indicates interstitial deletion of D13S272. Cells from case 26 were hybridized with (i) D13S1150, (ii) D13S272, and (iii) AFMA301wB5 probes (red) and the chromosome 10 probe (green). Note the loss of signal for D13S272 (ii), which occurred in 22% of the light chain-restricted plasma cells. Loss of signal was not observed for the other markers. (C) Clonal heterogeneity and dispersed interstitial deletions in myeloma. Bone marrow cells from case 50 were simultaneously hybridized with probes for D13S272 (red) and D13S31 (green). Approximately 5% of the cells were normal (i), 48% of the cells demonstrated monosomy (ii), 12% of the cells showed monosomy for D13S272 and complete loss of the D13S31 signal (iii), and 23% of the cells had no signal for either probe (iv).
Analysis of bone marrow cells.
(A) Representative cells from a myeloma bone marrow as analyzed by TRI-FISH. A normal plasma cell (blue cytoplasm) is surrounded by several nonplasma cells. Note that all nuclei contain 2 red and 2 green signals. The green signal is derived from the chromosome 10 reference probe and the red signal originates from the chromosome 13 probe D13S272. (B) TRI-FISH indicates interstitial deletion of D13S272. Cells from case 26 were hybridized with (i) D13S1150, (ii) D13S272, and (iii) AFMA301wB5 probes (red) and the chromosome 10 probe (green). Note the loss of signal for D13S272 (ii), which occurred in 22% of the light chain-restricted plasma cells. Loss of signal was not observed for the other markers. (C) Clonal heterogeneity and dispersed interstitial deletions in myeloma. Bone marrow cells from case 50 were simultaneously hybridized with probes for D13S272 (red) and D13S31 (green). Approximately 5% of the cells were normal (i), 48% of the cells demonstrated monosomy (ii), 12% of the cells showed monosomy for D13S272 and complete loss of the D13S31 signal (iii), and 23% of the cells had no signal for either probe (iv).
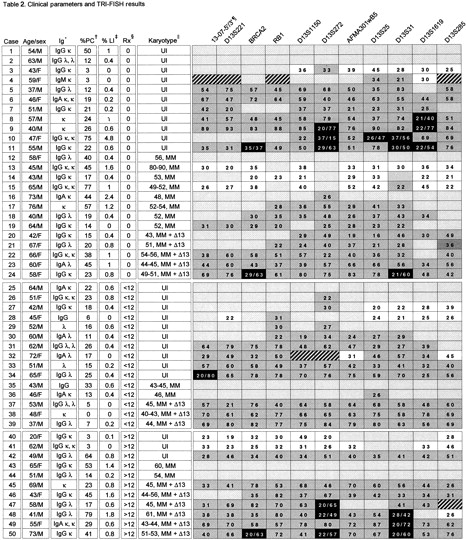
An organizational heirarchy (top to bottom) has been used to present the data in this table. The first level of organization is based on previous therapy (0,<12, and >12 months). Within each therapy group the cases are organized according to karotype (UI, MM, MM+Δ13). Within the each karyotype group the TRI-FISH data is organized based on the extent of molecular deletion.
Ig heavy chain isotype and light chain.
Percent plasma cells in bone marrow aspirate.
Percent myeloma cells incorporating BrdU.
Months of prior therapy as of the sample data.
UI = uniformative metaphases; MM = numeric and structural abnormalities consistent with myeloma without chromosome 13 abnormalities; MM+D13 myeloma karyotype plus alterations in chromosome 13. Number preceding MM or MM+Δ13 = numer of chromosomes in cell.
TRI-FISH results. Eleven TRI-FISH probes (top of each column) ordered from centromere to telomere (left to right). Number of FISH signals = box color: 3 = white; 2 = stippled; 1 = gray; 0 = black; not analyzed = hatched; Numbers in box = percent of cells with given abnormality; Two numbers = percent with 0 signal/percent with 1signal. Case #5 and #39 are from the same patient on different dates.
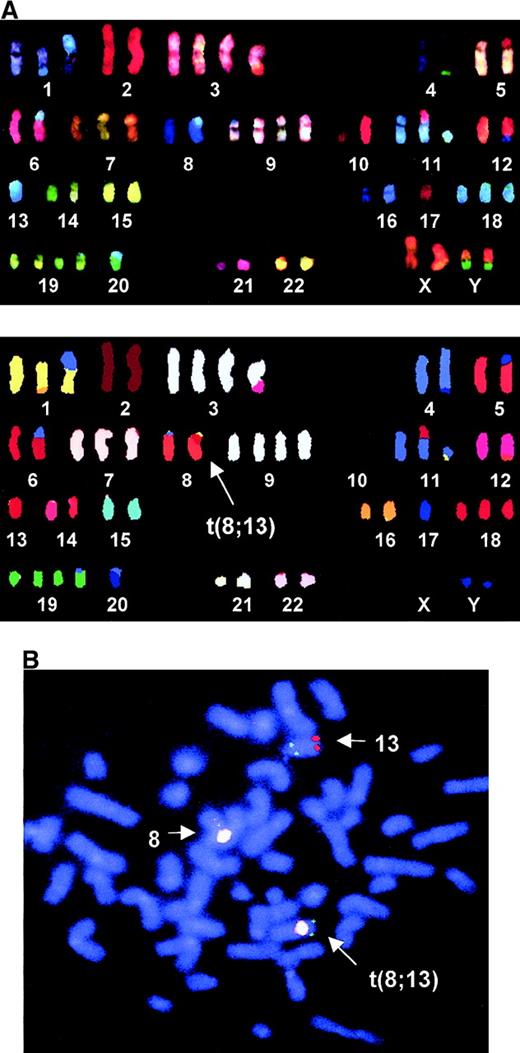
To demonstrate sensitivity of the TRI-FISH method, the results of TRI-FISH, SKY, and metaphase FISH from the same patient were compared. TRI-FISH analysis of case 46 showed that all probes from BRCA2 to the telomeric marker, D13S285, showed only one signal in more than 20% of the cells, whereas the probes 13-07-5′/3′ and D13S221 showed 2 signals in more than 80% (Table 2). The data suggested that although a large portion of one chromosome 13 had been deleted, the centromeric region remained disomic. SKY and metaphase FISH on chromosomes from the same patient showed the presence of an apparently normal chromosome 13 and a t(8;13) translocation that was not recognized by conventional G-banding (Figure 4). Eleven separate metaphase FISH experiments were performed using a commercial probe for the centromere of chromosome 8 labeled in orange, each of the chromosome 13 probes labeled in green, and the telomeric chromosome 13 probe labeled in red. The data showed that both 13-07-5′/3′ and D13S221 hybridized to a normal chromosome 13 (colocalization of a green and red signal on the same chromosome) and to chromosome 8 (colocalization of a green and orange signal) (Figure 4). Although all remaining probes along the chromosome showed colocalization of a green and red signal on the same chromosome (normal chromosome 13) they failed to cohybridize with the chromosome 8 probe. Thus, SKY and metaphase FISH confirm and extend the TRI-FISH data in this case, in that a q arm deletion had occurred, but that the centromeric region was disomic and involved in a subtle t(8;13) translocation.
Results from case 46.
(A) SKY of bone marrow chromosomes from case 46. Upper panel shows display of colors representation; lower panel shows karyotype of spectra-based classification chromosomes. Note the monosomy of chromosome 13 and the subtle t(8;13) translocation. (B) Metaphase-FISH of bone marrow chromosomes from case 46. The t(8;13) is identified by metaphase showing colocalization of D13S221 (green) and the chromosome 8 centromere-specific probe CEP8 (orange). The most centromeric marker in our panel, 13-07-5′/3′, also showed colocalization with the orange signal. Other markers along the chromosome were tested and none showed colocalization (data not shown). The normal chromosome 13 is identified by the colocalizing green and red signals. The red signal is derived from the telomeric probe D13S285. The normal chromosome 8 is identified by the single orange signal. The 2 orange signals derived from the sister chromatids are overlapping due to overexposure. All chromosomes are stained blue with DAPI.
Results from case 46.
(A) SKY of bone marrow chromosomes from case 46. Upper panel shows display of colors representation; lower panel shows karyotype of spectra-based classification chromosomes. Note the monosomy of chromosome 13 and the subtle t(8;13) translocation. (B) Metaphase-FISH of bone marrow chromosomes from case 46. The t(8;13) is identified by metaphase showing colocalization of D13S221 (green) and the chromosome 8 centromere-specific probe CEP8 (orange). The most centromeric marker in our panel, 13-07-5′/3′, also showed colocalization with the orange signal. Other markers along the chromosome were tested and none showed colocalization (data not shown). The normal chromosome 13 is identified by the colocalizing green and red signals. The red signal is derived from the telomeric probe D13S285. The normal chromosome 8 is identified by the single orange signal. The 2 orange signals derived from the sister chromatids are overlapping due to overexposure. All chromosomes are stained blue with DAPI.
Chromosome 13 deletion heterogeneity in MM
Because all TRI-FISH experiments used a single chromosome 13 probe, these analyses could not address the question of whether deletions of multiple probes occurred in the same or different tumor cells. Simultaneous hybridization of 2 probes carrying different fluorophores was therefore carried out in 3 patients. A representative case (case 50) revealed 5% of the clonotypic plasma cells to be diploid for both probes (colocalization of 2 probes in 2 distinct regions of the nucleus), 48% to be monosomic for both probes (a single red and green colocalizing signal), 12% monomeric for only D13S272 and nullisomic for D13S31 (single red signal, no green signal), and 23% to be nullisomic for both probes (no signals). Such heterogeneity in this and the other 2 cases (not shown) is consistent with the presence of distinct subpopulations of tumor cells with varying degrees of deletion in this chromosome.
Association of molecular chromosome 13 deletion and clinical parameters
There was no statistically significant association of molecular deletion to age, sex, Durie-Salmon tumor stage, immunoglobulin isotype, or serum β2-microglobulin (data not shown). Molecular deletions were seen in all 14 patients with cytogenetic chromosome deletions, in 21 of 24 of those with uninformative diploid karyotypes, and even in 8 of 12 with myeloma karyotypes without obvious abnormalities of chromosome 13 (Table 2). Molecular deletions were more frequently observed in case of higher tumor proliferative activity, because 9 of 25 patients with plasma call labeling indices more than 0.4% (above the median value) had homozygous deletions compared to 1 of 24 with values of 0.4% or less (P = .01). Deletion of chromosome 13 markers occurred with similar frequency regardless of prior therapy in 22 of 24 untreated patients, 13 of 15 treated for up to 12 months, and 9 of 11 treated for more than 12 months (P = 1.0) (Table 2).
Discussion
This first comprehensive analysis of molecular chromosome 13 deletions in MM, using an 11-probe panel spanning the long arm of this chromosome, has revealed a surprisingly high incidence of abnormalities. Although 86% of patients demonstrated at least one abnormality, 75% of the deletions were accounted for by the probe D13S272 and 66% by D13S31, both of which were concurrently deleted in 59%. Together with the observation of homozygous deletions of these 2 probes in 12%, our findings suggest a pathogenetic role of the loss of these 2 loci in MM, especially because deletion extent and frequency were not influenced by prior therapy and hence treatment induced. However, a more complex scenario with participation of other deletions in disease progression is likely because both RB1 andBRCA2 were also deleted in 60% and 48%, respectively.
The high incidence of deletion detected here (86%) poses a dilemma with regard to the adverse prognostic implications recently reported using single probe FISH for the RB1 and D13S319 loci (40% of patients).21 22 In this study, these 2 probes were deleted in 52% and 70%, respectively, of newly diagnosed patients (data confirmed in an additional 35 newly diagnosed patients, J.S., unpublished data). Prospective investigations are in progress to determine which of these and additional deleted regions, alone or in combination, confer the observed poor prognosis.
The analysis of concurrent deletions of 2 or more loci suspected in MM progression requires mutlicolor analysis. Preliminary data with the D13S272 and D13S31 probes indeed indicated marked deletion heterogeneity in individual patients comprising cells with homozygous deletions for one and both probes, monosomy, as well as complete lack of deletion. Given the higher proliferative activity in patients exhibiting homozygous chromosome 13 probe deletions, a proliferation hierarchy may also exist within an individual patient's tumor population, with a cell division advantage conferred by the extent of chromosome 13 deletion. To resolve this issue, concurrent cytokinetic analysis will be required, probably best performed by in vivo BrdU labeling to obtain authentic information. Contrary to other chromosomal aberrations, especially numeric anomalies (B.B., unpublished data), chromosome 13 deletion not only implies rapid disease recurrence but also initial drug resistance.26 Studies of apoptotic pathways in patients lacking chromosome 13 deletion and in those with varying deletion extent should be informative.
Studies are in progress to determine whether the observed chromosome 13 deletion frequency and intrapatient deletion heterogeneity is chromosome or disease specific, especially vis-a-vis chronic lymphocytic leukemia (CLL) in which chromosome 13 deletion has been shown not to confer a dismal prognosis. The divergent clinical implications of chromosome 13 loss in MM and CLL27,28 may thus result from the deletion(s) of additional gene(s) in MM. If the inactivated gene happens to be the same in these 2 diseases, several alternative explanations can be envisioned to explain the difference in clinical phenotype. The loss of one functional copy of a gene (haploinsufficiency) may be sufficient to impart the poor clinical phenotype seen in MM. It is also possible that redundant biochemical pathways functional in CLL, but absent in myeloma, compensate for the loss of function incurred by gene inactivation. Additionally, postgerminal center hypermutations, recently shown to also affect the BCL6proto-oncogene,29 are more frequent in MM than CLL,30 and may affect chromosome 13 genes. Thus, the stage of B-cell differentiation may play a critical role in modulating the effects of a commonly deleted gene, and chromosome 13 tumor suppressor gene(s) may be more prone to inactivating base substitution mutations in MM than in CLL.
The observation of a lack of molecular chromosome 13 deletion in 19% of patients with either uninformative diploid karyotypes (3 of 24 patients) or especially MM-typical chromosome abnormalities without chromosome 13 involvement (4 of 12 patients) (Table 2) is consistent with the existence of a distinct nonchromosome 13 deletion entity. However, interstitial deletions (involving additional chromosome 13 regions not covered by our 11-probe panel), microdeletions (not detectable but rather masked by the large probes used) or even more subtle mutations (eg, base substitutions) may exist. Additionally, patients with a seemingly normal diploid chromosome 13 may in fact harbor duplication of an abnormal chromosome that may be identifiable by loss of heterozygosity studies.
Based on our data, progression of chromosome 13 deletion-positive monoclonal gammopathies of undetermined significance (MGUS) to MM, as recently demonstrated by Avet-Loiseau,31 may require a deletion evolution. The observed heterogeneity in deletion extent in individual patients with frank MM may thus reflect different stages of disease progression, including the persistence of an original MGUS clone with minimal deletion. The lower proliferative activity of a MGUS subclone may account for its survival even in the face of high-dose therapy, thus explaining the persistence of low-level monoclonal gammopathy in remission. The molecular mechanism(s) of such deletion evolution may be traced to a genomic instability associated with DNA replication in terminally differentiated plasma cells or loss of genome fidelity maintenance genes such as P53, BRCA1,BRCA2, and ATM.
To address these critical issues, multiprobe chromosome 13 FISH will be applied in the following scenarios: (1) long-term MGUS without progression (Q: no chromosome 13 deletion?) versus MGUS to MM progression (Q: chromosome 13 deletion evolution?); (2) long-term MM survivors after high-dose therapy with minimal residual disease (Q: no or minimal chromosome 13 deletion?); and (3) early relapse after high-dose therapy (Q: marked deletion heterogeneity and evolution?). Systematic studies of molecular deletions of chromosome 13 in CLL (especially in the recently identified subsets of unmutated CD38+ and hypermutated CD38− entities with different prognoses)32 33 and of other chromosomes in MM will address the important issues of chromosome- and disease-specific deletion heterogeneity.
Acknowledgments
We would like to thank Todd Bell, Peter Kim, Janet Lukacs, Alian Li, Yan Xiao, and Elizabeth Williamson for their excellent technical assistance. We thank the members of the Myeloma and Transplantation Research Center clinical staff and data management group. Finally, we would like to thank the patients and their families for their support of our myeloma research program.
Supported in part by CA55819 from the National Cancer Institute, Bethesda, MD.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
John D. Shaughnessy Jr, Myeloma and Transplantation Research Center, University of Arkansas for Medical Sciences and Arkansas Cancer Research Center, 4301 West Markham Street, Little Rock, AR 72205.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal