Abstract
The monofunctional alkylating agent N-methyl-N-nitro-N-nitrosoguanidine (MNNG) is a widespread environmental carcinogen that causes DNA lesions, leading to cell death. However, MNNG can also trigger a cell-protective response by inducing the expression of DNA repair/transcription-related genes. We demonstrate that the urokinase-type plasminogen activator (uPA) gene product, a broad spectrum extracellular protease to which no DNA repair function has been assigned, is transcriptionally induced by MNNG in C2C12 and NIH3T3 cells. This induction required an AP1-enhancer element located at −2.4 kilobase (kb), because it was abrogated by deletion of this site. MNNG was found to induce the activation of JNK/SAPK and p38 mitogen-activated protein kinases (MAPKs). Accordingly, we attempted to assess the contribution of each of these MNNG-inducible MAPKs to uPA gene induction by this alkylating agent. Coexpression of dominant negative versions of kinases of the JNK pathway, such as catalytically inactive forms of MEKK1, MKK7, and JNKK, and of cytoplasmic JNK-inhibitor JIP-1, as well as treatment of cells with curcumin (which blocks JNK activation by MNNG), inhibited MNNG-induced uPA transcriptional activity. In contrast, neither dominant negative MKK6 nor SB203580, which specifically inhibit p38 MAP kinase activation, abrogated the MNNG-induced effect. Taken together, our results show that the JNK signaling pathway links external MNNG stimulation and AP1-dependent uPA gene expression, providing the first functional dissection of a transcription-coupled signal transduction pathway for MNNG.
Introduction
Urokinase-type plasminogen activator (uPA) is a secreted serine protease that converts the zymogen plasminogen to plasmin, a trypsin-like serine protease capable of degrading extracellular matrix components and of activating other proteinases (for revision see Irigoyen et al1). Analysis of the consequences of loss in uPA function of uPA-deficient mice has confirmed the participation of this protease in physiologic processes such as fibrinolysis and angiogenesis, as well as in pathologic events such as wound healing, inflammation, and tumor invasiveness.2-9 In addition to these proteolytic functions, recent studies have demonstrated mitogenic and chemotactic properties of uPA through interaction with its cell surface high-affinity receptor.10,11 Reflecting its wide spectrum of functions, uPA expression is regulated by numerous extracellular stimuli depending on the cell type. uPA gene transcription can be induced by growth factors, phorbol esters, cytokines, cytoskeletal reorganization, and several oncoproteins.12-22
We have recently shown that ultraviolet (UV) light irradiation induces the expression of urokinase-type plasminogen activator (uPA) gene in NIH3T3 fibroblasts.23 uPA was the first protein shown to be inducible in xeroderma pigmentosum cells at much lower UV doses than in parental heterozygotic cells,24 suggesting that DNA damage might be responsible for this induction. Damage to DNA can be inflicted by a broad spectrum of agents, including UV light, ionizing radiation (IR), and alkylating agents. Recent studies have demonstrated that extracellular uPA activity is inversely related to the cell capacity to repair the DNA lesions induced by alkylation agents such as N-methyl-N-nitro-N-nitrosoguanidine (MNNG).25 Monofunctional alkylating agents like MNNG and methyl-methanesulfonate (MMS) are widely distributed environmental mutagens and carcinogens that, on activation, react with DNA and proteins generating aducts.26-28 Among the aducts, O6-alkyl guanine (generated by N-alkylation of the DNA base) is the predominant cytotoxic and mutagenic lesion, because of its mispairing properties, which leads eventually to chromosomal aberrations, point mutations, and cell killing.29,30 This lesion also appears to be involved in tumor induction, in particular gastric carcinogenesis.31-34 However, monofunctional alkylating agents not only cause cell destruction, but also induce the transcription of many genes, including genes coding for transcriptions factors such as c-fos, c-jun, junB, and junD,35 for cell cycle regulatory proteins such as p53 and p21,36-38 for growth arrest and DNA damage (GADD) proteins39,40 and for DNA repair proteins such as O6-methylguanine-DNA-methyltransferase (MGMT) and DNA polymerase β (β-pol).41,42 Recent evidence suggests that the response to DNA-damaging agents may have a protective function other than DNA repair.43,44 The main target of genotoxic agents was believed to be chromosomal DNA damage, which in turn would provide the primary signal, triggering the response.45,46 However, the rapid UV light activation of Ras, Src, and other molecules located at or near the plasma membrane argues against DNA damage as the primary signal, suggesting that a nuclear signal is not always required for the UV response.43,47 Similarly, genomic DNA has been ruled out as a prerequisite for the induction of JNK activation in response to MMS, because the induction was also detected in enucleated cells.48 However, although the first cellular reaction detectable in UV-irradiated cells is the phosphorylation of different cell membrane growth factor receptors at tyrosine residues,49 the response to MMS seems to be independent of growth factor receptor activation, suggesting that the primary cellular target of alkylation-driven stimulation is different from that of UV.48 Altogether, the signaling cascades induced by alkylayting agents appear to be complex. Wherever generated, the alkylating signal, like the UV-induced signal, seems to activate different components of the mitogen-activated protein kinase (MAPK) family, which mediate transcription factor activation (reviewed in Bender et al50). Three MAPK cascades have been shown to transduce stress signals to the nucleus: ERK, JNK/SAPK, and p38. Although JNK and p38 are predominantly involved in the response to stresses and cytokines, ERK is mainly implicated in the regulation of cell growth and differentiation.43,51-71 The alkylating agents MMS and MNNG are potent inducers of JNK activation in 293 human cells.48 MMS induced p38 phosphorylation in these cells, but did not activate ERK. In contrast to MMS, ethylnitrosourea (ENU), the most powerful alkylating agent in inducing mutagenesis, failed to induce any of the 3 subgroups of MAPKs, indicating that activation of MAPKs is not a general response to alkylating agents.48
In this study, we show that the monofunctional alkylating agent MNNG is a potent inducer of uPA gene expression in 2 different murine cell lines. Specifically, we have examined the mechanism(s) of MNNG-induced uPA transcription. This induction required an AP1-enhancer element, which is located at −2.4 kilobase (kb) from theuPA transcription start site, and was mediated by the JNK signaling pathway.
Materials and methods
Cell culture
The murine C2C12 and NIH3T3 cell lines were obtained from the American Type Culture Collection and grown in DMEM containing 10% fetal bovine serum (FBS). For MNNG stimulation, cells were kept in DMEM containing 0.5% FBS for 16 hours. The next day, cells were treated with MNNG (70 μmol/L) for different periods. Alternatively, cells were treated with 20% FBS or UV irradiated at 254 nm (30 J/m2). When indicated, cells were pretreated for 30 minutes with 30 μmol/L curcumin, 50 μmol/L PD98059, or 10 μmol/L SB203580, before induction with MNNG. MNNG and curcumin are from Sigma, and PD98059 and SB203580 from Calbiochem. MNNG was dissolved according to Kaina et al.41
RNA analysis
Plasmids
The p-8.2Luc, a murine uPA-promoter luciferase reporter plasmid (kindly provided by Dr Y. Nagamine), contains 8.2 kb of murine uPA promoter region, as described.13 The p-2.0Luc (−2 kb of murine uPA promoter), p-4.9Luc (−4.9 kb of uPA promoter), and p-6.6Luc (−6.6 kb of uPA promoter) were derived from p-8.2Luc.13,23,72 the p-8.2(ΔAP1)Luc, p-6.6(ΔAP1)Luc, and p-4.9(ΔΑP1)Luc, lacking the AP1 enhancer element, are described elsewhere.13,23,72 Expression vectors pSRα-MEKK1(K432M), pcDNAIII-MKK6b(A), pcDNAIII-JIP-1, pcDNAIII-SEK1/MKK4(KR), pcDNAIII-MKK7(A) were kindly provided by Drs M. Karin, J. Han, R. Davis, and E. Nishida.57,70 73-77
Western blotting
Cells were cultured in 0.5% FBS and, at the indicated time points after treatment, whole cell extracts (WCE) were prepared as described in Miralles et al.23 Total JNK1 and ERK2 were detected in 30 μg WCE by immunoblotting using specific antibodies at 1:1000 dilution. JNK1 and ERK antibodies are from Santa Cruz Biotechnology (sc-474 and sc-154, respectively). Alternatively, phosphorylated p38 was detected using an antiphospho p38 antibody (New England Biolabs No 9211S). Immunoblots were developed using ECL detection system (Amersham).
Protein kinase assays
JNK and ERK were immunoprecipitated from WCE with anti-JNK1 and anti-ERK2 antibodies, respectively, and immunocomplexes were recovered with protein A-Sepharose and washed, as described previously.23 Phosphorylation reactions were performed in a 30-μL volume containing kinase buffer supplemented with 20 μmol/L ATP, 0.0185 MBq (0.5 μCi) γ-[32P]ATP and 1 μg GST-cJun1-79 or myelin basic protein (MBP) as substrates for JNK and ERK assays, respectively, at 30°C for 30 minutes. Reactions were stopped by the addition of 4 × Laemmli sample buffer and resolved by 10% or 12% SDS-PAGE for JNK or ERK assays, respectively.
Transfections assays
2.5 × 104 cells were cotransfected using the transfection reagent DOTAP (Boehringer Mannheim) with 300 ng of uPA-luciferase plasmid and 50 ng of RSV-βGal, as internal control. After transfection, cells were cultured in DMEM containing 0.5% FBS for 16 hours before MNNG stimulation (70 μmol/L), and reporter activities were analyzed after 8 hours. When indicated, cells were cotransfected with 150 ng of reporter plasmid and 150 ng of expression plasmids or empty vector alone, together with 50 ng of internal control. Inhibition of MEK and p38 kinase was performed by pretreating transfected cells with 50 μmol/L PD98059 and 10 μmol/L SB203580, respectively, for 30 minutes before MNNG treatment. Alternatively, transfected cells were pretreated with 30 μmol/L curcumin for 30 minutes before an 8-hour period of incubation with MNNG.Firefly luciferase activities were standardized for β-galactosidase activity, used as internal control. All transfection/reporter assays were repeated at least 3 times, showing less than 25% variability. A Student t test was used to validate the results.
C2C12 cell lines containing stables transfected the different uPA promoter-luciferase plasmids have recently been characterized.72
Electrophoretic mobility shift assays
Nuclear extracts were obtained from C2C12 cells before and after MNNG treatment. The extraction of nuclear proteins was performed as described by Miralles et al.23 For electrophoretic mobolity shift assays (EMSAs), 5 μg of nuclear extracts was incubated in 50 mmol/L Tris-HCl pH 7.9, 12.5 mmol/L MgCl2, 1 mmol/L EDTA, 1 mmol/L DTT, 20% glycerol, 0.5 mmol/L PMSF, and 2 μg of poly dI-dC for 10 minutes at room temperature to titrate out nonspecific binding before the addition of 15 000-20 000 cpm–labeled oligonucleotide and the reaction was further incubated for 20 minutes. When unlabeled competing olinucleotides were added, nuclear extracts were preincubated for 30 minutes at room temperature before the addition of the labeled probe. Samples were loaded on a prerun 5% polyacrylamide gel (29:1 in 0.25 × TBE) and electrophoresed at 200 V. Gels were dried and autoradiographed at −80° C. The sequences of the sense strands of the oligonucleotides used in EMSAs are as follows:
AP1A, 5′-GAGGAAATGAGGTCATCTTGCTCTG-3′;
AP1B, 5′-GGCCATGTGAATCACGACAGCCTG-3′;
IgkB, 5′-CAGAGGGGACTTTCCGAG-3′.
Results
MNNG induces murine uPA messenger RNA (mRNA) expression.
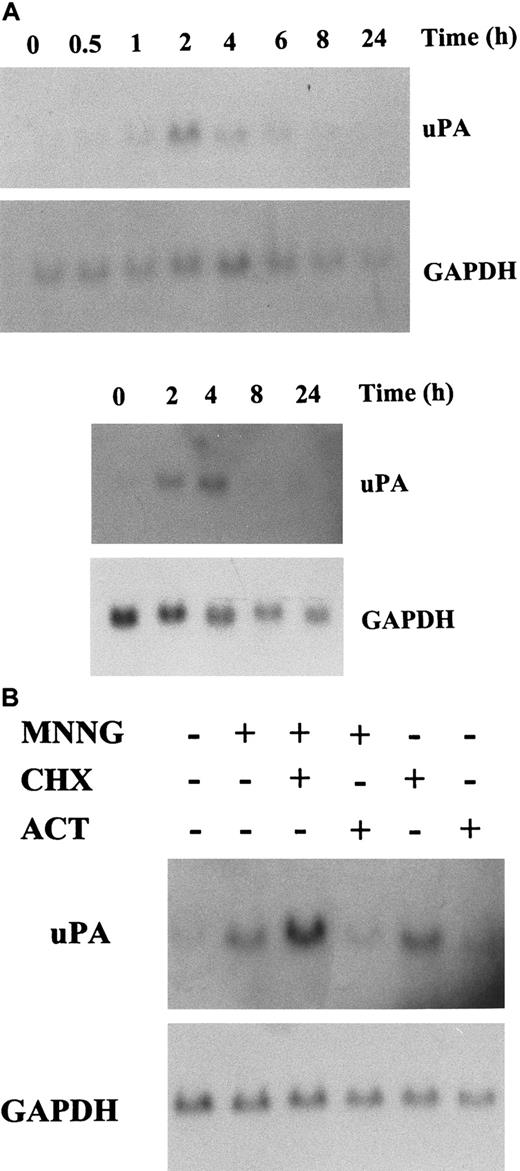
We have previously shown that uPA expression is induced during the UV response.23 To extend this observation, we analyzed whether other environmental mutagenic agents, such as the monofunctional alkylating agent MNNG, modulate the expression of theuPA gene. As shown in Figure1A, treatment of C2C12 and NIH3T3 murine cell lines with MNNG increase the expression of a 2.7-kb transcript corresponding to murine uPA mRNA, with respect to untreated cells. uPA mRNA induction was not due to an unspecific up-regulation of RNA synthesis, because MNNG did not significantly modify the levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA in either cell line. The increase in uPA gene induction was time-dependent, being first observed after 30 minutes after stimulation in C2C12 cells and reaching its maximum 2 hours after MNNG treatment; uPA mRNA induction in these cells decreased 4 hours after treatment, returning to basal levels after 8 hours (Figure 1A, top). Induction of uPA mRNA in NIH3T3 cells was maximal after 4 hours stimulation with MNNG, and was undetectable by 8 hours after treatment (Figure 1A, bottom). To gain an insight into the mechanisms leading to increased uPA mRNA expression in MNNG-treated cells, we studied the effects of RNA and protein synthesis inhibitors on the uPA transcript level in cells that were stimulated with the alkylating agent. The protein synthesis inhibitor cycloheximide did not inhibit uPA mRNA induction by MNNG in either cell type (Figure 1B, and data not shown); moreover, cells treated only with cycloheximide expressed higher levels of uPA transcripts, suggesting that MNNG stimulation of uPA mRNA did not require de novo protein synthesis. In contrast, uPA mRNA induction by MNNG was abrogated in cells treated with the RNA synthesis inhibitor actinomycin D (Figure 1B). These data suggested that increased uPA transcription, rather than message stabilization, was the mechanism responsible for the MNNG-induced effect.
uPA induction by MNNG treatment.
(A) Analysis of uPA mRNA expression in MNNG-stimulated C2C12 (top) and NIH3T3 (bottom) cells. Cells were cultured in 0.5% FBS for 16 hours and then treated with MNNG (70 μmol/L). Total RNA was extracted at the indicated time points (in hours) after MNNG stimulation and analyzed by Northern blotting using the mouse uPA and GAPDH cDNA probes as indicated. (B) Effect of RNA and protein synthesis inhibitors on MNNG-stimulated uPA expression. C2C12 cells were treated for 2 hours with MNNG as in (A), except that cultures were grown in the presence (+) or absence (−) of actinomycin D (5 μg/mL) or cycloheximide (10 μg/mL), which were added 30 minutes before the addition of MNNG.
uPA induction by MNNG treatment.
(A) Analysis of uPA mRNA expression in MNNG-stimulated C2C12 (top) and NIH3T3 (bottom) cells. Cells were cultured in 0.5% FBS for 16 hours and then treated with MNNG (70 μmol/L). Total RNA was extracted at the indicated time points (in hours) after MNNG stimulation and analyzed by Northern blotting using the mouse uPA and GAPDH cDNA probes as indicated. (B) Effect of RNA and protein synthesis inhibitors on MNNG-stimulated uPA expression. C2C12 cells were treated for 2 hours with MNNG as in (A), except that cultures were grown in the presence (+) or absence (−) of actinomycin D (5 μg/mL) or cycloheximide (10 μg/mL), which were added 30 minutes before the addition of MNNG.
An AP1-enhancer element is involved in uPA promoter induction by MNNG
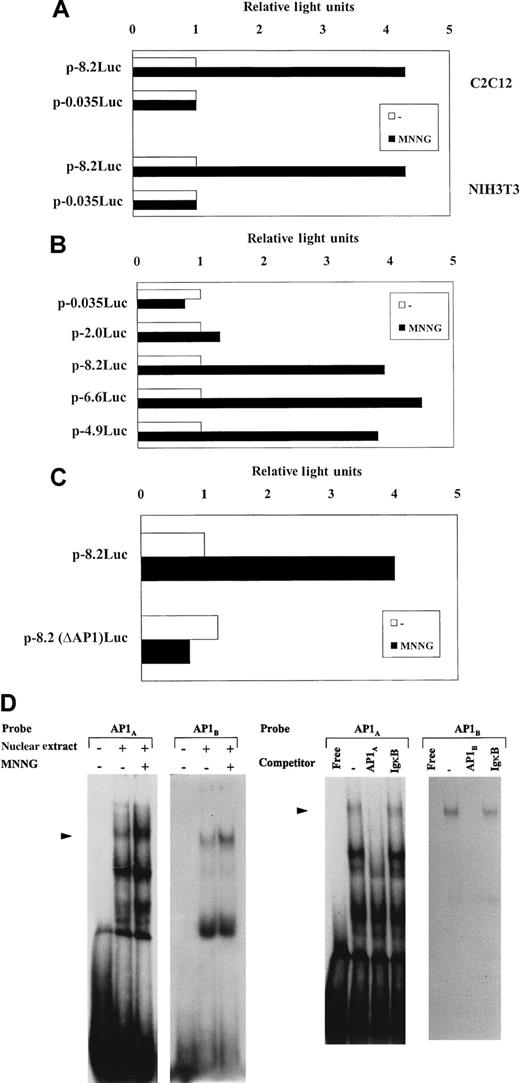
We next examined the effect of MNNG on the activity of the murineuPA gene promoter. A murine uPA genomic fragment (−8.2 kb to +398 base pairs [bp]), ligated upstream the luciferase reporter gene, p-8.2Luc,13 was assessed for luciferase activity after transient transfection in C2C12 and NIH3T3 cells. Comparison of luciferase activities generated byp-8.2Luc, between unstimulated and MNNG-stimulated cells, showed that the uPA promoter activity increased an average 4-fold after MNNG treatment in both cell types, whereas activity of p-0.035Luc (a plasmid including only 35-bp of the uPA minimal promoter sequence) was unaffected (Figure2A), indicating that the murine uPA promoter contains MNNG-responsive sequences that might account, at least in part, for the MNNG-mediated induction of uPA in these cells. To begin identifying the main regions involved in the uPA transcriptional response to MNNG, different murine uPA promoter-deletion luciferase constructs, generated from the full-length −8.2-kb promoter plasmid, were stably trasfected into C2C12 cells,72 and the corresponding uPA promoter-luciferase (uPA-Luc)–containing cell lines were analyzed for luciferase inducibility after treatment with MNNG. As shown in Figure 2B, deletion of 1.6 and 3.3 kb from the −8.2-kb construct (generating plasmids p-6.6Luc and p-4.9Luc, respectively) did not alter the luciferase induction of the full-length −8.2-kb plasmid (P > .05), suggesting that these upstream promoter regions were irrelevant for uPA transcriptional induction by MNNG in the uPA-Luc–containing cell lines. However, although C2C12 cells containing p-4.9Luc retained full MNNG-induced luciferase activity, C2C12 cells transfected with p-2.0Luc, a plasmid containing 2.0 kb of the murine uPA minimal promoter, showed no luciferase inducibility (P < .01) (Figure 2B), clearly indicating the location within this region of cis-element(s) relevant for MNNG-induced uPA transcription. As expected (according to transient transfection results shown in Figure 2A), cells stably transfected with p-0.035Luc showed no luciferase induction after stimulation with MNNG (Figure 2B).
MNNG induces uPA transcription in C2C12 cells: requirement of an AP1 enhancer element.
(A) Transcriptional activity of uPA promoter constructs in response to MNNG in C2C12 and NIH3T3 cells. C2C12 and NIH3T3 cells were transiently transfected with p-8.2Luc and p-0.035Luc, containing 8.2 kb and 35 bp, respectively, of murine uPA 5′-flanking region ligated upstream of the luciferase reporter gene, and luciferase activity was determined after 8 hours stimulation with 70 μmol/L MNNG. Three independent experiments, showing less than 25% variability in luciferase activity, were performed. Luciferase activity corresponding to each plasmid in the absence of MNNG stimulation was arbitrarily assigned a value of 1. (B) Luciferase induction of uPA promoter-containing cell lines by MNNG. C2C12 cells were stably transfected with different uPA 5′-deletion mutants, containing −0.035 kb, −2.0 kb, −4.9 kb, −6.6 kb, and −8.2 kb, respectively. For each uPA-luciferase plasmid transfection, neomycin-resistant colonies were pooled and chimerical plasmid insertion was normalized by Southern blot analysis with a luciferase DNA probe, as described in Miralles et al,72 and the corresponding cell lines were treated or not with MNNG for 8 hours. Luciferase activities for each cell line are expressed relative to the activity found in the corresponding untreated cells, which was assigned a value of 1. Values are the mean value of 4 experiments. (C) The uPA AP1-enhancer is required for MNNG transcriptional induction. C2C12 cells stably transfected with different uPA promoter-luciferase constructs containing (as in panel B) or lacking the AP1-enhancer (p-8.2(ΔAP1)Luc, p-6.6(ΔAP1)Luc, p-4.9(ΔAP1)Luc,72were treated with MNNG as described in (A). Only results obtained with p-8.2Luc– and p-8.2(ΔAP1)Luc–containing cell lines are shown. Luciferase activity for each cell line is expressed relative to the activity found in the untreated cells, which was assigned a value of 1. All normalized activities represent a minimum of 4 experiments, showing less than 25% variability. (D) MNNG treatment induces AP1-binding activity to the uPA enhancer element. Left: Induction of uPA 5′-TRE- and 3′-TRE-binding activities by MNNG treatment. C2C12 cells were grown in 0.5% FBS for 16 hours and either treated (+) or not (−) with MNNG (70 μmol/L) for 2 hours. Nuclear extracts were prepared at 2 hours after treatment and incubated with 20 000 cpm of the indicated32P-labeled probes, corresponding to the 5′-TRE (AP1A) and 3′-TRE (AP1B) elements of the uPA promoter, respectively. Right: Specificity of the induced uPA AP1-binding activity. Two-hour–induced C2C12 nuclear extract was incubated with the labeled AP1A or AP1Boligonucleotide, respectively, in the absence or presence of 150-fold molar excess of unlabeled competitors (described in the “Materials and methods” section): cold AP1A and AP1Bwere used as specific competitors for the corresponding labeled oligonucleotides, whereas cold IgκB (κB site of the Igκ enhancer) was used as unrelated competitor. The filled arrowhead indicates specific TRE-binding complexes.
MNNG induces uPA transcription in C2C12 cells: requirement of an AP1 enhancer element.
(A) Transcriptional activity of uPA promoter constructs in response to MNNG in C2C12 and NIH3T3 cells. C2C12 and NIH3T3 cells were transiently transfected with p-8.2Luc and p-0.035Luc, containing 8.2 kb and 35 bp, respectively, of murine uPA 5′-flanking region ligated upstream of the luciferase reporter gene, and luciferase activity was determined after 8 hours stimulation with 70 μmol/L MNNG. Three independent experiments, showing less than 25% variability in luciferase activity, were performed. Luciferase activity corresponding to each plasmid in the absence of MNNG stimulation was arbitrarily assigned a value of 1. (B) Luciferase induction of uPA promoter-containing cell lines by MNNG. C2C12 cells were stably transfected with different uPA 5′-deletion mutants, containing −0.035 kb, −2.0 kb, −4.9 kb, −6.6 kb, and −8.2 kb, respectively. For each uPA-luciferase plasmid transfection, neomycin-resistant colonies were pooled and chimerical plasmid insertion was normalized by Southern blot analysis with a luciferase DNA probe, as described in Miralles et al,72 and the corresponding cell lines were treated or not with MNNG for 8 hours. Luciferase activities for each cell line are expressed relative to the activity found in the corresponding untreated cells, which was assigned a value of 1. Values are the mean value of 4 experiments. (C) The uPA AP1-enhancer is required for MNNG transcriptional induction. C2C12 cells stably transfected with different uPA promoter-luciferase constructs containing (as in panel B) or lacking the AP1-enhancer (p-8.2(ΔAP1)Luc, p-6.6(ΔAP1)Luc, p-4.9(ΔAP1)Luc,72were treated with MNNG as described in (A). Only results obtained with p-8.2Luc– and p-8.2(ΔAP1)Luc–containing cell lines are shown. Luciferase activity for each cell line is expressed relative to the activity found in the untreated cells, which was assigned a value of 1. All normalized activities represent a minimum of 4 experiments, showing less than 25% variability. (D) MNNG treatment induces AP1-binding activity to the uPA enhancer element. Left: Induction of uPA 5′-TRE- and 3′-TRE-binding activities by MNNG treatment. C2C12 cells were grown in 0.5% FBS for 16 hours and either treated (+) or not (−) with MNNG (70 μmol/L) for 2 hours. Nuclear extracts were prepared at 2 hours after treatment and incubated with 20 000 cpm of the indicated32P-labeled probes, corresponding to the 5′-TRE (AP1A) and 3′-TRE (AP1B) elements of the uPA promoter, respectively. Right: Specificity of the induced uPA AP1-binding activity. Two-hour–induced C2C12 nuclear extract was incubated with the labeled AP1A or AP1Boligonucleotide, respectively, in the absence or presence of 150-fold molar excess of unlabeled competitors (described in the “Materials and methods” section): cold AP1A and AP1Bwere used as specific competitors for the corresponding labeled oligonucleotides, whereas cold IgκB (κB site of the Igκ enhancer) was used as unrelated competitor. The filled arrowhead indicates specific TRE-binding complexes.
The uPA promoter contains an AP1-enhancer element, located at −2.4 kb, known to respond to several extracellular stimuli (reviewed in Besser et al10). To assess whether this element might also be responsible for MNNG stimulation, we analyzed the transcriptional inducibility of C2C12 cell lines expressing uPA-Luc constructs containing or lacking the AP1-enhancer site.72 Figure 2C shows that deletion of the AP1 enhancer element in p-8.2Luc, p-8.2(ΔAP1)Luc totally abrogated luciferase induction by MNNG in C2C12 cells. Similar results were obtained with additional uPA-Luc-C2C12 cell lines containing a specific deletion of the AP1-enhancer element: p-6.6(ΔAP1)Luc- and p-4.9(ΔAP1)Luc-containing cell lines showed no luciferase inducibility by MNNG (data not shown). These results demonstrated the requirement of the AP1-enhancer for uPA transcriptional activation by MNNG. Moreover, the uPA AP1-enhancer showed increased AP1 binding activity after treatment of the cells with MNNG (Figure 2D). The uPA AP1-enhancer is composed of 2 phorbol ester responsive elements (TRE): an upstream octameric AP1A site (5′-TRE) and a downstream canonical AP1B site (3′-TRE), separated by an intervening element (COM). As shown in Figure 2D, both AP1A and AP1B sequences of the uPA AP1-enhancer bound a major nuclear protein complex, whose intensity increased after MNNG treatment of C2C12 cells, as assessed by EMSA (left). When each oligonucleotide was used as an unlabeled competitor, the formation of the corresponding DNA-protein complex was prevented; in contrast, excess of an unrelated competitor (the κB site of the Igκ enhancer, IgκB) sequence did not affect the formation of either complex (Figure2D, right), demonstrating the specificity of the DNA-protein interaction.
Induction of MAP kinases by MNNG in C2C12 and NIH3T3 cells
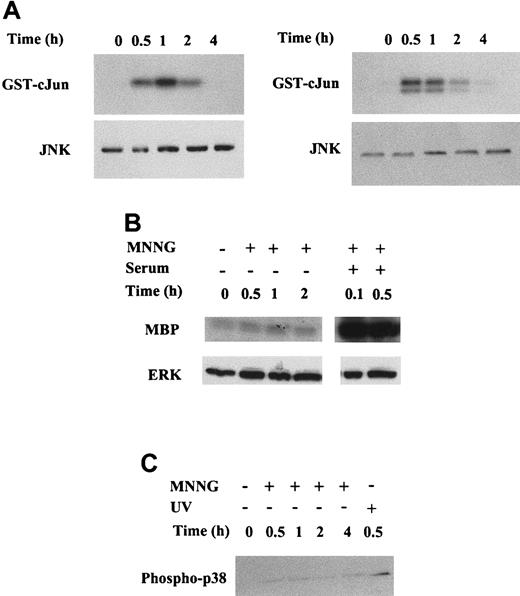
The alkylating agent MMS has been shown to promote the induction of JNK and p38 activities in human 293 cells.48 MNNG could also induce JNK activity in these cells. However, JNK activation is not a general response to alkylating drugs, because alkylating agents such as ENU cannot exert this effect. To determine the potential activation of the 3 main classes of MAPKs (JNK, ERK and p38) by MNNG in C2C12 and NIH3T3 murine cell lines, we measured the activities of JNK, ERK and p38, respectively, in response to MNNG treatment (Figure3). Accordingly, immune complex kinase assays with C2C12 and NIH3T3 cell extracts, using an antibody against JNK1 and GST-cJun as substrate, were performed to determine JNK activation by MNNG in these cells. MNNG induced a very strong activation of JNK activity, peaking 1 hour after stimulation, remaining high after 2 hours, and returning to basal levels 4 hours after treatment in C2C12 cells (Figure 3A, left). MNNG was also a very potent inducer of JNK activity in NIH3T3 cells, reaching maximal levels between 30 minutes and 1 hour after stimulation, decreasing after 2 hours, and returning to basal levels 4 hours after treatment (Figure3A, right). Similarly, we measured the activation of ERK using an antibody against ERK2 and myelin basic protein (MBP) as a substrate. As shown in Figure 3B, no activation of ERK2 was observed after MNNG treatment (up to 2 hours) in C2C12 cells, whereas ERK2 was potently activated in these cells within 6 minutes (0.1 hour) after serum (20% FBS) treatment. As in C2C12 cells, no ERK activation was observed after MNNG treatment of NIH3T3 cells (data not shown). Finally, activation of p38 by MNNG in C2C12 cells was assessed by Western blotting using an antibody specific for phospho-p38. As shown in Figure 3C, phosphorylation of p38 by MNNG was detected within 30 minutes after treatment of C2C12 cells, and still apparent after 4 hours. However, the signal corresponding to phosphorylated p38 in response to MNNG was weaker than that obtained in response to UV-C irradiation in C2C12 cells (Figure 3C, last lane). Furthermore, the activation of p38 induced by MNNG (Figure 3C) is almost negligible in comparison with the potent activation of JNK induced by the same alkylating agent (Figure3A). Similar differences in the activation of JNK and p38 by MNNG were obtained on transient transfection of epitope-tagged HA-JNK1 and HA-p38, followed by immunoprecipitation with anti-HA antibody and measurement of kinase activities using GST-cJun and MBP substrates, respectively (data not shown). As in C2C12 cells, MNNG induced a very weak phosphorylation of p38 in NIH3T3 cells (data not shown). Altogether, these results suggested that JNK is the most potently activated MAPK in response to MNNG in C2C12 and NIH3T3 cells, whereas p38, but not ERK, was weakly activated by MNNG in these cells.
Analysis of MAPK activation in response to MNNG.
Whole cell extracts (WCE) were prepared as detailed in “Material and methods” from C2C12 and NIH3T3 cells at the indicated time points after MNNG stimulation. (A) MNNG induces JNK activation in C2C12 (left) and in NIH3T3 (right) cells. (Top panel) JNK activity was determined by immunoprecipitation of JNK1 from WCE, followed by kinase assay using the substrate GST-cJun.1-79 (Bottom panel) Western blot showing total JNK1 amount present in each cell extract (30 μg/lane). (B) MNNG does not induce ERK activation in C2C12 cells. Cells were stimulated with either 70 μmol/L MNNG or 20% serum as indicated. (Top panel) ERK activity was determined by immunoprecipitation of ERK2 from WCE, followed by kinase assay using the substrate MBP. (Bottom panel) Western blot showing total ERK2 amount present in each cell extract (30 μg/lane). (C) MNNG induces phosphorylation of p38 in C2C12 cells. Cells were either treated with 70 μmol/L MNNG or irradiated with UV-C (254 nm, 30 J/m2) 70 μg WCE were resolved by SDS-PAGE and phospho-p38 was detected by Western blotting using an anti-phospho-p38 antibody.
Analysis of MAPK activation in response to MNNG.
Whole cell extracts (WCE) were prepared as detailed in “Material and methods” from C2C12 and NIH3T3 cells at the indicated time points after MNNG stimulation. (A) MNNG induces JNK activation in C2C12 (left) and in NIH3T3 (right) cells. (Top panel) JNK activity was determined by immunoprecipitation of JNK1 from WCE, followed by kinase assay using the substrate GST-cJun.1-79 (Bottom panel) Western blot showing total JNK1 amount present in each cell extract (30 μg/lane). (B) MNNG does not induce ERK activation in C2C12 cells. Cells were stimulated with either 70 μmol/L MNNG or 20% serum as indicated. (Top panel) ERK activity was determined by immunoprecipitation of ERK2 from WCE, followed by kinase assay using the substrate MBP. (Bottom panel) Western blot showing total ERK2 amount present in each cell extract (30 μg/lane). (C) MNNG induces phosphorylation of p38 in C2C12 cells. Cells were either treated with 70 μmol/L MNNG or irradiated with UV-C (254 nm, 30 J/m2) 70 μg WCE were resolved by SDS-PAGE and phospho-p38 was detected by Western blotting using an anti-phospho-p38 antibody.
Curcumin blocks JNK activation by MNNG
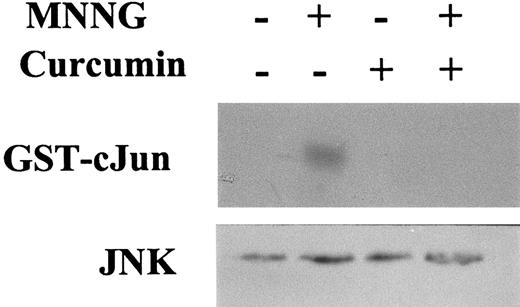
PD98059 and SB203580 are specific inhibitors of ERK and p38 activation, respectively. However, no synthetic inhibitor of JNK activation has yet been identified. The dietary pigment curcumin is a potent inhibitor of JNK activation by various agonists, including PMA plus ionomycin, anysomycin, UV-C, and TNFα.78 To test whether curcumin could also inhibit JNK activation in response to MNNG, C2C12 cells were pretreated with curcumin before their stimulation with MNNG, and cell extracts were immunoprecipitated with an antibody against JNK1, and the enzymatic activity assessed using GST-cJun as the substrate. As shown in Figure 4, activation of JNK activity after a 1-hour treatment with MNNG was completely abrogated by curcumin pretreatment, thus providing a useful tool for blocking JNK activation by this alkylating agent. As expected, neither 10 μmol/L SB203580 nor 50 μmol/L PD98059 affected JNK activation by MNNG, whereas these compounds inhibited activation of p38 by UV-C irradiation and of ERK2 by 100 nmol/L TPA, respectively, in NIH3T3 and C2C12 cells (results not shown).
Inhibition of MNNG-induced JNK activation by MNNG in the presence of curcumin.
C2C12 cells were treated or not with 70 μmol/L MNNG for 1 hour; 30 μmol/L curcumin was added to the medium 30 minutes before the addition of MNNG. JNK activity was determined by immunoprecipitation of JNK1 from WCE, followed by kinase assay using the substrate GST-cJun.1-79 Bottom panel: Western blotting showing total JNK1 amount present in each cell extract (30 μg/lane).
Inhibition of MNNG-induced JNK activation by MNNG in the presence of curcumin.
C2C12 cells were treated or not with 70 μmol/L MNNG for 1 hour; 30 μmol/L curcumin was added to the medium 30 minutes before the addition of MNNG. JNK activity was determined by immunoprecipitation of JNK1 from WCE, followed by kinase assay using the substrate GST-cJun.1-79 Bottom panel: Western blotting showing total JNK1 amount present in each cell extract (30 μg/lane).
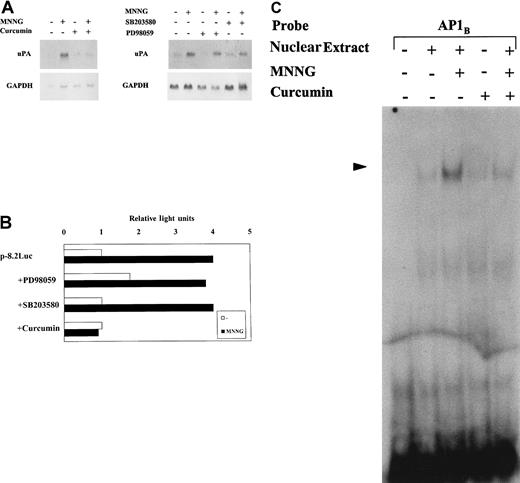
Curcumin abrogates induction of AP1 binding, uPA transcription, and uPA mRNA expression by MNNG
Once we had shown that both JNK and p38 MAPKs are induced in response to MNNG in C2C12 cells, we analyzed whether any of these MAPK signaling pathways were responsible for the induction of uPAgene expression by MNNG. We studied the effect of JNK and p38 inhibitors on uPA expression at different levels (Figure5). As shown by Northern blotting, uPA mRNA expression was induced in C2C12 cells after a 2-hour treatment with MNNG. Curcumin pretreatment of C2C12 cells resulted in a complete inhibition of uPA mRNA induction by MNNG (Figure 5A); in contrast, pretreatment of cells with p38 inhibitor SB203580 did not alter this induction and, as expected, ERK inhibitor PD98059 had no effect on uPA induction by MNNG (Figure 5A, right). These data suggested thatuPA gene induction by MNNG most likely occurred via the JNK signaling pathway. We next examined whether these MAPK inhibitors modify the MNNGtranscriptional response of uPA-Luc-containing C2C12 cell lines. As shown in Figure 5B, MNNG induced luciferase activity from the p-8.2Luc-C2C12 cell line an average of 4-fold. Although neither PD98059 nor SB203580 had any significant effect on uPA promoter induction by MNNG, curcumin fully abrogated uPA transcriptional induction in response to MNNG. Moreover, pretreatment of C2C12 cells with curcumin decreased AP1-binding activity to the uPA 3′-TRE (AP1B) after stimulation with MNNG (Figure 5C). In summary, curcumin specifically inhibits uPA induction by MNNG at 3 different levels: (1) AP1-binding to the uPA enhancer element, (2) uPA transcriptional activity, and (3) uPA mRNA expression.
JNK inhibitor curcumin, but not ERK- or p38-specific inhibitors, abrogates uPA induction by MNNG.
C2C12 cells were pretreated for 30 minutes with 30 μmol/L curcumin, 50 μmol/L PD98059, or 10 μmol/L SB203580 before stimulation with 70 μmol/L MNNG. Curcumin inhibits uPA mRNA induction by MNNG. Northern blot analysis of 20 μg of total RNA from C2C12 cells stimulated or not for 2 hours with MNNG. Cell pretreatments (curcumin, SB203580, and PD98059), and DNA probes (uPA and GAPDH) used for hybridization, are indicated in the Figure. (B) Curcumin inhibits uPA transcriptional induction by MNNG. C2C12 cell lines expressing different uPA promoter-luciferase contructs were pretreated for 30 minutes with 50 mmol/L PD98059, 10 μmol/L SB203580, and 30 μmol/L curcumin, before an 8-hour stimulation with 70 μmol/L MNNG. Only results obtained with the C2C12 cell line containing p-8.2-Luc are shown. Reporter activities correspond to the mean of 5 independent luciferase measurements. Luciferase activity is expressed relative to the activity found in untreated cells, which was given a value of 1. (C) Curcumin inhibits AP1-binding to the uPA-enhancer element. Electrophoretic mobility shift assays were performed with 5 μg of nuclear extracts from unstimulated or 2-hour MNNG-stimulated C2C12 cells (pretreated or not for 30 minutes with 30 μmol/L curcumin) and with32P-labeled uPA-AP1B site.
JNK inhibitor curcumin, but not ERK- or p38-specific inhibitors, abrogates uPA induction by MNNG.
C2C12 cells were pretreated for 30 minutes with 30 μmol/L curcumin, 50 μmol/L PD98059, or 10 μmol/L SB203580 before stimulation with 70 μmol/L MNNG. Curcumin inhibits uPA mRNA induction by MNNG. Northern blot analysis of 20 μg of total RNA from C2C12 cells stimulated or not for 2 hours with MNNG. Cell pretreatments (curcumin, SB203580, and PD98059), and DNA probes (uPA and GAPDH) used for hybridization, are indicated in the Figure. (B) Curcumin inhibits uPA transcriptional induction by MNNG. C2C12 cell lines expressing different uPA promoter-luciferase contructs were pretreated for 30 minutes with 50 mmol/L PD98059, 10 μmol/L SB203580, and 30 μmol/L curcumin, before an 8-hour stimulation with 70 μmol/L MNNG. Only results obtained with the C2C12 cell line containing p-8.2-Luc are shown. Reporter activities correspond to the mean of 5 independent luciferase measurements. Luciferase activity is expressed relative to the activity found in untreated cells, which was given a value of 1. (C) Curcumin inhibits AP1-binding to the uPA-enhancer element. Electrophoretic mobility shift assays were performed with 5 μg of nuclear extracts from unstimulated or 2-hour MNNG-stimulated C2C12 cells (pretreated or not for 30 minutes with 30 μmol/L curcumin) and with32P-labeled uPA-AP1B site.
Involvement of the JNK signaling pathway in uPA transcriptional induction by MNNG
Because only curcumin seemed to abrogate uPA induction by MNNG, we hypothesized that the JNK signaling cascade was the intracellular mediator of this effect. To determine the specific involvement of the JNK pathway in uPA gene induction by MNNG, we determined uPA transcriptional activation by this agent in the absence or presence of specific MAPK inhibitors. MEKK1(K432M),79 a dominant negative form of MEKK1, was overexpressed in transient cotransfection experiments with p-6.6Luc in C2C12 cells, with or without MNNG. As shown in Figure 6, the uPA promoter activity induced by MNNG was strongly reduced by the catalytically inactive mutant form of MEKK1. Furthermore, overexpression of JIP-1, a cytoplasmic protein known to cause retention of JNK in the cytoplasm and subsequent inhibition of the JNK pathway,57 and to act as a scaffold protein for JNK signaling,58 also down-regulated MNNG-induced uPA promoter activity in these cells. In addition, the expression of MKK7(A), dominant negative form of MKK7, a specific activator of JNK, but not of p38, in response to TNFα and UV,77 abrogated uPA transcriptional induction by MNNG. A similar inhibitory effect on uPA promoter induction by MNNG was caused by overexpression of SEK1(KR), a dominant negative form of SEK1 (JNK kinase, also known as MKK4) (Figure 6). These results, together with those showing the inhibitory effect of curcumin on uPA promoter-luciferase stimulation (Figure 5B), indicated that the transcriptional induction of the uPA gene by MNNG was mediated, at least in part, by the MEKK1/JNK pathway. In agreement with the results shown in Figure 5B, SB203580, a specific inhibitor of 2 p38 isoforms (p38α and p38β),68,80 did not block the MNNG-induced activation of the −8.2-kb promoter-containing C2C12 cell line. However, because 2 additional p38 isoforms (p38γ and p38δ)64-67,69 are activated by stresses such as UV but insensitive to SB203580,68,81 the potential involvement of these latter kinases in uPA induction by MNNG was assessed. Accordingly, MKK6b(A), a dominant negative form of MKK6b, which diminishes the activation of all p38 isoforms70 71 was cotransfected together with p-6.6Luc in C2C12 cells. As shown in Figure6, the uPA promoter activity induced by MNNG was not suppressed by the catalytically inactive mutant form of MKK6b. Taken together, these results indicated that the JNK pathway was directly involved in the transcriptional activation of the murine uPA gene by MNNG in C2C12 cells, via its AP1-enhancer element.
The MEKK1-JNK1 pathway is required for uPA induction by MNNG.
Effect of inhibitors of the JNK and p38 MAPK pathways on the activation of the murine uPA promoter by MNNG. C2C12 cells were transiently transfected with the p-6.6Luc reporter plasmid, together with expression vectors for dominant negative MEKK1, MKK7, SEK1 or MKK6b (MEKK1[K432M], MKK7[A], SEK1[KR], and MKK6b[A], respectively), as well as JIP-1, or vector alone. Luciferase activities are expressed relative to the activity of p-6.6Luc in unstimulated C2C12 cells, which has been given an arbitrary value of 1. Results obtained represent the average of at least 3 independent experiments.
The MEKK1-JNK1 pathway is required for uPA induction by MNNG.
Effect of inhibitors of the JNK and p38 MAPK pathways on the activation of the murine uPA promoter by MNNG. C2C12 cells were transiently transfected with the p-6.6Luc reporter plasmid, together with expression vectors for dominant negative MEKK1, MKK7, SEK1 or MKK6b (MEKK1[K432M], MKK7[A], SEK1[KR], and MKK6b[A], respectively), as well as JIP-1, or vector alone. Luciferase activities are expressed relative to the activity of p-6.6Luc in unstimulated C2C12 cells, which has been given an arbitrary value of 1. Results obtained represent the average of at least 3 independent experiments.
Discussion
Genotoxic agents like UV light irradiation and monofunctional alkylating carcinogens trigger a rapid, highly regulated adaptative response, known as the cellular stress response, which involves coordinate control of multiple signal transduction pathways, leading to the induction of many genes. The gene inductive response to UV has been analyzed extensively, and it is known to promote transcription of genes coding for transcription factors, growth factors, viral proteins, and proteases (reviewed in Bender et al50). However, less is known about the inductive response to alkylating agents such as MMS and MNNG. These agents induce the early expression of several proto-oncogens including c-fos, c-jun, junB, and junD, although to a different extent.35 They also induce the level of cell cycle regulatory/tumor suppressor proteins such as p53, p21, and adenomatous polyposis coli (APC),36-38,82 and of DNA repair proteins such as 06-methylguanine-DNA methyltransferase (MGMT) and β-pol,41,42 as well as growth arrest and DNA damage-inducible (GADD) proteins.39,40 However, no additional cellular targets of alkylating agents, other than those related with gene transcription or DNA repair processes, have been identified in mammalian cells. Interestingly, in 1986, Brdar reported the induction of plasminogen activator (PA) activity by alkylating agents in a DNA repair-defective human glioblastoma cell strain,83 although whether this enzyme was a direct target for alkylating agents, and which were the mechanisms responsible for PA induction remained to be solved. Here we show that the uPA(urokinase-type plasminogen activator) gene product, a broad-spectrum extracellullar protease, can be induced by the alkylating agent MNNG via the JNK signaling pathway. In particular, we found that an AP1-enhancer element, which is conserved in murine, porcine, and human uPA promoters, is required for the induction by MNNG, because the deletion of this element abrogated the induction. Moreover, this is the first functional dissection of a transcription-coupled signal transduction pathway for MNNG stimulation.
The cellular response to genotoxic agents is complex. UV irradiation activates different MAPKs (ERK, JNK, and p38) as well as NFκB in numerous cell systems (reviewed in Bender et al50). Induction of JNK and p38 activation by MMS has also been shown in the human embryonic cell line 293,48 although no attempts were made to relate these events to the expression of endogenous genes. We have recently demonstrated that UV irradiation induced uPAgene expression in mouse fibroblasts.23 On the basis of this observation, we hypothesized that other genotoxic agents, in particular, alkylating agents, might also induce uPAexpression during the gene inductive response. We have found that MNNG induces uPA mRNA expression in C2C12 and NIH3T3 murine cell lines. However, the time course of uPA mRNA accumulation was different with MNNG and UV; UV induction peaked at 24 hours, lasting up to 48 hours, whereas MNNG induction occurred much earlier, showing a maximal increase 2 to 3 hours after treatment. Induction by MNNG did not require ongoing protein synthesis, indicating that MNNG activates preexisting components of a signaling pathway. Inhibition of protein synthesis by itself enhanced the expression of uPA mRNA, which may be due to suppression of a protein with a short half-life involved either in the degradation of uPA mRNA or in repression of uPA gene transcription. Moreover, uPA promoter induction by MNNG was completely blocked by inhibitors of cytoplasmic signaling cascades (Figures 4 and5), which originate at or near the plasma membrane,43suggesting that the uPA gene could be activated, at least in part, by a cytoplasm-transduced signal. These results are consistent with recent reports showing that the activation of JNK by MMS, like the response to UV light, may not be mediated by DNA damage per se; in fact, the MMS response is independent of a nuclear signal, because JNK induction can be observed in enucleated cells.48 We have shown that JNK and p38, but not ERK, MAP kinases are activated in response to MNNG treatment of C2C12 and NIH3T3 murine cell lines. However, JNK is more potently activated than p38 by MNNG. The induction kinetics of JNK (or p38) activation in response to MNNG are slower than in response to UV-C irradiation in C2C12 and NIH3T3 cells, because the peak of JNK activity in response to the former stress occurs around 1 hour after stimulation (Figure 3A), whereas this activity is maximal 15 minutes after UV-C irradiation.23 In addition, the MNNG-induced activation of JNK in both C2C12 and NIH3T3 cells was more transient than in 293 cells; whereas in C2C12 and NIH3T3 cells no JNK activity could be detected 4 hours after MNNG treatment (Figure 3A), JNK activation levels were still high in 293 cells at the same time after treatment.48
Promoter deletion analysis revealed that the murine AP1-enhancer element located at −2.4 kb was required for uPA promoter induction by MNNG, because its deletion abrogated the induction. Transient transfection assays with coexpression of specific inhibitors for proteins involved in distinct MAPK signaling pathways suggested that JNK, but not p38, is involved in MNNG-dependent uPA gene induction. Expression of MEKK1 results in efficient JNK activation without a significant increase in ERK1/2 or p38 activities.54,76 Our results show that overexpression of MEKK1(K432M), a catalytically inactive form of MEKK1, abrogated the MNNG-induced up-regulation of the murine uPA promoter in C2C12 cells. The involvement of JNK in MNNG-induced uPA transcription was further shown by the inhibitory effect caused by overexpression of JIP-1, a cytoplasmic inhibitor of JNK,57 which also functions as a scaffold protein for the JNK signaling pathway.58Similarly, overexpression of dominant mutant versions of SEK1 (MKK4), which activates both JNK and p38,55,56 or MKK7, a MAPK kinase isoform specific for JNK activation by TNFα, as well as by environmental stresses,77 down-regulated uPA transcriptional induction by MNNG. Moreover, we have previously shown that constitutively active MEKK1 activates uPA gene transcription in NIH3T3 cells, in the absence of MNNG stimulation, suggesting that the uPA gene is a target for this MAPK pathway.23
Reports from over a decade ago indicate that the alkylating agents mechlorethamine and MNNG could induce the production of plasminogen activator in U-87MG cells, an alkylation repair-deficient (Mer−) human glioblastoma strain, at much higher levels than in alkylation repair-proficient (Mer+) U-178MG cells.83 It was concluded that plasminogen activator induction in alkylation repair-deficient human cells is caused by unrepaired DNA damage and may represent an eukaryotic SOS-like function. In fact, most of the MNNG-inducible genes identified in mammalian cells appear to be involved in DNA repair in a way similar to that of the bacterial SOS response. However, several reports have suggested that the mammalian stress response to genotoxic agents was involved in a protective function other than DNA repair. In particular, the inhibition of the UV response by tyrosine kinase inhibitors has confirmed its protective role.43Furthermore, c-fos(−/−) cells and c-jun(−/−) cells are hypersensitive to UV,84 suggesting that the UV-inducible cJun and cFos are essential components of the cellular defense mechanisms against the cytotoxic effects of UV. Similarly, cells deficient in cFos are also hypersensitive to a broad spectrum of DNA-damaging carcinogens, including MMS and MNNG, showing that cFos/AP1 plays a decisive role in cellular defense against genotoxic agents.44 The results shown in this study indicate that the alkylating carcinogen MNNG induces the expression of a broad spectrum extracellular protease, uPA, with no DNA repair properties. Moreover, we demonstrated that the induction of uPA gene expression by MNNG occurs in the first hours after treatment and is mediated by a cytoplasmic signaling pathway, independent of DNA damage. These results contrast with previous reports suggesting that the induction of extracellular plasminogen activator activity, whose peak occurred 36 to 48 hours after alkylation treatment, is inversely related to the cell capacity to repair the DNA lesions induced by alkylating agents.25,83 However, the mechanism involved in this late DNA repair-dependent uPA induction by MNNG remains to be identified. It could be speculated that in DNA repair-proficient cells (this study), the early induction of uPA gene expression in response to MNNG is originated and mediated by cytoplasmic events, independently of DNA damage. In contrast, in DNA repair-deficient cells, the unrepaired DNA damage caused by MNNG might also induceuPA gene expression through a mechanism that has yet to be explained. The physiologic significance of the uPA induction by genotoxic agents is unknown. Exposure of cells to MNNG and most other DNA-damaging agents results in damage of biomembranes, proteins and nucleic acids, either directly or after oxidative stress.85 A simple protective mechanism against damage to such components would consist of replacing them with newly synthesized ones. As already mentioned, exposure of cells to DNA-damaging agents can increase DNA repair capacity and activate cell-cycle checkpoints, but such exposures may also induce enzymes that metabolize toxins to facilitate their elimination from the organism or may activate programmed cell death (apoptosis) to eliminate highly damaged cells. Accordingly, the increased expression of the proteolytic enzyme uPA after MNNG treatment may facilitate the clearance of the damaged cell components. This protease, in conjunction with other extracellular proteinases, has a function in the degradation of the extracellular matrix, in conditions like wound healing, inflammation, and cancer. Recent work with uPA-deficient mice has demonstrated that uPA is required for the inflammatory response to Cryptococcus neoformans, because lack of uPA resulted in inadequate cellular recruitment, uncontrolled infection and death.6 In contrast, using uPA-mutant mice, it has been shown that the PA system is a causal component of polyoma middle T-induced vascular tumor formation, implying a role for uPA in vascular oncogenesis.86 Taken together, these studies suggest thatuPA may have both beneficial and deleterious roles during the response to different pathologic situations. The functional significance of uPA induction by alkylation agents remains to be determined. At present, uPA-deficient mice are available. The sensitivity of uPA−/− cells to MNNG treatment is expected to clarify the role of this protease in the cellular response to alkylating agents.
Acknowledgments
We are grateful to Drs Y. Nagamine, M. Karin, J. Han, R. Davis, and E. Nishida for generously providing us with various plasmids. We also thank A. Martin, G. Aniorte, and D. Fernandez for assistance.
Supported by DGES (PM97-0088) and Fundació La Marató-TV3.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Pura Muñoz-Cánoves, Institut de Recerca Oncològica (IRO), Aut. Castelldefels, km 2.7, E-08907-L'Hospitalet Ll. (Barcelona), Spain; e-mail: pmunoz@iro.es.

![Fig. 6. The MEKK1-JNK1 pathway is required for uPA induction by MNNG. / Effect of inhibitors of the JNK and p38 MAPK pathways on the activation of the murine uPA promoter by MNNG. C2C12 cells were transiently transfected with the p-6.6Luc reporter plasmid, together with expression vectors for dominant negative MEKK1, MKK7, SEK1 or MKK6b (MEKK1[K432M], MKK7[A], SEK1[KR], and MKK6b[A], respectively), as well as JIP-1, or vector alone. Luciferase activities are expressed relative to the activity of p-6.6Luc in unstimulated C2C12 cells, which has been given an arbitrary value of 1. Results obtained represent the average of at least 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/4/10.1182_blood.v96.4.1415/4/m_h81600059006.jpeg?Expires=1767928050&Signature=TVVPAmPVtyWkEAqZ7S~N-1uvPym21BWVejrDpBNiQ9ODjsHXjTrbxxW8A89JAdfe03kY10VYnbXWwd6C1DNeh7cl52sw2PqHOoQHKIshK5gc-ILGJOmiIjeC0bnm5FLJfvhTkV7Y9QjIvpQnwwAxaCDUPPpS-IVZmcnjdnbpJxkEaRMJt3Y2j3CRf97itWbZc0qIJJbjiyi0nahAZGtt~jwD51UZ9h0eWLzcg6k8zam3cZCq~rR-CUlK~3y~XEkQGWSzUjwihiNW~E-sDfAtOcA2IXUdeGtgix0g3rwmAvLgT0z1pCzPxjjqDLbh2Q7JTfXxm8D6Apgy-4E5ZeFXhQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal