Abstract
The granulocyte macrophage–colony-stimulating factor receptor (GM-CSF-R) is a heterodimer composed of 2 subunits, and β, and ligand binding to the high-affinity receptor leads to signalling for the multiple actions of GM-CSF on target cells. In order to explore the role of the subunit in signalling, we used a yeast-2-hybrid system to identify proteins interacting with the intracellular domain of the GMR-. A cDNA encoding a predicted protein of 198 amino acids, designated GRAP (GM-CSFreceptor subunit-associatedprotein), was isolated in experiments using the intracellular portion of GMR- as bait. The interaction between GRAP and GMR- was confirmed by coimmunoprecipitation in mammalian cells. GRAP mRNA is widely expressed in normal human and mouse tissues and in neoplastic human cell lines, but it is not restricted to cells or tissues that express GM-CSF receptors. Three discrete GRAP mRNA species were detected in human tissues and cells, with estimated sizes of 3.3, 3.1, and 1.3 kb. GRAP is highly conserved throughout evolution, and homologues are found in yeast. The GRAP locus in Saccharomyces cerevisiae was disrupted, and mutant yeast cells showed an inappropriate stress response under normal culture conditions, manifested by early accumulation of glycogen during the logarithmic growth phase. GRAP is, therefore, a highly conserved and widely expressed protein that binds to the intracellular domain of GMR-, and it appears to play an important role in cellular metabolism.
Granulocyte macrophage–colony-stimulating factor (GM-CSF) is an important regulator of the growth, differentiation, and maturation of myeloid precursor cells, and it enhances the function of mature neutrophils and mononuclear phagocytes.1 GM-CSF exerts its effect by interacting with its cognate receptor on the cell surface. The GM-CSF receptor is a heterodimer composed of 2 subunits, α and β.1-4 The mature α and β subunits are glycoproteins of 85 and 120 kd, respectively, that span the plasma membrane once. The isolated α subunit (GMR-α) binds GM-CSF with low affinity (Kd approximately 2-5 nmol/L), whereas the isolated β subunit (GMR-β) does not bind GM-CSF but participates in the formation of an α/β complex that binds GM-CSF with high affinity (Kd approximately 30-100 pmol/L). Responsive hematopoietic cells express high-affinity receptors that transduce signals leading to intracellular protein phosphorylation.5-7 Because neither of the subunits of the GM-CSF receptor has intrinsic kinase activity, GM-CSF signal transduction appears to involve activation of cytosolic tyrosine kinases such as Lyn, Fes, and Jak2. Jak2 has been shown to associate with the β subunit of the GM-CSF receptor on ligand binding.8-10 The β subunit is central to signal transduction propagated by high-affinity GM-CSF receptors, and mutagenesis experiments have identified distinct intracellular domains of the β subunit required to activate MAP kinase andfos/jun pathways.11 Although the cytoplasmic domain of the α subunit is required for β subunit-mediated signaling, the precise role of the α subunit in signal transduction in cells expressing the high-affinity receptor is not well understood. We previously found that the isolated α subunit was able to signal for increased glucose uptake in different systems, including human melanoma cells that naturally express only low-affinity receptors and Xenopus laevis oocytes injected with GMR-α mRNA to express the low-affinity receptor.12,13 Signaling from the α subunit for substrate transport through the facilitative glucose transporters does not appear to involve activation of the usual kinase pathways.12 Experiments in βc-null mice, however, indicated that GM-CSF was not able to stimulate glucose uptake in bone marrow cells, implying that under those circumstances the low-affinity GMR was unable to signal for transport.14
The mechanism by which signals may be delivered through the isolated α subunit is unknown, and no proteins, other than GM-CSF and GMR-β, have been shown to associate with GMR-α. We used a yeast 2-hybrid system to identify proteins that interact with GMR-α, and we isolated a novel cDNA designated GRAP (GM-CSF receptor α subunit-associated protein). We propose that GRAP may play a role in GM-CSF receptor α subunit-mediated signal transduction and that it also has an important function in cellular metabolism.
Materials and methods
Materials
The enhanced chemiluminescence Western blot detection system, [α-32P]dCTP and secondary antibodies (horseradish peroxidase-linked antirabbit and antimouse) were from Amersham (Arlington Heights, IL). The random primer labeling kit was from Boehringer (Indianapolis, IN). Human tissue and cancer cell line Northern blots, mouse total RNA, ExpressHyb hybridization solution, human β-actin cDNA probe, pEGFP-C vector, monoclonal antibodies to green fluourescent protein (GFP), leukocyte GAD library and the human muscle cDNA library were from Clontech (Palo Alto, CA). The Dextran–sulfate transfection kit and TNT Quick coupled in vitro transcription translation kit were from Promega (Madison, WI). Antibodies to GMR-α were from Santa Cruz Biotechnology (Santa Cruz, CA). Protein G beads and the Coomassie plus protein assay reagent were from Pierce (Rockford, IL). COS cells were purchased from ATCC (Rockville, MD), and Nytran + membranes were from Schleicher & Schuell (Keene, NH).
Strains and yeast genetic methods
The following Saccharomyces cerevisiae strains were used in this study: R846 MATa his3-δ200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4.HIS3 URA3::(lexAop)8.lacZ gal4? gal80?, obtained from R. Rothstein (Columbia University, New York, NY) and AMP109 MATa/MATa leu2/leu2 trip1/trip1 ura3/ura3 lys2/lys2 ho:LYS2), obtained from A. Mitchell (Columbia University).Escherichia coli strains HB101 and XL1-Blue were used as plasmid hosts. Standard genetic methods were followed.15Yeast cells were grown in medium YEP (1% yeast extract, 2% bacto-peptone) or synthetic complete medium (SC) lacking appropriate amino acid supplements to maintain selection for plasmids. Growth on different carbon sources was scored as described previously.16 To assess sporulation proficiency, diploid cells were grown on YEP-2% glucose plates overnight, patched on sporulation medium15 incubated at room temperature for 1 to 6 days, and then examined microscopically for the presence of asci.
Plasmids
The construct pLexA-GMR-α was made by inserting the C-terminal (last 54 amino acids) GMR-α SmaI–BamHI fragment, obtained by PCR, into the multicloning sites of pBTM116.17 The PCR primers were: 5′ primer, 5′-CGCGGATCCCGGGCTTTAAAAGGTTCCTTAGGATACAG-3′; 3′ primer, 5′-CGCGGATCCGTCGACTACACCCTCTGGGTCTCAGG-3′. The construct pLexA-IL3R-α was made in a similar manner using the following primers: 5′ primer, 5′-CGCGGATCCCGGGCTGCAGAAGGTATCTGGTGATGCAG-3′; 3′ primer, 5′-CGCGGATCCGTCGACTTCCTGGCAGCTTCGGACGA-3′. The construct pEGFP-GRAP was made by cloning theHindIII–BamHI PCR fragment of GRAP C-terminal region (the last 73 amino acids) into the multicloning sites of pEGFP-C (Figure 1A). The PCR primers used for amplification were: 5′ primer, 5′-CCCCAAGCTTGCGGCCGCGTCGACGAAATATG-3′; 3′ primer, 5′-CGGGATCCCCTTGGCCATATCAACAACTC-3′. Sequences of clones recovered from the 2-hybrid screen were determined by primer extension from the GAL4 region.18
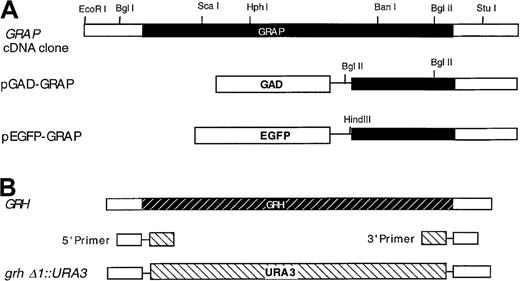
Restriction maps of the GRAP gene, theGRH gene, and related constructs.
Genes and constructs are described in “Materials and methods,” and the direction of transcription in all maps is from left to right. (A) GRAP gene-coding region is shown in solid shading. GAD extends from residues 768 to 881. Both pGAD-GRAP and pEGFP-GRAP are fused to GRAP at codon 126. (B) GRH gene-coding region is shown in the shaded striped bar. URA3 gene is shown in the striped bar.
Restriction maps of the GRAP gene, theGRH gene, and related constructs.
Genes and constructs are described in “Materials and methods,” and the direction of transcription in all maps is from left to right. (A) GRAP gene-coding region is shown in solid shading. GAD extends from residues 768 to 881. Both pGAD-GRAP and pEGFP-GRAP are fused to GRAP at codon 126. (B) GRH gene-coding region is shown in the shaded striped bar. URA3 gene is shown in the striped bar.
2-hybrid screen for GMR-–interacting proteins
The yeast LexAop-lacZ reporter strain R846 was transformed with LexAp DNA-binding hybrid pLexA-GMR-α, which expresses the C-terminal end of GMR-α (last 54 amino acids) fused to LexA. A single transformant was selected to grow on SC-Trp medium and then was transformed with a human leukocyte GAD library that expresses fusions between the Gal4 activation domain (GAD) and human leukocyte cDNA. The transformants containing the 2 hybrids were selected on SC-Trp-Leu-His plates plus 25 mmol/L 3-AT. Positive clones were further selected for β-galactosidase activity.19 The plasmid DNA from the positive clones was recovered from yeast19 and amplified in the E coli strain HB101. Each plasmid from the positive clones was retested for interaction with LexA-GMR-α, LexA, and LexA-lamin.20 The plasmids that interacted with LexA-GMR-α and not with LexA or LexA-lamin were mapped and sequenced. A 0.2-kb open reading frame DNA (BglII fragment) isolated from pGAD-GRAP was used as a probe to screen a human muscle cDNA library by hybridization. A 1.9-kb EcoRI fragment was recovered, subcloned into the pUC19 vector, and sequenced. Both DNA strands were sequenced.
Transfections and immunoprecipitations
COS cells were co-transfected with vector containing the EGFP-GRAP fusion protein or vector pEGFP alone, with the expression vector pMX-GMR-α encoding the full-length GMR-α.12 Cells were transfected with a dextran-sulfate method according to the manufacturer's instructions. Proteins were extracted as described previously21 using a sodium dodecyl sulfate (SDS)-free RIPA buffer (150 mmol/L NaCl, 1% deoxycholine, 1% Triton X-100 and 50 mmol/L Tris, and pH 7.2). Protein concentration was determined spectrophotometrically, and approximately 100 μg of total protein extract was used for each immunoprecipitation experiment. Anti-GM-CSFR-α antibodies were added (dilution 1:100) and incubated at 4°C for 2 hours, and this was followed by a 1-hour incubation with protein G beads. After washing with RIPA buffer, the immunoprecipitate was resuspended in SDS–polyacrylamide gel electrophoresis (PAGE) sample buffer, fractionated by 10% SDS-PAGE, and immunoblotted. The primary antibodies were polyclonal anti-GMR-α C-18 (against the C-terminal 18 amino acids) and monoclonal anti-GFP antibody. Secondary antibodies were horseradish peroxidase-linked antirabbit and antimouse immunoglobulin. Immunoprecipitated proteins were detected by enhanced chemiluminescence.
Northern and dot blot analyses of GRAP in human and mouse tissues
Human Northern blots and RNA master dot blots contained poly A+ RNA (Clontech, Palo Alto, CA). For the mouse Northern blot, 5 μg total RNA was fractionated on a 1% formaldehyde–agarose gel and transferred to Nytran + membrane. A BglII fragment from pGAD-GRAP or a BglI–BglII fragment of GRAP cDNA clone (Figure 1A) was radiolabeled using a random primer kit and [α-32P]dCTP. The Northern and the dot blot membranes were hybridized with ExpressHyb hybridization solution (Clontech, Palo Alto, CA) containing 2 × 106 cpm/mL radiolabeled probe and washed according to the manufacturer's protocol. Human β-actin (Northern blot) and ubiquitin (dot blot) probes were used as controls. The blots were visualized by exposure of Kodak X-Omat film (Eastman Kodak, Rochester, NY).
Disruption of the GRAP-related homologue in yeast
A BLAST search revealed a yeast homologue of GRAP (GRAP-related homologue; GRH) located in the SPC1-ILV3 intergenic region. The hypothetical protein, YJRO14W, weighs 22.5 kd, and the function is unknown. The GRH gene in S. cerevisiae was disrupted at the chromosomal locus. Primers containing 5′ and 3′ untranslated sequences were fused with 5′ and 3′ flanking sequences of the URA3 gene, respectively (Figure1B): The 5′ primer was 5′-CACCCCAAGGAAACAGTTCAAGAGCTAAACTA- AAGAAAAGCATATTGCATAAAATGTTAAGAGAATTCATCGATATC- TAGATCTCGAGC-3′. The 3′ primer was 5′-CTCCCAGAACGGTGCTATTACATATTTATGGATTGCTTACTTGGCAGCTCCTTCTGCAGCAGG- CCAGTGAATTCCCGGGGATCCG-3′. Using these 2 primers and pIC19R22 as a template, the PCR product containing the URA3 gene and GRH1 flanking sequences was used to disrupt the GRH locus of the wild-type diploid strain AMP109 selecting for uracil prototrophs.23Tetrad analysis was performed by standard genetic methods.15 The absence of the GRH gene in haploid disrupters was confirmed by PCR analysis with primers containing 5′ and 3′ flanking sequences: 5′ primer, 5′-CGTGAACTTGCACCATGTACATC-3′; 3′ primer, 5′-CTAACCTACACGCCTGGTTACCAAG-3′. The resultant allele was designated grhδ1::URA3 (Figure 1B).
β-galactosidase and glycogen assays
To test for β-galactosidase expression, the transformants from the positive clones were patched on selective SC-2% glucose plates, grown for 1 day, replicated onto nitrocellulose filters, permeabilized (−70°C for 10 minutes), and incubated with X-Gal overnight.24 To test for glycogen accumulation, yeast cells were plated onto YEP-2% glucose plates, incubated at 30°C overnight, and stained with iodine vapor.25
Results
Identification of proteins that interact with GMR- using the yeast 2-hybrid system
We used a yeast 2-hybrid system to isolate proteins that interact with the cytoplasmic portion of GMR-α. The construct LexA-GMR-α was used as “bait” to screen a human leukocyte cDNA library fused to the GAD. From 2 million independent transformants, we isolated 107 positive clones that grew on SC-Trp-Leu-His plus 25 mmol/L 3-AT. Only 1 of the 107 clones activated the LexAop-lacZ reporter gene in combination with the LexA-GMR-α fusion protein. This positive clone failed to activate the reporter lacZ gene in combination with lexA or LexA-lamin (negative control; Table1). Partial DNA sequencing from the GAD junction revealed an in-frame fusion of a 0.2-kb open reading frame fragment. We designated this new gene GM-CSF receptor α-associated protein (GRAP).
Interactions of GMR- and IL-3- with GRAP in a yeast 2-hybrid system
| DNA-binding hybrid . | Activation partner . | Color* . |
|---|---|---|
| LexA-GMR-α | GAD | White |
| LexA-GMR-α | GAD-GRAP | Blue |
| LexA | GAD-GRAP | White |
| LexA-Lamin | GAD-GRAP | White |
| LexA-IL-3-α | GAD-GRAP | White |
| DNA-binding hybrid . | Activation partner . | Color* . |
|---|---|---|
| LexA-GMR-α | GAD | White |
| LexA-GMR-α | GAD-GRAP | Blue |
| LexA | GAD-GRAP | White |
| LexA-Lamin | GAD-GRAP | White |
| LexA-IL-3-α | GAD-GRAP | White |
Four transformants were patched on selective SC-2% glucose plates and grown for 1 day, and the color of the transformants was determined by filter assay.
Because the GM-CSF and IL-3 receptors share the same β subunit, we tested whether GRAP associates with the α subunit of the IL-3 receptor. We constructed a LexA-IL3R-α fusion protein containing the cytoplasmic portion of the IL3R-α receptor (confirmed by Western blot analysis; data not shown) and tested its association with the original GAD-GRAP clone. We found that the IL3-α receptor did not interact with C-terminal 73aa of GRAP (Table 1), which indicates the selectivity in the interaction of GRAP with the GMR-α.
GRAP is a novel protein that interacts with GMR-
To clone the full-length cDNA of GRAP, we conducted a preliminary Northern blot analysis that showed GRAP was highly expressed in skeletal muscle. Therefore, we screened a human skeletal muscle cDNA library using the original 0.2-kb GRAP DNA in the GAD clone as a probe. A 1.9-kb cDNA clone was isolated and sequenced. An open reading frame of 594 codons, encoding a putative 22.5-kd GRAP protein, was identified. The original GAD clone corresponds to the C-terminal 73 amino acids of GRAP (Figure 1A). A GRAP cDNA clone was also isolated from a cDNA library derived from the human myeloid leukemia cell line, HL-60, and was identical to the GRAP open reading frame in the cDNA clone of skeletal muscle library (data not shown).
BLAST sequence analysis revealed that GRAP is almost identical to the human drp gene (Gene Bank accession number AF038554), which is described as a density-regulated protein.26 The expression of drp was shown to increase in cultured cells at high density. We identified 3 extra G residue positions at −50, −33, and −30 in the 5′ region upstream of GRAP compared with drp.
The cDNA sequence of GRAP contains 2 putative translational start sites within the first 103 bp. The first ATG has a short open reading frame of 22 amino acids. The second ATG is surrounded by nucleotide sequences known to increase the translation initiation efficiency (Kozak consensus sequences, 5′(A/G)NNATGG),27 28 and it is followed by an open reading frame encoding a 198-amino acid protein with an apparent molecular weight of 22.5 kd (Figure2). In vitro translation of GRAP generated a single band that migrated at approximately 30 kd on an SDS-PAGE gel (data not shown). The disparity between the apparent and the expected molecular weights of GRAP may be due to its amino acid composition. GRAP is highly charged and such proteins, for example certain histones, can have atypical migration on SDS-PAGE gels.
Sequence analysis of GRAP cDNA.
Nucleotide and amino acid numbers are indicated. The open reading frame is shown in uppercase letters, and the 5′- and 3′-noncoding regions are shown in lowercase letters. The atg and tag codons of the small ORF are underlined. The Kozak consensus sequence is in bold.
Sequence analysis of GRAP cDNA.
Nucleotide and amino acid numbers are indicated. The open reading frame is shown in uppercase letters, and the 5′- and 3′-noncoding regions are shown in lowercase letters. The atg and tag codons of the small ORF are underlined. The Kozak consensus sequence is in bold.
Evidence that GMR- and GRAP interact in vivo
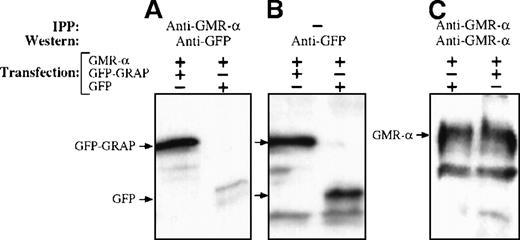
To provide biochemical evidence of the interaction between GRAP and GMR-α, we tested whether GRAP and GMR-α can be coimmunoprecipitated with antibodies that recognize the C-terminal portion of the GMR-α. We first constructed a fusion protein (EGFP-GRAP) that contained the last 73 amino acids of GRAP fused in-frame with the C terminal end of enhanced green fluorescent protein (EGFP). COS cells cotransfected with the plasmids pMX-GMR-α and pEGFP-GRAP, or pMX-GMR-α and pEGFP (control), were lysed under nondenaturing conditions. Cellular extracts were collected and immunoprecipitated with anti-GMR-α antisera. The immunoprecipitated proteins were analyzed by Western blotting with anti-GMR-α and anti-GFP antibodies (Figure3). An intense signal corresponding to EGFP-GRAP fusion protein was detected in COS cells cotransfected with pEGFP-GRAP and pMX-GMR-α (Figure 3A). When the pEGFP-GRAP vector was replaced by pEGFP, only a weak background signal corresponding to EGFP was identified that likely represented nonspecific binding between EGFP and GMR-α. The coimmunoprecipitation of GMR-α protein and the fusion protein EGFP-GRAP supported the notion that GRAP and GMR-α associate in vivo. The immunoprecipitation data also confirmed the interaction between GMR-α and GRAP observed in the yeast 2-hybrid system. In parallel experiments, anti-IL3R-α antibodies failed to coprecipitate EGFP-GRAP, suggesting that the interaction between GRAP and GMR-α is selective (data not shown).
Interaction of GMR- with GRAP.
COS cells were cotransfected with plasmids encoding the wild type of GMR-α and the fusion protein EGFP-GRAP or EGFP as indicated. Cells were lysed, and proteins were immunoprecipitated with anti-GMR-α sera. Immunoprecipitates were fractionated by 10% SDS-PAGE and immunoblotted. Primary antibodies were polyclonal anti-GMR-α and monoclonal EGFP. The secondary antibody was horseradish peroxidase-linked antirabbit or antimouse immunoglobulin. (A) Cell lysates from cells transfected with GMR-α and EGFP-GRAP or EGFP. Immunoprecipitation with anti-GMR-α and immunoblotting with anti EGFP. (B) No immunoprecipitation. Immunoblotting was with anti-EGFP. (C) Immunoprecipitation and immunoblotting with anti-GMR-α.
Interaction of GMR- with GRAP.
COS cells were cotransfected with plasmids encoding the wild type of GMR-α and the fusion protein EGFP-GRAP or EGFP as indicated. Cells were lysed, and proteins were immunoprecipitated with anti-GMR-α sera. Immunoprecipitates were fractionated by 10% SDS-PAGE and immunoblotted. Primary antibodies were polyclonal anti-GMR-α and monoclonal EGFP. The secondary antibody was horseradish peroxidase-linked antirabbit or antimouse immunoglobulin. (A) Cell lysates from cells transfected with GMR-α and EGFP-GRAP or EGFP. Immunoprecipitation with anti-GMR-α and immunoblotting with anti EGFP. (B) No immunoprecipitation. Immunoblotting was with anti-EGFP. (C) Immunoprecipitation and immunoblotting with anti-GMR-α.
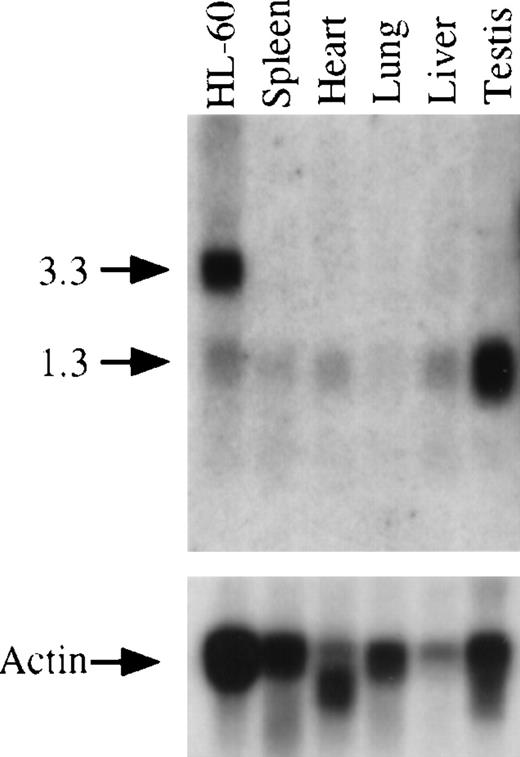
Expression of GRAP mRNA in human cells and tissues
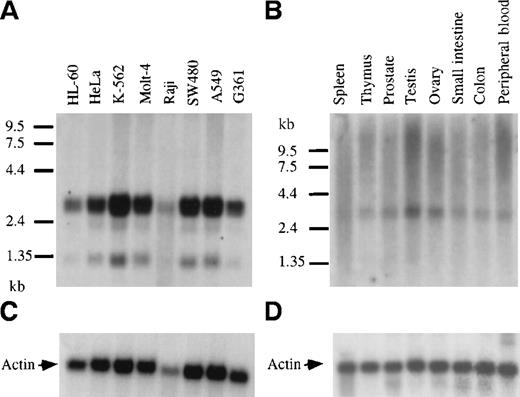
Northern blots containing polyA+ RNA from a panel of human cancer cell lines were hybridized with a 32P-labeled GRAP probe (Figure 4). The analysis revealed the presence of GRAP mRNA in myeloid leukemia cells (HL-60, K-562, Molt4, and Raji) and also in tumor cell lines from ovary, colon, lung, and melanoma (HeLa, SW480, A549, and G361) (Figure 4A). Three RNA species were detected in all cell lines—a doublet at 3.1 and 3.3 kb with intense hybridization signals and a fainter band at 1.3 kb. GRAP mRNA was detected in cells known to express GM-CSF receptors, such as HL-60, and was also found in cells lacking GM-CSF receptors, such as Molt4 and K-562.29 Similarly, the expression of GRAP was detected in normal human tissues, including spleen, thymus, prostate, testis, ovary, small intestine, colon, and peripheral blood (Figure4B). Because of the higher background on Northern blots from normal human tissues, expression of the 1.3-kb mRNA species could not be assessed. To investigate whether the 3 mRNA species detected on the Northern blots encoded the same protein, we sequenced GRAP cDNA clones isolated from HL-60 cells (3.0, 2.8, and 1.3 kb). All 3 clones had identical ORF but variable 3′ ends, suggesting that the GRAP mRNA species on the Northern blots encoded the same protein but that alternative splicing might have resulted in different mRNA species (data not shown).
Northern blot analysis and expression of GRAP mRNA in tumor cell lines and normal human tissues.
Northern blots containing 2.5 μg human polyA+RNA extracted from cancer cell lines or normal human tissues (panels A and B, respectively). Blots were hybridized with GRAP sequences. RNA size markers are shown on the left. Panels A and B correspond to blots. Panels C and D correspond to β-actin controls.
Northern blot analysis and expression of GRAP mRNA in tumor cell lines and normal human tissues.
Northern blots containing 2.5 μg human polyA+RNA extracted from cancer cell lines or normal human tissues (panels A and B, respectively). Blots were hybridized with GRAP sequences. RNA size markers are shown on the left. Panels A and B correspond to blots. Panels C and D correspond to β-actin controls.
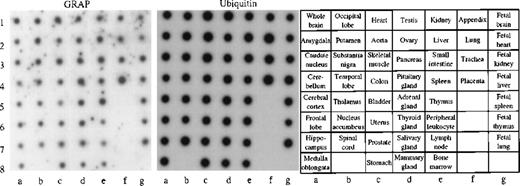
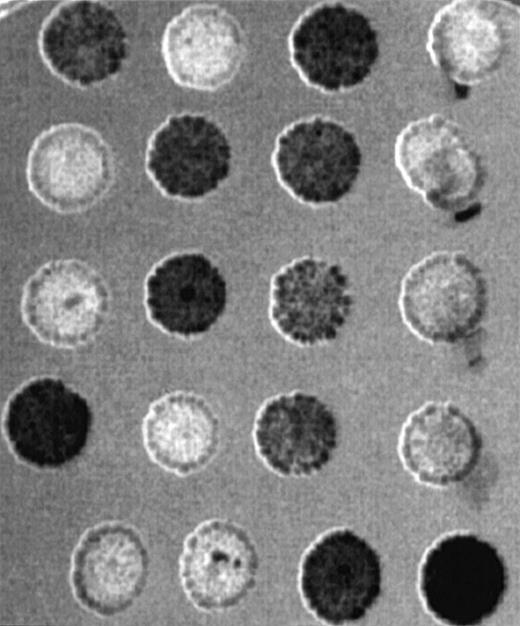
To obtain a more complete picture of the expression pattern of GRAP in human tissues, we used a human multi-tissue dot blot containing polyA+ RNA from 50 different normal human tissues. We found that GRAP is widely expressed in all human tissues, though the intensity of the signal is variable (Figure5). To confirm the wide distribution and expression of GRAP in human tissues, we used the ORF of GRAPand searched the BCM search launcher web site (Human Genome Center, Baylor College of Medicine) for sequence similarities betweenGRAP and the database of Expressed Sequence Tags (dbEST). Human ESTs with high homology to GRAP (98%-100%) were expressed in normal adult, embryonic, and neonatal tissues, and also in colon carcinoma (Table 2). Based on the above evidence, GRAP appears to be widely expressed in normal and neoplastic tissues, and its expression is not restricted to cells and tissues containing GM-CSF receptors.
Human RNA master dot blot.
The human RNA dot blot contains polyA+ RNA extracted from 50 different human tissues. The dot blot was hybridized with GRAP sequences. As a control, the same dot blot was hybridized with ubiquitin cDNA sequences (middle panel). The dot blot diagram is shown on the right panel.
Human RNA master dot blot.
The human RNA dot blot contains polyA+ RNA extracted from 50 different human tissues. The dot blot was hybridized with GRAP sequences. As a control, the same dot blot was hybridized with ubiquitin cDNA sequences (middle panel). The dot blot diagram is shown on the right panel.
Human and mouse EST sequence homologues ofGRAP
| Species . | Homology tissue . | Accession no. . | % . |
|---|---|---|---|
| Human | Colon carcinoma | aa314224 | 98-100 |
| Germ cell tumor | aa738379 | ||
| Germinal center B cell | aa740763 | ||
| Adrenal gland | aa550797 | ||
| Fetal heart | n89402 | ||
| Neonate human brain | t03910 | ||
| 8-week embryo | aa332544 | ||
| Mouse | Kidney | aa986955 | 91-92 |
| Carcinoma/embryonic | aa073109 | ||
| Blastocyst/pre-implant | aa544398 | ||
| Total fetus | aa41562 |
| Species . | Homology tissue . | Accession no. . | % . |
|---|---|---|---|
| Human | Colon carcinoma | aa314224 | 98-100 |
| Germ cell tumor | aa738379 | ||
| Germinal center B cell | aa740763 | ||
| Adrenal gland | aa550797 | ||
| Fetal heart | n89402 | ||
| Neonate human brain | t03910 | ||
| 8-week embryo | aa332544 | ||
| Mouse | Kidney | aa986955 | 91-92 |
| Carcinoma/embryonic | aa073109 | ||
| Blastocyst/pre-implant | aa544398 | ||
| Total fetus | aa41562 |
GRAP shares homology with proteins in mouse
Using the ORF of GRAP, we searched the BCM search launcher web site for sequence similarities between GRAP and other ESTs. Several mouse EST sequences were homologous to GRAP (Table 2). The mouse ESTs were translated, and the consensus mouse GRAPsequence was aligned with the human GRAP protein. As shown in Figure6, the human and mouse GRAP proteins are highly conserved with 96% identity. The sequence similarity between GRAP and its mouse homologue was confirmed by Northern blot analysis. Total RNA from several mouse tissues and HL-60 cells (positive control) was transferred to a nylon membrane and hybridized with human GRAP sequences (nucleotides 71-673). The human GRAP sequences hybridized with a mouse homologue of GRAP message (mGRAP) (Figure 7). Mouse tissues express a single mRNA band at approximately 1.3 kb. The 2 prominent bands of 3.3 and 3.1 kb, seen on the Northern blots containing human RNA from HL-60 cells, are absent in the mouse, suggesting that the shorter transcript of GRAP contains the entire ORF (Figure 6).
Amino acid alignment of GRAP in human, mouse, and yeast.
Shaded boxes indicate the amino acids that differ between the human and mouse GRAP. The underlined sequence shows the part of GRAP that was used to make the EGFP-GRAP fusion protein. Amino acid residues not present in the GRAP sequence homologues are shown as dashes.
Amino acid alignment of GRAP in human, mouse, and yeast.
Shaded boxes indicate the amino acids that differ between the human and mouse GRAP. The underlined sequence shows the part of GRAP that was used to make the EGFP-GRAP fusion protein. Amino acid residues not present in the GRAP sequence homologues are shown as dashes.
Disruption of the GRAP homologue in S cerevisiae causes early accumulation of glycogen
GRAP homologue genes are also found in S pombe andS cerevisiae. At the protein level, the homology between GRAP,S cerevisiae, and S pombe is 45% and 48%, respectively. The predicted molecular weight of the S cerevisiae GRAP homologue protein GRH is the same as the human, 22.5 kd, but its function in yeast is unknown. To determine the phenotype of a GRH null mutation, we disrupted the GRAPhomologue locus in a wild-type diploid strain of SK-1 genetic background.30 Tetrad analysis of 2 disrupted diploids heterozygous for the grhδ1::URA3 allele (Figure 1B), showed 2:2 segregations for the URA3 marker. The grhδ1 mutant showed no obvious defects in growth on glucose, galactose, or glycerol at 30° or in growth on glucose at 37°. The mutants also showed glucose-regulated invertase expression and sensitivity to the glucose analogue 2-deoxyglucose (data not shown). To examine the effect on sporulation, we crossed 2 grhδ1::URA3mutant strains and found that the resultant mutant diploid strains could sporulate. Iodine vapor stain for glycogen accumulation after overnight growth showed 2:2 co-segregation with URA3 (Figure8). The grhδ1 mutant (URA3-positive) cells start to accumulate glycogen earlier. Overgrowing cells (2-day incubation) showed little difference in glycogen accumulation between wild-type and mutant cells (data not shown). Early accumulation of glycogen in yeast lacking the GRAP homologue represented an aberrant response to stress and suggested a role for GRAP in glucose or glycogen metabolism.
Northern blot analysis and expression of GRAP mRNA in mouse tissues and HL-60 cells.
The upper panel shows the expression of GRAP in various mouse tissues. Total RNA from human promyelocytic cells (HL-60) is used as a positive control. RNA size markers are shown on the left. The lower panel shows the same blot hybridized to β actin sequences.
Northern blot analysis and expression of GRAP mRNA in mouse tissues and HL-60 cells.
The upper panel shows the expression of GRAP in various mouse tissues. Total RNA from human promyelocytic cells (HL-60) is used as a positive control. RNA size markers are shown on the left. The lower panel shows the same blot hybridized to β actin sequences.
Glycogen accumulation of wild-type and mutant yeast cells with a disrupted GRAP homologue.
Five tetrads dissected from heterozygous diploids for thegrhδ1::URA3 allele were patched on the YED-2% glucose plate, incubated at 30°C overnight, and stained with iodine vapor. Dark-stained patches contain cells positive for glycogen accumulation as well as the URA3 marker (data not shown).
Glycogen accumulation of wild-type and mutant yeast cells with a disrupted GRAP homologue.
Five tetrads dissected from heterozygous diploids for thegrhδ1::URA3 allele were patched on the YED-2% glucose plate, incubated at 30°C overnight, and stained with iodine vapor. Dark-stained patches contain cells positive for glycogen accumulation as well as the URA3 marker (data not shown).
Discussion
The cytoplasmic portion of the GMR-α is important for GM-CSF–mediated signaling through the high-affinity GM-CSF receptor and in certain systems the α subunit appears capable of inducing increased uptake of glucose through the facilitative glucose transporters. Proteins associating with GMR-α that might mediate these signals, however, have not been identified. We used a yeast 2-hybrid system to identify proteins interacting with GMR-α. A cDNA encoding a novel protein designated GRAP was isolated, and the protein was shown to interact with the cytoplasmic segment of GMR-α. This interaction was verified biochemically by coimmunoprecipitation experiments in which anti-GMR-α antibody coprecipitated GMR-α and the EGFP-GRAP fusion protein. The isolation of GRAP, which directly interacts with GMR-α, offers an opportunity to explore signaling mediated by GMR-α.
A previously identified gene, drp, was isolated by differential screening of a human teratocarcinoma cell line, PA-1; drp is a partial clone thought to encode a 70-kd protein. Based on evidence derived from our studies, which includes in vitro coupled transcription translation, Kozak consensus sequence, and sequencing of cDNA clones isolated from HL-60 cells representing mRNA species detected on Northern blots, we believe that we have the entire ORF of GRAP. The GRAP cDNA clone has an open reading frame of 198 amino acids and a Kozak consensus sequence at position −3 to + 4 (gaaATGG). The predicted molecular weight of GRAP is in agreement with the predicted molecular weight of the S cerevisiae homologue. A GRAPhomologue identified in mouse tissues has 96% identity with the human GRAP and lacks the 3.1- and 3.3-kb mRNA species. Therefore, the smallest 1.3-kb mRNA detected in human and mouse Northern blots likely contains the entire ORF. The significance of the 3.1- and 3.3-kb GRAP messages in human cells is unknown. Analysis of the expression at the RNA level revealed that GRAP is widely expressed and is present in all tissues examined, though the degree of expression is variable. There is a GRAP homologue in Saccharomyces, and GRAP mRNA is expressed in cells that do not have GMR-α (eg, K562 and IM-9 cells).
To elucidate the function of GRAP in vivo, we disrupted GRH inS cerevisiae. The disruption did not interfere with normal yeast growth but did result in early accumulation of glycogen. Glycogen accumulation in wild-type yeast cells starts when cells enter the stationary phase, reflecting the stress response for yeast to nutrient limitation. The early accumulation of glycogen in the grh1mutant cells indicates an inappropriate stress response. Other known mutant yeast cells with similar phenotype include mutation of genes that cause defects in glucose signaling, such as reg1,decreased cyclic adenosine monophosphate-dependent kinase activity, such as ras2, cdc35/cyr1, and decreased glycogen synthase activity, such as pho85. The role of GRH in the regulation of glycogen accumulation requires further investigation, however. Glycogen metabolism and signal transduction in mammals and yeast share extensive similarities.31 We speculate that GRAP may play a role in glucose or glycogen metabolism.
We have identified a novel protein that binds to the cytoplasmic portion of GMR-α. The GRAP protein is widely expressed in human tissues and is highly conserved phylogenetically, indicating that its function is not restricted to cells expressing GM-CSF receptors. Phenotypic abnormalities characteristic of a stress response are observed in yeast cells when the GRAP homologue is deleted. GRAP is the first intracellular protein reported to interact directly with GMRα, though it is likely that the protein plays a wider role in cellular metabolism.
Acknowledgments
We thank Marian Carlson, Rodney Rothstein, and Aaron P. Mitchell for generously providing strains and plasmids, Maria Soushko for assisting with analysis of the GRH1 clones, and Nina Chicharoen for technical assistance.
Supported by grants RO1 CA30388, RO1 HL42107, and P30 CA08748 from the National Institutes of Health and by a research grant from the Leukemia and Lymphoma Society.
J.T. and N.K. contributed equally to this work.
Reprints:David W. Golde, Memorial Sloan–Kettering Cancer Center, 1275 York Avenue, New York, NY 10021; e-mail:d-golde@ski.mskcc.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal