Abstract
To begin to study the sequence variations identified in the 5′ flanking genomic DNA of the ankyrin gene in ankyrin-deficient hereditary spherocytosis patients and to provide additional insight into our understanding of the regulation of genes encoding erythrocyte membrane proteins, we have identified and characterized the erythroid promoter of the human ankyrin-1 gene. This compact promoter has characteristics of a housekeeping gene promoter, including very high G+C content and enzyme restriction sites characteristic of an HTF-island, no TATA, InR, or CCAAT consensus sequences, and multiple transcription initiation sites. In vitro DNAseI footprinting analyses revealed binding sites for GATA-1, CACCC-binding, and CGCCC-binding proteins. Transfection of ankyrin promoter/reporter plasmids into tissue culture cell lines yielded expression in erythroid, but not muscle, neural, or HeLa cells. Electrophoretic mobility shift assays, including competition and antibody supershift experiments, demonstrated binding of GATA-1, BKLF, and Sp1 to core ankyrin promoter sequences. In transfection assays, mutation of the Sp1 site had no effect on reporter gene expression, mutation of the CACCC site decreased expression by half, and mutation of the GATA-1 site completely abolished activity. The ankyrin gene erythroid promoter was transactivated in heterologous cells by forced expression of GATA-1 and to a lesser degree BKLF.
Ankyrins are a homologous family of multifunctional proteins involved in the local segregation of integral membrane proteins within functional domains on the plasma membrane.1,2 This important cellular localization of membrane proteins is provided by the relative affinities of the many different tissue-specific and developmentally regulated isoforms of ankyrin for target proteins. Ankyrin isoform diversity arises from both different gene products and from differential, alternative splicing or alternate polyadenylation of the same gene product. The complementary DNAs (cDNAs) for 3 different ankyrins, ankyrin-1, -2, and -3, have been cloned and their gene products studied.3-12
Ankyrin-1, first discovered in preparations of erythrocyte membranes, provides the primary linkage between the spectrin-actin–based erythrocyte membrane skeleton and the plasma membrane by attaching tetramers of spectrin to the cytoplasmic domain of band 3, the anion exchanger. Ankyrin-1 deficiency is one of the most common abnormalities found in the erythrocyte membranes of patients with hereditary spherocytosis (HS).14-16 Genetic studies have revealed that ankyrin-1 gene defects, primarily frameshift or nonsense mutations, are the most common cause of typical, dominant HS.17,18 One molecular mechanism that could lead to ankyrin deficiency is a mutation in the ankyrin-1 erythroid promoter, leading to decreased ankyrin synthesis. Sequence variations in the 5′ flanking DNA of the erythrocyte ankyrin gene have been identified in several kindreds with both dominant and recessively inherited ankyrin-deficient HS.17 19 Whether these are disease-causing mutations or are merely polymorphisms in linkage disequilibrium with the as yet, unidentified mutation is unknown.
To provide additional insight into our understanding of the regulation of genes encoding erythrocyte membrane proteins and to obtain the tools necessary for study of the sequence variations identified in the 5′ flanking genomic DNA of the ankyrin-1 gene in patients with ankyrin-deficient HS, we have identified and characterized the erythroid promoter of the human ankyrin-1 gene. It is a compact, housekeeping gene-like promoter with a single GATA-1 site responsible for its erythroid specificity.
Materials and methods
Genomic cloning
A human ankyrin-1 cDNA fragment corresponding to the 5′ end of the coding region, ANK58,3 was used as hybridization probe to screen a human genomic DNA library. The library is a Charon4A bacteriophage library, containing fragments of genomic DNA partially digested with AluI and HaeIII withEcoRI linkers added. Selected recombinants that hybridized to the screening probe were purified and subcloned into pGEM-7Z plasmid vectors (Promega Corp, Madison, WI). Subcloned fragments were analyzed by restriction endonuclease digestion, Southern blotting, and nucleotide sequencing.
Nucleotide sequencing
Nucleotide sequencing was performed using the dideoxy chain termination method of Sanger et al20 with T7 DNA polymerase (Sequenase, US Biochemical Corp, Cleveland, OH). The sequencing primers were the Sp6 or T7 vectors of the pGEM-7Z plasmid vector or, for some reactions, synthetic oligonucleotides corresponding to known cDNA sequences. Deoxyinosine triphosphate was substituted for deoxyguanosine triphosphate to resolve band compressions and ambiguities.
RNA preparation
Total RNA was prepared from human fetal liver tissue, human bone marrow, or from the human tissue culture cell lines K562 (chronic myelogenous leukemia in blast crisis with erythroid characteristics, ATCC, CCL 243), HEL (human erythroleukemia, ATCC, TIB 180), HeLa (epithelial carcinoma, cervix, CCL 2), or HL60 (promyelocytic leukemia, CCL 240) as described.21
Primer extension analyses and ribonuclease protection assays
The transcription initiation site of the ankyrin-1 cDNA was determined using primer extension analysis and RNase protection assays. Primers A or B (Table 1) were used in primer extension reactions as described.22 Templates in these reactions were 10 μg of total human fetal liver RNA or 10 μg of total RNA from the human cell lines K562, HEL, HeLa, and HL60, or 10 μg of transfer RNA (tRNA). For ribonuclease protection assays (RPAs), a 32P-labeled antisense RNA probe was synthesized by transcription with T7 polymerase of a 584-base pair (bp)HindIII-ScaI fragment corresponding to the first exon and 5′ flanking sequences of the human ankyrin-1 gene. The probe (1 × 105 cpm per assay) was hybridized to template RNA at 42°C for 16 hours. Templates in these reactions were 20 μg of total human fetal liver RNA, 20 μg of total RNA from the human cell lines HEL, K562, HeLa, and HL60, or 20 μg of tRNA. Hybrids were digested with a mixture of the nucleases RNase A and RNase T1 (0.125 and 5 μ, respectively, per assay) at 37°C for 30 minutes. After digestion, protected fragments were detected by autoradiography after electrophoresis in 6% polyacrylamide—7 mol/L urea gels. Further increases in nuclease concentration or length of incubation did not alter the pattern of the protected fragment (not shown).
Oligonucleotide primers
| A | 5′-CGC AAGCTTCCGGACCACCTCATCCTGCC-3′ |
| B | 5′-CTA CCCCATTCCGCAGGTGA-3′ |
| C | 5′-CTTGCTCTCAGAAAGCTGGTAGC-3′ |
| D | 5′-CCATCCTAATACGACTCACTATAGGGC-3′ |
| E | 5′-GGCGGGCGCCACGCCTCCGCC-3′ |
| F | 5′-GGCCTCGAGGGGCCCGCCGAAGGGCCTTGGGG-3′ |
| G | 5′-AACTCTGGGCTCTGAACTCAGCTC-3′ |
| H | 5′-CCCTCGGGGGCCTGGCCCGCACGT-3′ |
| I | 5′-CGCCTCTGGGGCCGATAAGGCCCTCGGGGG-3′ |
| J | 5′-CCCCCGAGGGCCTTATCGGCCCCAGAGGCG-3′ |
| K | 5′-CCCAACACCTGCTGCCTCAG-3′ |
| L | 5′-CTCAGGCAGCAGGTGTTGGG-3′ |
| M | 5′-CCCCTCCGCCCGCCCGTGCCG-3′ |
| N | 5′-CGGCACGGGCGGGCGGAGGGG-3′ |
| O | 5′-GGCGGAGGCGTGGCGCCCGCC-3′ |
| P | 5′-TAAATGAAGCCCGCCCCAGTCGATC-3′ |
| Q | 5′-GATCGACTGGGGCGGGCTTCATTTA-3′ |
| R | 5′-AGCTTCAGGGTGTGGCTAGCT-3′ |
| S | 5′-AGCTAGCCACACCCTGAAGCT-3′ |
| T | 5′-TAGAGCCACACCCTGGTAAG-3′ |
| U | 5′-CTTACCAGGGTGTGGCTCTA-3′ |
| A | 5′-CGC AAGCTTCCGGACCACCTCATCCTGCC-3′ |
| B | 5′-CTA CCCCATTCCGCAGGTGA-3′ |
| C | 5′-CTTGCTCTCAGAAAGCTGGTAGC-3′ |
| D | 5′-CCATCCTAATACGACTCACTATAGGGC-3′ |
| E | 5′-GGCGGGCGCCACGCCTCCGCC-3′ |
| F | 5′-GGCCTCGAGGGGCCCGCCGAAGGGCCTTGGGG-3′ |
| G | 5′-AACTCTGGGCTCTGAACTCAGCTC-3′ |
| H | 5′-CCCTCGGGGGCCTGGCCCGCACGT-3′ |
| I | 5′-CGCCTCTGGGGCCGATAAGGCCCTCGGGGG-3′ |
| J | 5′-CCCCCGAGGGCCTTATCGGCCCCAGAGGCG-3′ |
| K | 5′-CCCAACACCTGCTGCCTCAG-3′ |
| L | 5′-CTCAGGCAGCAGGTGTTGGG-3′ |
| M | 5′-CCCCTCCGCCCGCCCGTGCCG-3′ |
| N | 5′-CGGCACGGGCGGGCGGAGGGG-3′ |
| O | 5′-GGCGGAGGCGTGGCGCCCGCC-3′ |
| P | 5′-TAAATGAAGCCCGCCCCAGTCGATC-3′ |
| Q | 5′-GATCGACTGGGGCGGGCTTCATTTA-3′ |
| R | 5′-AGCTTCAGGGTGTGGCTAGCT-3′ |
| S | 5′-AGCTAGCCACACCCTGAAGCT-3′ |
| T | 5′-TAGAGCCACACCCTGGTAAG-3′ |
| U | 5′-CTTACCAGGGTGTGGCTCTA-3′ |
5′ rapid amplification of cDNA ends
The 1 μg of total human fetal liver RNA was reverse transcribed using primer A (Table 1) and avian myeloblastosis virus (AMV) reverse transcriptase (Promega Corp). Single-stranded oligonucleotide ligation and polymerase chain reaction (PCR) amplification were carried out as described using primers B+D and C+D.23 24 Amplification products were subcloned and sequenced.
Cell culture
The tissue culture cell lines K562 and HEL (erythroid), SH-SY5Y (neural), and HeLa (nonerythroid) were used to study expression of the putative promoter of the ankyrin-1 gene. K562, HEL, and SH-Sy5Y cells were maintained in RPMI 1640 medium, containing 10% fetal calf serum. HeLa cells were maintained in Eagle's minimal essential media, supplemented with 10% fetal calf serum.
Preparation of nuclear extracts
Nuclear extracts were prepared from K562, HEL, MEL (murine erythroleukemia, NIGMS GM00086E), and HeLa cells by hypotonic lysis, followed by high salt extraction of nuclei as described by Andrews and Faller.25
DNase I footprinting in vitro
Probes for DNAse I footprinting were by produced by PCR amplification of plasmid p656 (see below) as template and 1 of 2 pairs of oligonucleotide primers, E+F and G+H. One oligonucleotide in each reaction was 5′ end labeled with 32P-ATP using polynucleotide kinase before use in PCR. Footprinting reaction mixes contained 1 to 20 μg MEL cell nuclear extracts, 20 000 cpm of labeled probe, and 1 μg of poly (dI-dC). After digestion with DNase I, samples were electrophoresed in 6% polyacrylamide gels, the gels were dried and subjected to autoradiography.
Preparation of promoter-reporter plasmids for transfection assays
Test plasmids were prepared by inserting an approximately 2270-bp fragment of the 5′ flanking ankyrin-1 genomic DNA upstream of the firefly luciferase reporter gene in the plasmid pGL2B (Promega Corp). Serial truncations of this 2270-bp fragment in the pGL2B plasmid were constructed using convenient restriction enzyme sites or PCR amplification. One promoter fragment, p656, was inserted into pGL2B in both orientations. All test plasmids were sequenced to exclude cloning or PCR-generated artifacts.
Transient transfection analyses
All plasmids tested were purified using Qiagen columns (Qiagen, Inc, Chatsworth, CA) or cesium chloride plasmid purification and at least 2 preparations of each plasmid were tested. The 107 K562, HEL, and SH-SY5Y cells were transfected by electroporation with a single pulse of 300 V at 960 microfarad (μF) with 20 μg of test plasmid and 0.5 μg of pCMVβ, a mammalian reporter plasmid expressing β-galactosidase driven by the human cytomegalovirus immediate early gene promoter (Clontech). The 105 HeLa or C2C12 (murine myoblast, ATCC 1772-CRL) cells were transfected with 2.0 μg test plasmid and 0.25 μg of the pCMVβ plasmid by lipofection using 4 μL Lipofectamine (Gibco BRL Life Technologies, Inc, Gaithersburg, MD). Twenty-four hours after transfection, cells were harvested, lysed, and the activity of both luciferase and β-galactosidase activity determined in cell extracts. All assays were performed in triplicate. Differences in transfection efficiency were determined by cotransfection with the pCMVβ plasmid. For transactivation assays, HeLa cells were transfected using 1 μg of reporter plasmid and varying amounts of GATA-1, BKLF (basic Kruppel-like factor), or EKLF (erythroid Kruppel-like factor) cDNA expression plasmids26-28 (see below) and the reporter gene activity assayed.
Electrophoretic mobility shift analyses
Binding reactions were carried out as described.26Competitor oligonucleotides were added at molar excesses of 10- or 100-fold. Resulting complexes were separated by electrophoresis through 6% polyacrylamide gels in 0.5X tris-borate-EDTA at 21°C at 200W for 2 hours. Gels were dried and subjected to autoradiography.
COS cells (107) were transfected with 20 μg of the expression plasmids pMT/BKLF (a kind gift of Drs M. Crossley and S. Orkin.) or pSG5/EKLF (a kind gift of Dr J. Bieker.) as described above. Forty-eight hours after transfection, nuclear extracts were prepared for use in gel shift analyses. Antibodies to GATA-1 and Sp1 were obtained from Santa Cruz Biotechnologies (Santa Cruz, CA). Antibodies to BKLF and EKLF were a kind gift of Drs M. Crossley and S. Orkin.
Computer analyses
Computer-assisted analyses were performed using the sequence analysis software package of the University of Wisconsin Genetics Computer Group (UW GCG; Madison, WI) and the BLAST algorithm, National Center for Biotechnology Information (Bethesda, MD).29 30
The sequences reported in this paper have been deposited in the GenBank database (accession number U50092).
Results
Cloning of chromosomal gene: isolation and analysis of recombinant clones
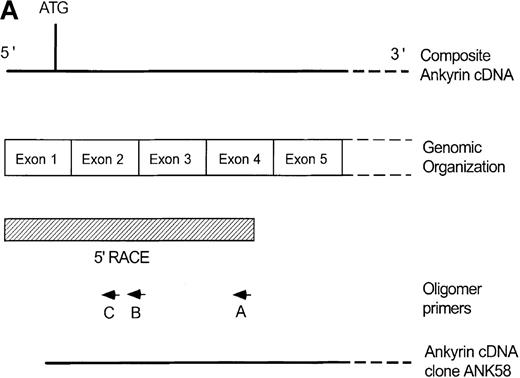
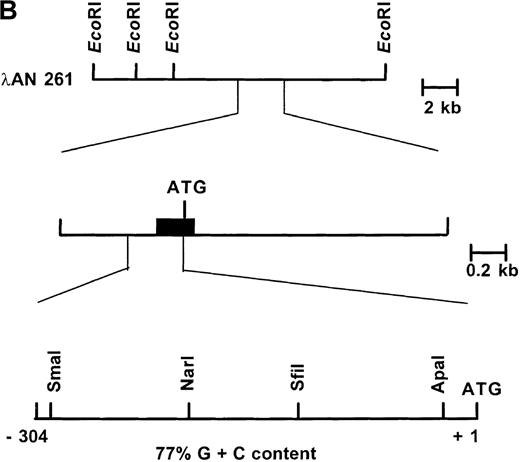
Primary screening of a human genomic DNA library with the ankyrin-1 cDNA probe ANK58 (Figure 1A) yielded 5 hybridization-positive plaques. Selected recombinants were analyzed and 1 clone identified, λAN261, that spanned about 16-kilobase (kb) of DNA containing the ankyrin-1 gene. A limited restriction map of this region is shown in Figure 1B.
Structure of the 5′ end of the human ankyrin-1 erythroid cDNA.
(A) A diagram of the 5′ end of the human ankyrin-1 cDNA is shown. The composite ankyrin cDNA is denoted by the line and the location of the initiation codon is shown (top). The genomic organization of the 5′end of the ankyrin-1 gene is shown, with the exons denoted by the boxes. Sequences obtained by 5′ RACE are denoted by the hatched box. Oligonucleotide primers used in 5′ RACE experiments are denoted by the arrows. The location of the probe used in genomic library screening, ANK58, is shown (bottom). (B) The 5′ end of the human ankyrin-1 chromosomal gene. A genomic DNA clone, λAN 261, spanning approximately 16-kb of DNA, was isolated from a human genomic DNA library. An EcoRI restriction map of clone λAN 261 is shown (top). This clone contained exon 1 of the ankrin-1 erythroid gene in a 2-kb EcoRI fragment (middle). The core ankyrin-1 promoter had very high G+C content and contained restriction enzyme sites found in HTF islands (bottom). See text for details.
Structure of the 5′ end of the human ankyrin-1 erythroid cDNA.
(A) A diagram of the 5′ end of the human ankyrin-1 cDNA is shown. The composite ankyrin cDNA is denoted by the line and the location of the initiation codon is shown (top). The genomic organization of the 5′end of the ankyrin-1 gene is shown, with the exons denoted by the boxes. Sequences obtained by 5′ RACE are denoted by the hatched box. Oligonucleotide primers used in 5′ RACE experiments are denoted by the arrows. The location of the probe used in genomic library screening, ANK58, is shown (bottom). (B) The 5′ end of the human ankyrin-1 chromosomal gene. A genomic DNA clone, λAN 261, spanning approximately 16-kb of DNA, was isolated from a human genomic DNA library. An EcoRI restriction map of clone λAN 261 is shown (top). This clone contained exon 1 of the ankrin-1 erythroid gene in a 2-kb EcoRI fragment (middle). The core ankyrin-1 promoter had very high G+C content and contained restriction enzyme sites found in HTF islands (bottom). See text for details.
The 5′-flanking genomic DNA sequence of the human ankyrin-1 gene exhibits features of a housekeeping gene promoter.
The nucleotide sequence of the 5′ flanking genomic DNA of the human ankyrin-1 gene is shown in Figure 2. Inspection of the sequence reveals features characteristic of a housekeeping gene promoter, including lack of consensus TATA, InR, or CCAAT sequences.31,32 In addition, this region appears to be a HpaII tiny fragment (HTF) island, based on a high G+C content (77% between positions −1 to −306) (all numbering is relative to the A of the translation initiator ATG) and a cluster of characteristic restriction enzyme sites, ApaI, SfiI,SmaI, and NarI (Figure 1B).33 34 Consensus sequences for a number of potential DNA-binding proteins, including GATA-1, CACCC-binding proteins, and CGCCC-binding proteins, are present in the 5′ flanking sequence (Figure 2).
The 5′ flanking genomic DNA sequence.
The nucleotide sequence of the 5′ flanking genomic DNA of the human ankyrin-1 gene is shown. Consensus sequences for potential DNA-protein binding sites are underlined. The location of the initiator methionine codon is underlined and in bold. The junction between exon 1 and intron 1 is shown by the inverted triangle.
The 5′ flanking genomic DNA sequence.
The nucleotide sequence of the 5′ flanking genomic DNA of the human ankyrin-1 gene is shown. Consensus sequences for potential DNA-protein binding sites are underlined. The location of the initiator methionine codon is underlined and in bold. The junction between exon 1 and intron 1 is shown by the inverted triangle.
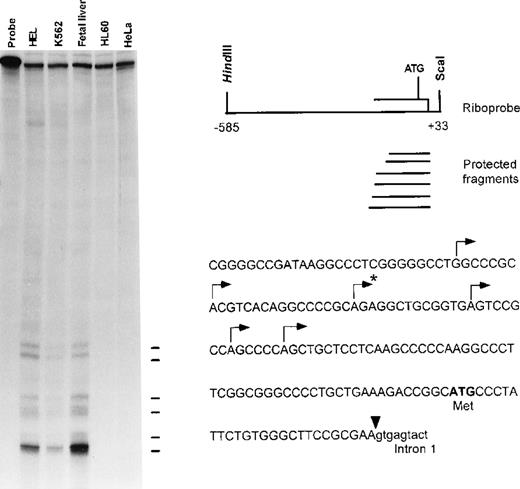
Mapping the human ankyrin-1 erythroid messenger RNA transcription initiation sites and identification of 5′ cDNA sequences
To identify the 5′ end of the human ankyrin-1 cDNA, primer extension analyses and RNase mapping with RNase A and RNase T1 nucleases were performed. These experiments identified 6 transcription initiation sites. The longest fragment obtained by RNase protection (Figure 3) and primer extension (not shown) predicted the presence of an additional 23-bp in the messenger RNA (mRNA) upstream of the 5′ end of the sequence obtained from cDNA cloning. These additional 23-bp of upstream 5′ untranslated sequence were obtained by 5′ rapid amplification of cDNA ends (RACE) (Figure 2) and verified by comparison to corresponding genomic DNA sequences. No additional ATGs were present in the 5′ untranslated sequences. Taken together, these data suggest that these sequences are at or very near the 5′ end of the human ankyrin-1 erythroid cDNA.
Mapping the 5′ end of the human ankyrin-1 cDNA.
RNase mapping was carried out using an ankyrin-1 riboprobe (upper right). This probe contains sequences corresponding to exon 1 of the ankyrin-1 gene, 5′flanking DNA, and part of intron 1. Ten micrograms of K562, HEL, human fetal liver, HL60 total RNA, or tRNA were used as template. Six transcription initiation sites, noted by the lines (left) and annotated by an arrow above the corresponding sequences (lower right), were identified. The 5′ end of the previously reported sequence obtained from cDNA cloning is denoted by the asterisk.
Mapping the 5′ end of the human ankyrin-1 cDNA.
RNase mapping was carried out using an ankyrin-1 riboprobe (upper right). This probe contains sequences corresponding to exon 1 of the ankyrin-1 gene, 5′flanking DNA, and part of intron 1. Ten micrograms of K562, HEL, human fetal liver, HL60 total RNA, or tRNA were used as template. Six transcription initiation sites, noted by the lines (left) and annotated by an arrow above the corresponding sequences (lower right), were identified. The 5′ end of the previously reported sequence obtained from cDNA cloning is denoted by the asterisk.
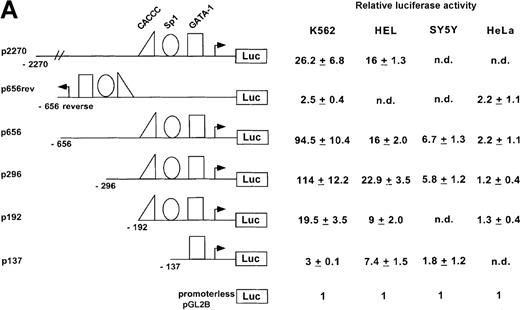
An ankyrin-1 gene promoter fragment is active in erythroid cells.
To investigate whether the region from −656 to −15 was capable of directing expression of a reporter gene in cultured mammalian cells, test plasmids p656 or p656-reverse were transiently transfected into erythroid (K562 or HEL) or nonerythroid cells. The relative luciferase activity was determined 48 hours after transfection and compared with the activity obtained with pGL2B, a negative control, the promoterless plasmid, and pGL2P, a positive control, the luciferase reporter gene under control of the SV40 early promoter. The ankyrin-1 promoter plasmid, p656, directed high-level expression of the luciferase reporter gene in erythroid cells, but not HeLa, SH-Sy5Y (Figure 4A), or C2C12 (not shown, relative luciferase activity = 1.23 ± 0.23) cells. These cell lines were chosen because ankyrin-1 expression has previously been demonstrated in muscle and neural tissues.1 2 The plasmid with the promoter in reverse orientation, p653rev, did not direct expression of the reporter gene in any of the cell lines. Transient transfection analysis of deletions of this ankyrin-1 gene erythroid promoter fragment, as well as a promoter fragment with an additional approximately 1400-bp upstream, identified a 286-bp minimal promoter fragment, p296, that directed ankyrin-1 gene erythroid-specific expression (Figure 4B).
Activity of the ankyrin-1 gene erythroid promoter in erythroid and nonerythroid cell lines in transient transfection assays.
(A) Plasmids containing 5′ flanking DNA of the ankyrin-1 gene upstream of a firefly luciferase reporter gene were transfected into K562, HEL, and SY5Y (human neuroblastoma), or HeLa cells and relative luciferase activity determined as described. The data are means ± SD of at least 6 independent transfection experiments. (B) Promoter/reporter plasmids containing mutations in consensus DNA-protein binding sites of the ankyrin-1 gene promoter, denoted by the “X” and listed to the right, were transiently transfected into K562 cells and relative luciferase reporter gene expression determined as described.
Activity of the ankyrin-1 gene erythroid promoter in erythroid and nonerythroid cell lines in transient transfection assays.
(A) Plasmids containing 5′ flanking DNA of the ankyrin-1 gene upstream of a firefly luciferase reporter gene were transfected into K562, HEL, and SY5Y (human neuroblastoma), or HeLa cells and relative luciferase activity determined as described. The data are means ± SD of at least 6 independent transfection experiments. (B) Promoter/reporter plasmids containing mutations in consensus DNA-protein binding sites of the ankyrin-1 gene promoter, denoted by the “X” and listed to the right, were transiently transfected into K562 cells and relative luciferase reporter gene expression determined as described.
The ankyrin-1 erythroid promoter contains binding sites for GATA-1, Sp1, and CACCC-related binding proteins.
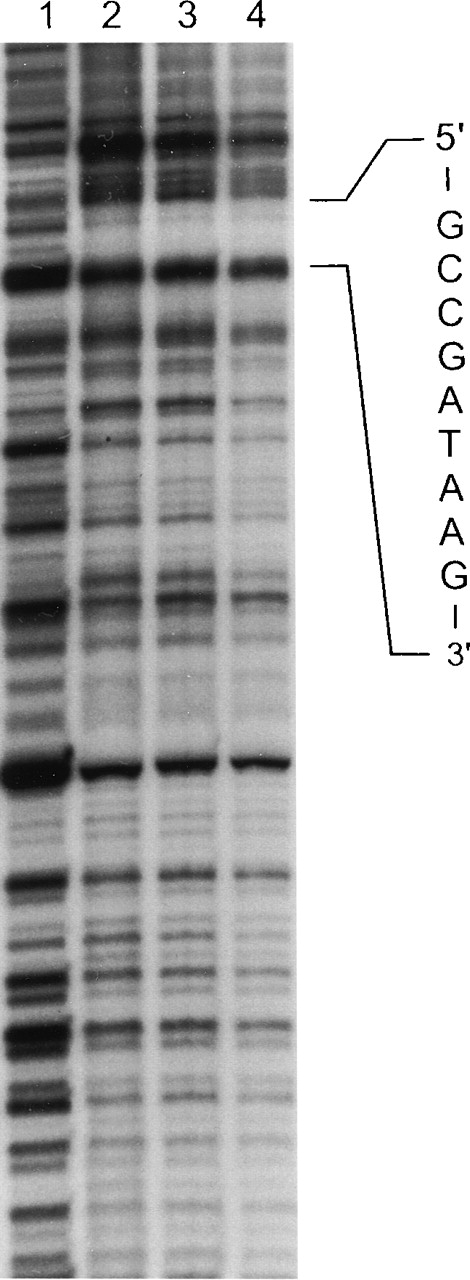
The 286-bp minimal promoter fragment, p296, contains consensus binding sequences for GATA, CACCC, and CGCCC-binding proteins. To identify binding sites for transcription factors within the ankyrin-1 promoter, in vitro DNase I footprinting analysis with nuclear extracts from K562 cells was performed in 2 steps. Footprints at 2 protected sites were observed. Site 1, GCCGATAAG, contained consensus binding sequences for GATA-1 (Figure 5). GATA-1 is a transcription factor that plays a critical role in erythropoiesis via its binding to the sequence GATA of the promoters and/or enhancers of nearly all erythroid-expressed genes. The second site, GCCACCCCTCCGCCC, consists of sequences recognized by members of the Kruppel-like family of transcription factors and other CACCC-related proteins (not shown). These include Sp1, EKLF, and BKLF. EKLF, erythroid Kruppel-like factor, is a transcription factor that is important in β-globin expression via binding to the CACCC sequence of the β -globin gene promoter. BKLF, basic Kruppel-like factor, is a widely, but not ubiquitously expressed transcription factor that binds to the CACCC sequence of many erythroid gene promoters and enhancers.
In vitro DNase I footprinting of the human ankyrin-1 promoter.
In vitro DNAse I footprinting of the human ankyrin-1 gene promoter was performed using erythroid (K562) extracts. Lane 1 reaction mixture does not contain nuclear extracts. Lanes 2, 3, and 4 contain nuclear extracts and increasing amounts of DNaseI. (See text for details.) A protected site, GCCGATAAG, corresponding to a GATA-1 consensus binding site was identified. In additional experiments, a second protected site, GCCACCCCTCCGCCC, corresponding to binding motifs for Sp1 and CACCC-related proteins was identified in the upstream sequence (not shown).
In vitro DNase I footprinting of the human ankyrin-1 promoter.
In vitro DNAse I footprinting of the human ankyrin-1 gene promoter was performed using erythroid (K562) extracts. Lane 1 reaction mixture does not contain nuclear extracts. Lanes 2, 3, and 4 contain nuclear extracts and increasing amounts of DNaseI. (See text for details.) A protected site, GCCGATAAG, corresponding to a GATA-1 consensus binding site was identified. In additional experiments, a second protected site, GCCACCCCTCCGCCC, corresponding to binding motifs for Sp1 and CACCC-related proteins was identified in the upstream sequence (not shown).
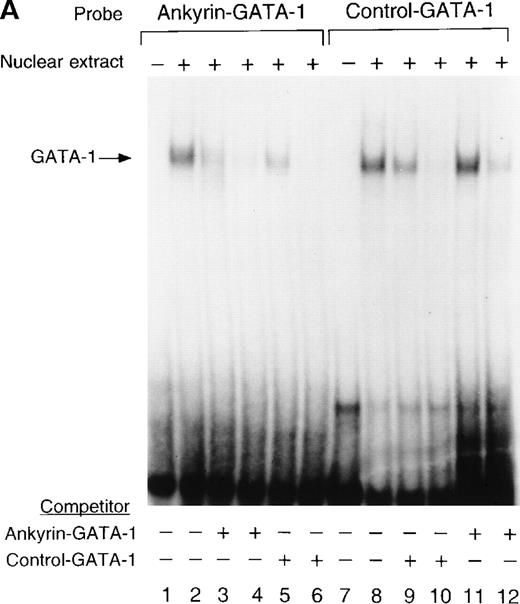
GATA-1 binds the ankyrin-1 gene promoter site in vitro.
To determine whether nuclear proteins could bind this GATA-1 site in vitro, double-stranded (DS) oligonucleotides containing the corresponding ankyrin-1 promoter GATA-1 sequences (I+J, Table 1) or control sequences (K+L; Table 1)35 were prepared and used in gel shift analyses. When DS oligonucleotides containing the footprinted GATA-1 sequences were used in gel shift analyses, a single retarded species was observed in K562 extracts (Figure6A), but not in HeLa extracts (not shown). These species migrated at the same location as a control oligonucleotide containing a GATA-1 consensus sequence. This species was effectively competed both by an excess of unlabeled homologous oligonucleotide and by an excess of unlabeled control GATA-1 oligonucleotide. The inclusion of GATA-1 antisera abolished most or all of the DNA binding (Figure 6B). These data indicate that GATA-1 binds to this site in the ankyrin-1 gene promoter in vitro.
Electrophoretic mobility shift assays of the footprinted GATA-1 site of the human ankyrin-1 gene promoter.
(A) Gel mobility shift assays using oligonucleotides corresponding to the ankyrin-1 promoter footprinted GATA-1 site or control GATA-1 site and nuclear extracts from erythroid (K562) cells. Increasing amounts of unlabeled, DS oligonucleotide (10 molar and 100 molar excess) corresponding to the ankyrin-1 GATA-1 site or a control GATA-1 site were added to the reactions as competitor where indicated. (B) Gel mobility shift assays using oligonucleotides corresponding to the ankyrin-1 promoter footprinted GATA-1 site or control GATA-1 site and K562 cell nuclear extracts. GATA-1 antibody was added to the reaction mixtures where indicated.
Electrophoretic mobility shift assays of the footprinted GATA-1 site of the human ankyrin-1 gene promoter.
(A) Gel mobility shift assays using oligonucleotides corresponding to the ankyrin-1 promoter footprinted GATA-1 site or control GATA-1 site and nuclear extracts from erythroid (K562) cells. Increasing amounts of unlabeled, DS oligonucleotide (10 molar and 100 molar excess) corresponding to the ankyrin-1 GATA-1 site or a control GATA-1 site were added to the reactions as competitor where indicated. (B) Gel mobility shift assays using oligonucleotides corresponding to the ankyrin-1 promoter footprinted GATA-1 site or control GATA-1 site and K562 cell nuclear extracts. GATA-1 antibody was added to the reaction mixtures where indicated.
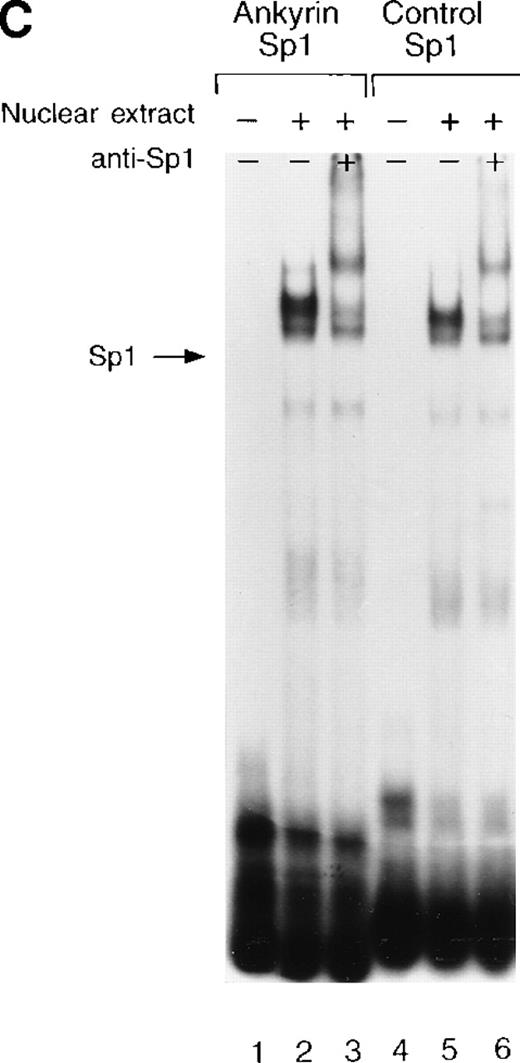
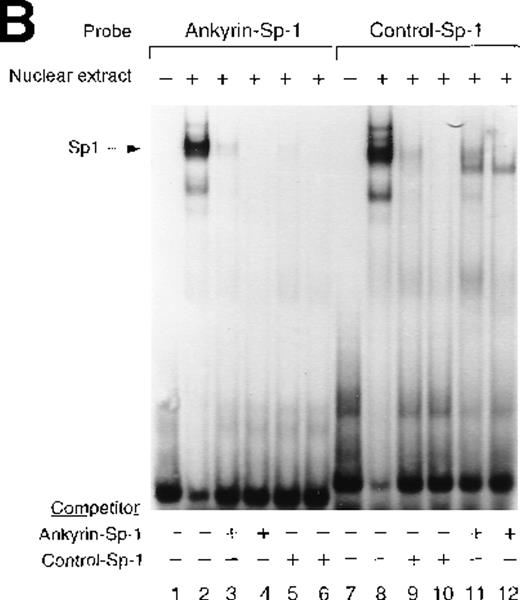
Sp1 and BKLF bind to the ankyrin-1 gene promoter CACCC site in vitro.
Site 2 identified by DNase I footprinting contains CACCC and CGCCC, consensus binding sites for CACCC-related binding proteins and members of the Kruppel family of transcription factors. Although CACCC-binding proteins and Kruppel-like proteins both bind CACCC and CGCCC sequences, they show distinct binding preferences. To determine whether the nuclear proteins Sp1, BKLF, or EKLF bind these sites in vitro,electrophoretic mobility shift assays were performed. When DS oligonucleotides containing the corresponding ankyrin-1 promoter site Sp1 (M+N; Table 1) or CACCC (E+O, Table 1) sequences or control sequences (Sp1: P+Q;36,37 CACCC: R+S,28 Table1) were used in gel shift analyses with K562 extracts, 1 larger, slower-migrating species and 2 smaller, faster-migrating species were detected. These species migrated at the same location as those obtained using control oligonucleotides containing either Sp1 (Figure7A) or CACCC consensus binding sequences (Figure 7B). All 3 species were effectively competed by an excess of unlabeled homologous oligonucleotide and an excess of unlabeled Sp1 or CACCC control oligonucleotides. The inclusion of Sp1 antisera supershifted the larger, slower migrating species in the gel when either the ankyrin Sp1 oligonucleotide (Figure 7C) or the CACCC oligonucleotide (not shown) was used.
Electrophoretic mobility shift assays of the footprinted Sp1 site in the human ankyrin-1 gene promoter.
(A) Gel mobility shift assays using oligonucleotides corresponding to the ankyrin-1 promoter footprinted Sp1 site or control Sp1 site and nuclear extracts from erythroid (K562) cells. Increasing amounts of unlabeled, DS oligonucleotide (10 molar and 100 molar excess) corresponding to the ankyrin-1 Sp1 site or a control Sp1 site were added to the reactions as competitor where indicated. (B) Gel mobility shift assays using ankyrin-1 promoter oligonucleotides corresponding to the footprinted CACCC site and K562 cell nuclear extracts. Increasing amounts of unlabeled, DS oligonucleotide corresponding to the ankyrin-1 CACCC site, a control CACCC site (10 molar and 100 molar excess), and a control Sp1 site (100 molar excess) were added to the reactions as competitor where indicated. (C) Gel mobility shift assays using oligonucleotides corresponding to the ankyrin-1 promoter footprinted Sp1 site or control Sp1 site and K562 cell nuclear extracts. Sp1 antibody was added to the reaction mixtures where indicated.
Electrophoretic mobility shift assays of the footprinted Sp1 site in the human ankyrin-1 gene promoter.
(A) Gel mobility shift assays using oligonucleotides corresponding to the ankyrin-1 promoter footprinted Sp1 site or control Sp1 site and nuclear extracts from erythroid (K562) cells. Increasing amounts of unlabeled, DS oligonucleotide (10 molar and 100 molar excess) corresponding to the ankyrin-1 Sp1 site or a control Sp1 site were added to the reactions as competitor where indicated. (B) Gel mobility shift assays using ankyrin-1 promoter oligonucleotides corresponding to the footprinted CACCC site and K562 cell nuclear extracts. Increasing amounts of unlabeled, DS oligonucleotide corresponding to the ankyrin-1 CACCC site, a control CACCC site (10 molar and 100 molar excess), and a control Sp1 site (100 molar excess) were added to the reactions as competitor where indicated. (C) Gel mobility shift assays using oligonucleotides corresponding to the ankyrin-1 promoter footprinted Sp1 site or control Sp1 site and K562 cell nuclear extracts. Sp1 antibody was added to the reaction mixtures where indicated.
To determine whether the CACCC-box binding transcription factors BKLF or EKLF could bind the ankyrin-1 gene promoter CACCC site in vitro, gel shifts using nuclear extracts prepared from COS cells transfected with expression plasmids containing either BKLF or EKLF cDNAs, and the ankyrin-1 gene CACCC site DS oligonucleotide or a control β-globin CACCC oligonucleotide (T+U)38 were performed. A major complex was identified in BKLF-transfected cells (Figure 8). This complex migrated at the same location as the control β-globin CACCC control oligonucleotide. The complexes obtained with both the ankyrin-1 and β-globin oligonucleotides were supershifted with an anti-BKLF antibody. When extracts from EKLF-transfected cells were used in similar experiments, a major complex was identified using the β-globin CACCC consensus sequence oligonucleotide. No complex was identified using the ankyrin-1 CACCC oligonucleotide (not shown). Together, these data indicate that Sp1 and BKLF, but not EKLF, bind to the ankyrin-1 gene promoter in vitro.
Electrophoretic mobility shift assays of the footprinted CACCC site of the human ankyrin-1 gene promoter.
Gel mobility shift assays using oligonucleotides corresponding to the ankyrin-1 promoter footprinted CACCC site or a control β-globin CACCC site and nuclear extracts from COS cells transfected with a BKLF cDNA expression plasmid. BKLF antibody was added to the reaction mixtures where indicated.
Electrophoretic mobility shift assays of the footprinted CACCC site of the human ankyrin-1 gene promoter.
Gel mobility shift assays using oligonucleotides corresponding to the ankyrin-1 promoter footprinted CACCC site or a control β-globin CACCC site and nuclear extracts from COS cells transfected with a BKLF cDNA expression plasmid. BKLF antibody was added to the reaction mixtures where indicated.
GATA-1 and CACCC-related proteins are both major activators of the human erythroid ankyrin-1 gene promoter.
To assess the relative importance of these transcription factor binding sites in promoter function, mutations were introduced into each of the 3 consensus binding sites protected in DNase I footprinting experiments and the affect assayed in mutant promoter/reporter plasmids on expression in transient transfections (Figure 4B). Mutation of the GATA site to GTTA, a mutation shown to disrupt GATA-1 binding,39 reduced promoter activity nearly to background, indicating that this site is of major importance in the ankyrin-1 gene promoter. Mutation of the CACCC site to CACGC, a mutation of the human β-globin gene CACCC site associated with β thalassemia because of decreased EKLF binding,40 had no effect on ankyrin promoter activity. When the CACCC site was completely abolished, CACCC to TTTTC, which also abolished an overlapping Sp1 site, ankyrin promoter activity was decreased by over half (43%). Mutation of the downstream Sp1 site, CCGCCCGCCC to CCGTTTTCCCG, had no effect on promoter activity.
Transactivation of the ankyrin-1 gene erythroid promoter in heterologous cells
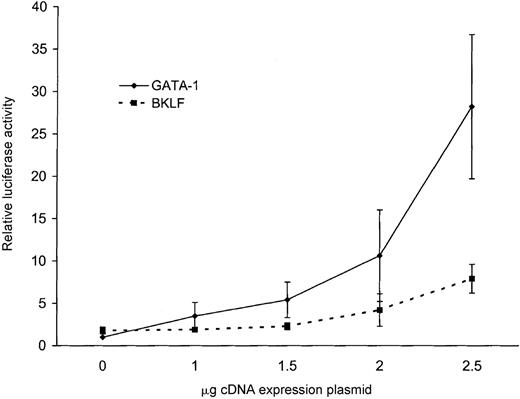
None of the ankyrin-1 promoter fragments directed expression of a reporter gene in HeLa cells, but the addition of GATA-1 by cotransfection conferred promoter activity to an ankyrin-1 promoter fragment. Cotransfection of 1 μg of an ankyrin-1 erythroid promoter fragment, p296, and increasing amounts of a GATA-1 cDNA expression plasmid resulted in increasing promoter activity with increasing amounts of GATA-1 plasmid (Figure 9). The ability of GATA-1 to transcriptionally activate the ankyrin-1 erythroid promoter in these cells, which do not contain this erythroid-specific factor, correlates with the inability of the ankyrin-1 erythroid promoter to function in these cells.
Forced expression of the human ankyrin-1 gene promoter in HeLa cells.
Cotransfection of the ankyrin-1 gene promoter/luciferase reporter plasmid p296 with increasing amounts of either GATA-1 (solid line) or BKLF (dotted line) cDNA expression plasmids in HeLa cells.
Forced expression of the human ankyrin-1 gene promoter in HeLa cells.
Cotransfection of the ankyrin-1 gene promoter/luciferase reporter plasmid p296 with increasing amounts of either GATA-1 (solid line) or BKLF (dotted line) cDNA expression plasmids in HeLa cells.
Cotransfection of 1 μg of an ankyrin-1 erythroid promoter fragment, p296, and increasing amounts of a BKLF cDNA expression plasmid resulted in a mild increase (4-fold) in promoter activity (Figure 9). Cotransfection of 1 μg of the ankyrin-1 erythroid promoter fragment, p296, and increasing amounts of an EKLF cDNA expression plasmid resulted in no change in promoter activity with increasing amounts of EKLF plasmid (not shown).
Discussion
These studies show that the human ankyrin-1 erythroid promoter is compact, ie, a very short fragment of DNA directs high-level expression in erythroid cells. Like other erythroid gene promoters, the combination of GATA-1 and CACCC-binding proteins appears to be essential for high-level expression of the ankyrin-1 gene.41-43 The consensus binding sites for GATA-1 and CACCC-binding proteins are present in very close proximity in the ankyrin-1 promoter. This combination may lead to cooperation between GATA-1 and CACCC-binding proteins to enhance transcription, as has been shown in several reports.44-48 CACCC-box binding proteins and members of the Kruppel-family of transcription factors both bind CACCC and CGCCC sequences; however, they show distinct binding preferences. Although Sp1 and BKLF were found to bind to the CACCC site of the ankyrin gene erythroid promoter in vitro, it is unknown what transcription factors bind this site in vivo. The interactions of Sp1 and/or BKLF with a broad spectrum of erythroid gene promoters make them likely candidates for binding to this site.
Analysis of HS-associated mutations in the ankyrin promoter reveals that 2 of the reported mutations are located in potential DNA-transcription factor binding sites. The −108 T to C mutation is located in a potential AP2 site, a widely expressed transcription factor. The −204 C to G mutation is located in a potential PEBP2/PEA2 site, a member of a family of transcription factors homologous to the Drosophila runt gene and the humanAML1 gene. Neither of these regions revealed protein binding in DNAse I in vitro footprinting assays. It will be important to analyze the effects of these mutations on ankyrin gene expression in vivo.
The diversity of ankyrin isoforms appears to be critical for specific cellular functions.1,2 Various tissue- and developmental stage-specific isoforms of ankyrin-1 are generated by both complex patterns of alternative splicing and alternate polyadenylation.8-12 As a muscle tissue-specific ankyrin-1 gene promoter has already been described,12 these observations extend the molecular basis of ankyrin-1 isoform diversity to include the use of tissue-specific, alternate promoters. Remarkably, the ankyrin-1 erythroid and muscle promoters are remote, more than 100 kb apart. This is similar to another membrane-associated gene, dystrophin, where 5 autonomous promoters, spread over a greater than 100-kb region, direct expression of cell type- and developmentally regulated-transcripts.49
The identification of an ankyrin-1 erythroid promoter with characteristics of a housekeeping gene promoter is surprising. Another erythroid gene, ferrochelatase, has a promoter with characteristics of a housekeeping gene, including lack of TATA or CAAT recognition sequences and a very high G+C content.50,51 However, unlike other erythroid-specific genes such as the globins, ferrochelatase must be expressed in all cell types to supply the heme required for respiratory cytochromes. Ankyrin-1 isoforms have been shown to be expressed in erythroid, neural, and muscle cells,1,2,8-12and it has been hypothesized that these isoforms are regulated by tissue-specific alternate promoters.8 It is possible that, like ferrochelatase, transcripts of ankyrin-1 are expressed at a low level in all cells and this promoter, in combination with other regulatory elements, controls their expression.
Acknowledgments
We thank Drs Crossley, Orkin, and Bieker for sharing reagents with us and we thank C. Wong for skilled technical assistance.
Supported in part by grants from the National Institutes of Health, the March of Dimes Birth Defects Foundation, and the American Heart Association-Connecticut Affiliate.
Reprints:Patrick G. Gallagher, Department of Pediatrics, Yale University School of Medicine, 333 Cedar St, PO Box 208064, New Haven, CT 06520-8064; email: patrick.gallagher@yale.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal