Abstract
The erythroid isoform of aminolevulinate synthase (eALAS) protein is a major control point in erythroid heme synthesis and hemoglobin formation. Erythroid cells were extracted from mouse blood and bone marrow and metabolically labeled with 35S-methionine. This was followed by immunoprecipitation of eALAS protein products. The results show that the N-terminus of the expected full-length 59-kd form of the eALAS protein is truncated in bone marrow erythroid cells by approximately 7 kd. More differentiated erythroid cells in the peripheral blood exhibit very little of this protein truncation. Erythroid cells from the bone marrow were isolated using monoclonal antibody TER-119 and were shown to contain a unique endoprotease activity that could cleave the eALAS protein to the shorter form in vitro. With or without the mitochondrial signal sequence, the eALAS protein could serve as a substrate for the cleavage. This cleavage renders a functional eALAS protein and only removes a domain of unclear function, which has previously been reported to vary in size as a result of alternative RNA splicing. The protease activity was enriched from the membranes of mitochondria from bone marrow cells and was shown to be different from mitochondrial processing peptidase, medullasin, and other known proteases. Apart from the mitochondrial processing peptidase that cleaves the import signal sequence, this is the first description of a mitochondrially located site-specific processing protease activity.
Adult hemoglobin is made up of 2α- and 2β-globin polypeptides, each of which contains a prosthetic heme group. Heme (a porphyrin chelate of iron known as Fe-protoporphyrin IX) controls translation of the globin messenger RNAs (mRNAs) by using a heme-controlled kinase1 to phosphorylate the translation initiation factor eIF2-alpha as well as the expression of other erythroid genes.2,3 Formation of heme itself initially requires the protoporphyrin IX skeleton, which is made in 7 enzymatic steps, starting with the rate-limiting aminolevulinate synthase (ALAS) [Enzyme Catalogue 2.3.1.37] catalyzed reaction between glycine and succinyl-coenzyme A (succinyl-CoA). Erythroid cells express an isoform of ALAS (eALAS or ALAS2) in addition to the ubiquitously expressed housekeeping form of ALAS (hALAS or ALAS1).4-9
eALAS and other erythroid-specific genes are induced in nucleated erythroid progenitor cells via specific transcription factors. Even after expulsion of the nucleus, a tight posttranscriptional control of heme synthesis maintains iron and protoporphyrin compounds at nontoxic levels despite the fact that the rate of heme production is at least 1 order of magnitude greater in erythroid cells than in nonerythroid cells. eALAS is believed to be the key control point for regulating heme synthesis in reticulocytes in response to heme, iron, or other physiological stimuli.8 Part of the regulation of eALAS and heme synthesis has been attributed to the iron-responsive element (IRE) in the 5′ untranslated region of the eALAS mRNA.10,11 Under low-iron conditions, iron regulatory proteins (IRP 1 and 2) will bind to the IRE in the eALAS mRNA and repress translation. However, increased iron or heme will convert IRP into a nonbinding form and allow translation.12,13 IRP has also been shown to respond to other physiological stimuli in mammalian erythroid cells, such as nitric oxide and erythropoeitin,14,15 making it an important control point in hemoglobin production. Heme also exerts a negative feedback effect on its own synthesis; increasing heme levels appear to block import of the eALAS protein into the mitochondria by binding to a specific cysteine-proline rich sequence in the signal peptide.16Heme may also exert a negative effect on iron release from transferrin in red cells.8
Previous work on hemin and iron regulation of erythroid heme synthesis has been mainly limited to erythroid cell lines. The levels of hemoglobin are considerably lower in these cell lines, and the genes of the heme synthesis pathway are not fully induced compared with natural hemoglobin-producing cells in the body.17 During studies on eALAS regulation in erythropoietic cells, we observed that 2 forms of the protein were sometimes detected by immunoprecipitation. Because there is no previous evidence of eALAS heterogeneity, we decided to investigate the origins of these 2 forms.
Materials and methods
Preparation of cell lysates
Balb/C mice (6-8 weeks old) were killed by cervical dislocation following local ethical guidelines. Bone marrow cells were extracted from the femurs and added to minimal essential medium (MEM) with Earle salts or Dulbecco modified Eagle medium (DMEM) without methionine (Gibco BRL, Paisley, UK) using a 27-and-3/4–gauge needle. Suspensions of spleen or liver cells were prepared by gently mashing the nonperfused excised organ on a nylon net and discarding sedimented debris. Peripheral blood was collected into heparinized plastic tubes. All cells were washed by low-speed centrifugation in serum-free medium and routinely applied to a Ficoll-Hypaque gradient (Ficoll-Hypaque Plus; Pharmacia, Uppsala, Sweden) to enrich reticulocytes. The cells were diluted to 3 mL in MEM, carefully overlaid on an equal volume of Ficoll-Hypaque in a 15-mL plastic tube, and centrifuged at 30g for 90 minutes at 4°C. The high- and low-density fractions of cells were removed with a Pasteur pipette, washed 3 times, and resuspended in IMDM without methionine with 10% fetal calf serum (FCS), L-glutamine, and streptomycin/penicillin G (Gibco).
The cells from blood and bone marrow were stained with May-Grünwald Giemsa (MGG) or methylene blue stain and examined by microscopy. Peripheral blood contained 2.7% reticulocytes. Following Ficoll-Hypaque separation, the lower fraction contained 4.0% reticulocytes, whereas the upper fraction was virtually depleted of reticulocytes. Bone marrow cells contained 32% erythropoietic cells; of the erythroblasts, 30% were proerythropoietic; 49%, basophilic; 12%, polychromatic; and 9%, orthochromatic. Although we routinely used the lower fraction from the Ficoll-Hypaque gradient in our experiments, separation of bone marrow cells does not give a clear enrichment of the protease activity, which may also be present in several erythroblast stages that separate into the upper Ficoll-Hypaque fraction. To acquire labeled extracts, we used the following concentrations of 35S-L-methionine as indicated: 3.7 × 1013 Bq or 0.37 MBq/μL (1000 Ci/mmol or 10 μCi/μL) (NEG-009A; New England Nuclear, Boston, MA). Labeled cells were washed twice in ice-cold phosphate-buffered saline (PBS) and resuspended for 30 minutes on ice in ratio immunoprecipitation assay (RIPA) detergent buffer containing 0.15 mol/L sodium chloride (NaCl); 1% Nonidet P-40; 0.5% deoxycholate; 0.1% sodium dodecyl sulfate (SDS); 50 mmol/L Tris HCl (trishydroxymethyl aminomethane hydrochloride) (pH 7.5); and either the LPPA protease inhibitors, 1 μmol/L leupeptin, 1 μmol/L pepstatin, 2 mmol/L Pefabloc, and 0.2 μmol/L aprotinin, or Complete Mini tablets (all from Boehringer-Mannheim, Mannheim, Germany). After suspension on ice, the cells were centrifuged at 15 000g at 4°C to remove debris. Nonlabeled lysates of tissues were prepared without prior labeling. The protein content of lysates was analyzed by standard methods, and comparable amounts were used in experiments.
To isolate erythroid and granulocyte subpopulations, respectively, from bone marrow cell suspension, we used the rat monoclonal antibodies (mAbs) TER-11918 (PharMingen, San Diego, CA) and CL8991AP19 (Cedarlane, Quebec, Canada). Bone marrow cells from 2 mice (60 × 106 cells) were diluted in complete DMEM and incubated with 4 μL TER-119 (0.5 mg/mL) or CL8991AP (1 mg/mL) for 60 minutes at 4°C in a rotator. A 120-μL aliquot of a 50% suspension of Dynabeads (No. 110.07; Dynal AS, Oslo, Norway) that recognize rat immunoglobulin G (IgG) were added, and the samples were incubated for another 30 minutes at 4°C. The beads with attached cells were isolated and washed 3 times in complete DMEM using the supplied magnetic device, and the lysates were prepared as described above. Aliquots of separated cell populations were stained with MGG, and microscopic inspection confirmed a strong enrichment of the specific cell type.
Immunoprecipitation
Lysates were routinely precleared of endogenous protein A–binding Ig chains by incubation on a rotator with 50-μL of a 50% suspension of protein A CL-4B Sepharose beads (Pharmacia) for 1 hour at 4°C in a total volume of 500 μL. Titrated amounts of specific antisera against eALAS20 were rotated for 3 hours at 4°C in a total volume of 500 μL. This was followed by incubation with 50 μL beads for 1 hour. In the indicated experiments, cold lysates were included in the reaction. Thereafter, beads were washed at low speed 3 times in 1 mL RIPA, resuspended in Laemmli loading buffer, heated at 95°C for 10 minutes to elute proteins, and loaded on 12% SDS-PAGE (polyacrylamide gel electrophoresis), which was fixed and exposed to X-ray film or phosphor imager screens. The protein sizes were determined by parallel electrophoresis of commercial molecular weight standards. The presence of the shorter form of eALAS could not be explained by nonspecific protein degradation. No changes in the ratio between the different eALAS forms were observed when parallel reactions were subjected to different lengths of incubation, different temperatures, or the addition of protease inhibitors. Using this antiserum, we did not detect cross-reaction with hALAS or any other proteins of nonerythroid cells.20
Subfractionation of mitochondria
Bone marrow or blood cells were separated on 3-mL Ficoll-Paque cushions (Pharmacia) by centrifugation at 900g for 1 hour at 4°C. The high-density fraction was washed 3 times and resuspended in 1 mL methionine-free DMEM. During labeling experiments, the cells were incubated with 35S-methionine for 3 hours at 37°C, and the cells were then washed in PBS. The cells were subsequently washed twice in ice-cold buffer A (250 mmol/L sucrose, 50 mmol/L Tris HCl [pH 7.4], and 1 mmol/L EDTA [ethylenediamine tetraacetic acid]) with protease inhibitors. The cells were resuspended in 3 mL buffer A and then lysed by sonication (Branson Sonic-Power B-15P Sonifier, Danbury, CT) with 2 bursts of 10 seconds each (setting, 5). Lysis was confirmed by microscopic inspection. To remove debris, the lysate was centrifuged twice at 900g for 20 minutes at 4°C. To pellet mitochondria, the supernatant was subsequently centrifuged at 12 000g for 25 minutes at 4°C. The resulting supernatant was defined as “cytosol fraction.” The mitochondrial pellet was washed twice in ice-cold buffer B (250 mmol/L sucrose, 10 mmol/L Tris HCl [pH 7.4], and 1 mmol/L EDTA) and resuspended in RIPA (“crude mitochondrial lysate”).
For further subfractionation, the mitochondrial pellet was resuspended in 1 mL buffer B and sonicated on ice 3 times for 30 seconds each time (setting, 5) followed by a 12 000gcentrifugation for 15 minutes to sediment intact mitochondria. The suspension or sonicate was centrifuged at 250 000g for 60 minutes at 4°C. The supernatant was recentrifuged to remove debris and was defined as the “matrix fraction” (although it might also include intermembrane space proteins). The pellet was washed in buffer B and defined as the “membrane fraction.” Fractions were tested by Western blot analysis for purity by using antisera against the membrane-bound succinate dehydrogenase21 or the matrix-bound mitochondrial transcription factor A (MTA) protein22 (data not shown). To further enrich the mitochondrial population, we loaded the mitochondrial pellet (see above) on a gradient containing 30% Percoll (Pharmacia, Uppsala, Sweden), 0.25 mol/L sucrose, 5 mmol/L Tris (pH 7.4), and 0.5 mmol/L EDTA. This was followed by centrifugation at 60 000g for 60 minutes at 4°C, and then the specific mitochondrial fraction (“pure mitochondria”) was collected.
In vitro reconstitution
Plasmids pPre–ALAS-E-minor and pPre–ALAS-E-major express 2 natural variants of murine pre-eALAS under the control of a phage T3 promoter in the pBluescript vector.16 The protein derived from pPre–ALAS-E-minor is identical to the protein derived from pPre–ALAS-E-major except that it lacks amino acids 61-75 of the pre-eALAS sequence. Plasmids were linearized with BamHI and transcribed and translated in vitro in the presence of 35S-methionine in the Flexi Rabbit reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer's instructions. The products were dialyzed against 10 mmol/L HEPES-KOH (4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid) (pH 7.4) and 0.2 mmol/L DTT. We mixed 0.2 μg dialyzed 40%-60% ammonium sulfate precipitation fraction of a bone marrow crude mitochondria lysate with 9 μL in vitro translated eALAS protein in the above buffer (with 0.5 mmol/L MnCl2). This was completed in a total volume of 20 μL and incubated at 27°C for 60 minutes before gel separation.
Results
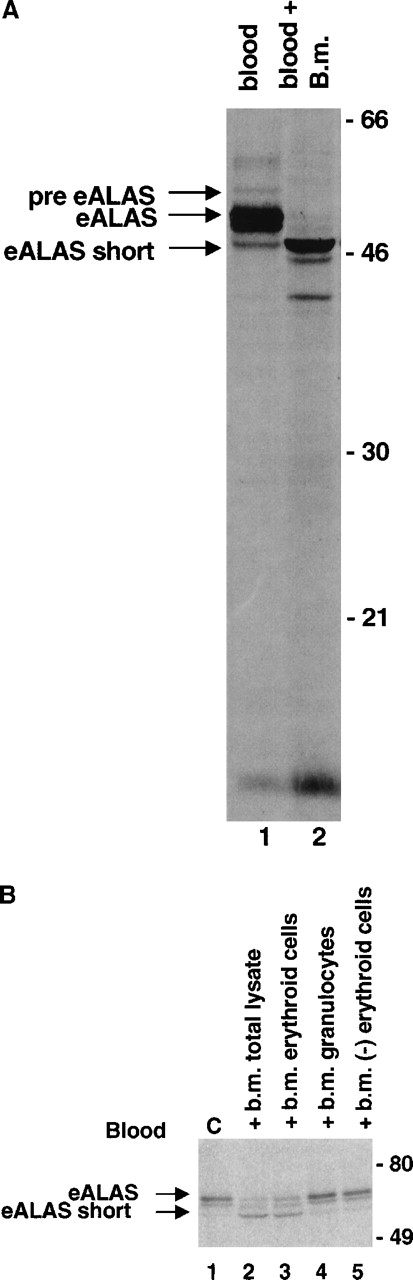
To study eALAS during differentiation, we metabolically labeled ex vivo cells from mouse peripheral blood and bone marrow with35S-methionine. As expression of eALAS is limited to erythroid cells, detergent lysates were prepared (in the strong ionic detergent RIPA) and used directly, without further enrichment of erythroblasts, for immunoprecipitation with an antiserum against murine eALAS. Immunoprecipitation of eALAS from lysates of peripheral blood (Figure 1) revealed an expected 59-kd double band (see below) representing the full-length forms of eALAS. A weaker 65-kd band, presumably corresponding to the precursor before cleavage of the mitochondrial signal peptide, was also revealed. Both have been previously demonstrated in mouse erythroleukemia (MEL) cells with this antiserum.20
eALAS protein synthesis in different erythroid tissues.
Peripheral blood and bone marrow were diluted with DMEM without methionine to 3 mL, separated on 3 mL Ficoll-Hypaque gradients, resuspended in 1 mL medium (blood, 1.5 × 108 cells; bone marrow, 1.9 × 107 cells), and incubated with 7.4 MBq (200 μCi) 35S-methionine for 3 hours. Detergent lysates were prepared, and eALAS and gel separation were immunoprecipitated. The arrows indicate the presumed pre-eALAS, the 59-kd form, and the herein described 52-kd eALAS-short.
eALAS protein synthesis in different erythroid tissues.
Peripheral blood and bone marrow were diluted with DMEM without methionine to 3 mL, separated on 3 mL Ficoll-Hypaque gradients, resuspended in 1 mL medium (blood, 1.5 × 108 cells; bone marrow, 1.9 × 107 cells), and incubated with 7.4 MBq (200 μCi) 35S-methionine for 3 hours. Detergent lysates were prepared, and eALAS and gel separation were immunoprecipitated. The arrows indicate the presumed pre-eALAS, the 59-kd form, and the herein described 52-kd eALAS-short.
In addition, a specific shorter form of the eALAS (eALAS-short) protein, approximately 52 kd in size, was precipitated from blood cells. This was consistently the only form to be expressed in bone marrow cell lysates. In blood cell lysates the relative amount of the eALAS-short protein varied between experiments, ranging from undetectable to being equimolar to the 59-kd form. This observed pattern thus suggested a loss of eALAS-short and the appearance of the 59-kd form with the maturation of the erythroid cells. It is important to note that the ex vivo metabolic labeling enables us to detect only de novo formation of proteins and not steady state levels of the 2 forms of eALAS in the blood. However, a de novo pool of the 59-kd form of eALAS in the peripheral blood reticulocyte could rapidly form, provided the eALAS protein turns over at a similar speed as the nonerythroid form of ALAS (half-life, approximately 35 minutes).23
There was no obvious difference in the eALAS mRNA pattern in blood and bone marrow (data not shown), as seen using high-resolution Northern blot gels and reverse transcriptase–polymerase chain reactions (RT-PCRs). This indicates that the eALAS-short protein arose by a posttranslational event. To determine whether posttranslational proteolytic cleavage might be responsible for the short form, we coincubated a “cold” bone marrow lysate and a radiolabeled blood cell lysate during immunoprecipitation. Figure2 demonstrates that we can immunoprecipitate a shorter form of eALAS, identical in size to the 52-kd eALAS-short protein observed in bone marrow cells, which originates from the 59-kd form of the protein in the blood cells. In addition, a shorter band of 5-10 kd accumulated, although it remains unclear if this product is related to the cleavage process.
Cleavage of eALAS by a proteolytic activity in bone marrow erythroid cells.
(A) Blood (1 mL) was separated on a 3-mL Ficoll-Hypaque gradient. The high-density fraction was resuspended in medium and divided into 2 tubes to which 7.4 MBq (200 μCi)35S-methionine was added. A 300-μL nonlabeled bone marrow lysate (1.5 × 108 cells) (lane 2) or medium alone (lane 1) was added. Both samples were incubated at 37°C for 3 hours, and eALAS proteins were subsequently immunoprecipitated and gel-separated. (B) Erythroid and granulocytic cells were isolated from the bone marrow by mAbs coupled to magnetic beads (“Materials and methods”). We incubated 80-μg lysates from these fractions with a labeled blood lysate for 30 minutes at 37°C, and eALAS proteins were subsequently immunoprecipitated and gel-separated. The far-right lane represents a lysate after depletion of erythroid cells.
Cleavage of eALAS by a proteolytic activity in bone marrow erythroid cells.
(A) Blood (1 mL) was separated on a 3-mL Ficoll-Hypaque gradient. The high-density fraction was resuspended in medium and divided into 2 tubes to which 7.4 MBq (200 μCi)35S-methionine was added. A 300-μL nonlabeled bone marrow lysate (1.5 × 108 cells) (lane 2) or medium alone (lane 1) was added. Both samples were incubated at 37°C for 3 hours, and eALAS proteins were subsequently immunoprecipitated and gel-separated. (B) Erythroid and granulocytic cells were isolated from the bone marrow by mAbs coupled to magnetic beads (“Materials and methods”). We incubated 80-μg lysates from these fractions with a labeled blood lysate for 30 minutes at 37°C, and eALAS proteins were subsequently immunoprecipitated and gel-separated. The far-right lane represents a lysate after depletion of erythroid cells.
To test if this protease activity was present in erythroid cells, we next isolated such cells from the bone marrow by employing magnetic beads and the erythroid-specific mAb TER-119.18 In parallel we isolated granulocytes with this technique using the mAb CL8991AP,19 as such cells contain potent lysosomal proteases that could be released during lysis and cleave other proteins. When we tested equal amounts of lysates from erythroid and granulocyte cells for protease activity, it clearly showed that erythroid cells contained this protease activity, whereas there was no activity detected in granulocytes (Figure 2B). The claim of an erythroid-specific activity was further supported by the fact that there was no activity retained in a lysate from bone marrow cells depleted of erythroid cells.
To localize the protease activity to a cell compartment, bone marrow cells were lysed by sonication and further subfractionated by differential centrifugation into a mitochondrial and a cytosolic fraction. To exclude the possibility that the protease activity resided in other organelles cosedimenting with the mitochondria, crude mitochondria were also separated on a density gradient, and the specific mitochondrial fraction was collected. The testing of this and other fractions for protease activity showed clearly that the activity resides in the mitochondria (Figure 3A). We also sonicated the mitochondria and divided them into a membrane and a matrix fraction. From testing these for the ability to cleave the 59-kd form of the eALAS protein, we could further localize the protease activity to the membrane fraction (Figure 3B). The mitochondrial membrane was additionally washed with up to 1 mol/L of chaotrophic salts without loss of protease activity, indicating that it was an integral part of the membranes (data not shown).
Fractionation of protease activity.
(A) Lysates of 35S-methionine–labeled blood cells were prepared (“Materials and methods”). Equal amounts were incubated alone or mixed with nonlabeled lysates of bone marrow cells and subfractions of 80 μg each. The mixtures were immunoprecipitated for 3 hours at 4°C with standard protease inhibitors before gel separation. (B) Bone marrow crude mitochondria (mit.) were further subfractionated (80 μg each) into a membrane and matrix fraction and were analyzed the same way. (C) Nonlabeled 80-μg lysates each of bone marrow, spleen, and liver tissue were mixed with the labeled blood lysate and analyzed the same way.
Fractionation of protease activity.
(A) Lysates of 35S-methionine–labeled blood cells were prepared (“Materials and methods”). Equal amounts were incubated alone or mixed with nonlabeled lysates of bone marrow cells and subfractions of 80 μg each. The mixtures were immunoprecipitated for 3 hours at 4°C with standard protease inhibitors before gel separation. (B) Bone marrow crude mitochondria (mit.) were further subfractionated (80 μg each) into a membrane and matrix fraction and were analyzed the same way. (C) Nonlabeled 80-μg lysates each of bone marrow, spleen, and liver tissue were mixed with the labeled blood lysate and analyzed the same way.
We also prepared lysates of cells from the liver and spleen and tested them for the presence of proteolytic activity in a similar way. No protease activity could be detected in these tissues under the conditions we used (Figure 3C). The bone marrow protease activity was potent in a strong detergent (RIPA) and insensitive to several inhibitors of serine, cysteine, and aspartic proteases or metalloproteases as well as high levels of EDTA when these agents were added to lysates at the indicated concentrations. We could only observe inhibition of cleavage with high amounts of alpha-1-antitrypsin (Figure4A). A dose response to alpha-1-antitrypsin, which showed a proportional increase in the substrate and a decrease in the product, was observed (Figure 4B). To exclude the possibility that cleavage was due to a proteolytic activity that was released from another cell type (or organelle) during lysis of the bone marrow cells, we included alpha-1-antitrypsin in the lysis buffer. But we could still only demonstrate the cleaved form (Figure4B), which shows that the short form was not an artifact during preparation. This conclusion was further consolidated by mixing labeled blood and bone marrow cells before lysis in RIPA with alpha-1-antitrypsin, as in the latter experiment. There was no observed cleavage in the 59-kd blood eALAS protein, which served as an internal probe in the lysate to exclude the presence of a protease (data not shown).
Inhibition of protease activity.
(A) Bone marrow cells were aliquoted into 6 different tubes containing RIPA and the indicated inhibitors (50 μg/mL elastatinal, 0.5 mg/mL alpha-1-antitrypsin, 5 mmol/L EDTA, and leupeptin/Pefabloc/pepstatin/aprotinin [LPPA]). A lysate of35S-methionine–labeled blood cells (300 μL each) was added, and the reaction was immunoprecipitated for 3 hours at 4°C before gel separation. (B) The experiment was repeated with varying amounts of alpha-1-antitrypsin (0.1, 0.25, 0.5, and 1 mg/mL, shown in lanes 3-6, respectively). 35S-methionine–labeled bone marrow cells were lysed in RIPA with or without 0.5 mg/mL alpha-1-antitrypsin followed by immunoprecipitation for 1 hour at 4°C without any preclearing (lanes 7 and 8).
Inhibition of protease activity.
(A) Bone marrow cells were aliquoted into 6 different tubes containing RIPA and the indicated inhibitors (50 μg/mL elastatinal, 0.5 mg/mL alpha-1-antitrypsin, 5 mmol/L EDTA, and leupeptin/Pefabloc/pepstatin/aprotinin [LPPA]). A lysate of35S-methionine–labeled blood cells (300 μL each) was added, and the reaction was immunoprecipitated for 3 hours at 4°C before gel separation. (B) The experiment was repeated with varying amounts of alpha-1-antitrypsin (0.1, 0.25, 0.5, and 1 mg/mL, shown in lanes 3-6, respectively). 35S-methionine–labeled bone marrow cells were lysed in RIPA with or without 0.5 mg/mL alpha-1-antitrypsin followed by immunoprecipitation for 1 hour at 4°C without any preclearing (lanes 7 and 8).
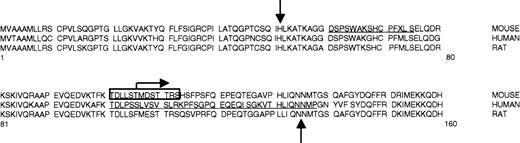
Murine eALAS mRNA can be alternatively spliced to a longer form (85% of the eALAS mRNA amount) and a shorter form. The shorter form results in a preprotein that lacks amino acids 61-75.24 We have not performed experiments specifically aimed to demonstrate these 2 mRNAs in our system, but we believe that the double band observed in the immunoprecipitations of the 59-kd form of eALAS on high-resolution gels (Figure 1) is likely to correlate to this difference of 15 amino acids. We translated in vitro these 2 pre-eALAS mRNAs in a rabbit reticulocyte lysate in the presence of 35S-methionine and determined 2 slightly different 65-kd bands after SDS-PAGE (Figure5). Both preproteins could be cleaved by lysates from bone marrow mitochondria to the same size band, identical to the shorter form of eALAS, because the 15 amino acids are in the part which is cleaved off (Figure 5). This demonstrated that cleavage occurs at the N-terminal region and not the C-terminal end of the protein. The N-terminal region of the human eALAS protein is also variable as a result of alternative splicing25 (Figure 6). The reason behind the alternative splicing in mouse and human has not been elucidated, but it is notable that this region seems to be the subject of more than one type of regulation. This N-terminal region is unique to the erythroid form of ALAS, unlike the rest of the protein, which is highly homologous with the housekeeping isoform. The N-termini of the eALAS protein are, however, homologous between mouse, human, and rat (Figure 6), which suggests that a similar type of cleavage could occur in other mammals. A recent report demonstrated the presence of a fully active shorter form of eALAS, approximately 49 kd, in human reticulocytes, but this was not investigated further.26
Proteolytic cleavage of in vitro translated eALAS.
In vitro translated eALAS proteins representing the 2 natural splice variants pre–ALAS-E minor and pre–ALAS-E major were incubated alone (lanes 1 and 2) or with a bone marrow (b.m.) mitochondrial lysate (lanes 3 and 4) for 60 minutes at 27°C before gel separation. A labeled blood lysate was included as a control (lane 5).
Proteolytic cleavage of in vitro translated eALAS.
In vitro translated eALAS proteins representing the 2 natural splice variants pre–ALAS-E minor and pre–ALAS-E major were incubated alone (lanes 1 and 2) or with a bone marrow (b.m.) mitochondrial lysate (lanes 3 and 4) for 60 minutes at 27°C before gel separation. A labeled blood lysate was included as a control (lane 5).
The N-termini of the pre-eALAS protein from mouse, human, and rat.
The downward arrow indicates mitochondrial processing peptidase (MPP) cleavage of the signal peptide.16 The reported cleavage is estimated to be within the boxed region. Horizontal bars show amino acids affected by alternative splicing: 15 amino acids (positions 61-75) for mouse eALAS24 and 37 amino acids (positions 102-138) for human eALAS.25 The bent arrow marks the N-terminus of the functional protein expressed from plasmid pTAD-ALAS2,27 and the upward arrow marks the N-terminus for a functional rat eALAS in vitro.28
The N-termini of the pre-eALAS protein from mouse, human, and rat.
The downward arrow indicates mitochondrial processing peptidase (MPP) cleavage of the signal peptide.16 The reported cleavage is estimated to be within the boxed region. Horizontal bars show amino acids affected by alternative splicing: 15 amino acids (positions 61-75) for mouse eALAS24 and 37 amino acids (positions 102-138) for human eALAS.25 The bent arrow marks the N-terminus of the functional protein expressed from plasmid pTAD-ALAS2,27 and the upward arrow marks the N-terminus for a functional rat eALAS in vitro.28
The 52-kd eALAS-short protein in the bone marrow must be functional, as it is the only form of eALAS made in this tissue, and bone marrow erythroid cells can synthesize heme. The catalytic activity is confined to the C-terminal part of the mammalian eALAS protein.4-8 Deletion of the first 56 N-terminal amino acids of the murine eALAS still allows an active protein in Escherichia coli,27 and truncation of the 83 N-terminal amino acids of the rat eALAS protein does not impede function in vitro28 (Figure 6). The observed cleaved form of eALAS is indistinguishable in size from the truncated protein27expressed in E coli on high-resolution gels (data not shown), and it is therefore very likely to be enzymatically active. The localization of the shorter form of eALAS to the mitochondria also indicates that it may be functional.
The eALAS protein domains involved in the binding (or catalysis) of the cofactor pyridoxal 5′-phosphate (Vitamin B6) are not in the N-terminal region and should not be directly affected by cleavage.29 Although the part which is cleaved off is not typically lipophilic, it may serve, directly or via another protein, as an anchor to the membrane. Electron microscopy of the housekeeping form of ALAS suggests it is associated with the mitochondrial inner membrane facing the matrix,30 possibly by being anchored to the succinyl-CoA synthase beta-subunit.31 To investigate the location of the 59-kd and eALAS-short forms of eALAS, we labeled erythroid cells from blood with 35S-methionine, ruptured them by sonication, and isolated the mitochondria, which were divided into a membrane and a matrix fraction. The subsequent immunoprecipitation of eALAS clearly demonstrated that both forms of eALAS were present in the mitochondrial membrane fraction (Figure7). The location of both the protease activity and its product in the membrane suggests that cleavage takes place in the mitochondrial membrane in conjunction with the import of the preprotein. This would predict that cells harboring protease activity would not produce a 59-kd form of eALAS, and this could be the reason that we were unable to detect any of this form in bone marrow cells.
The short eALAS is located in the mitochondrial membrane.
Blood (4 mL) was separated on a Ficoll-Hypaque gradient. The high-density fraction was collected and labeled with 22.2 MBq (600 μCi) 35S-methionine for 3 hours. Suborganelles were prepared, and the lysates were used for immunoprecipitation of the eALAS protein followed by gel separation. Lysates of membrane and matrix fractions contained 270 μg protein each.
The short eALAS is located in the mitochondrial membrane.
Blood (4 mL) was separated on a Ficoll-Hypaque gradient. The high-density fraction was collected and labeled with 22.2 MBq (600 μCi) 35S-methionine for 3 hours. Suborganelles were prepared, and the lysates were used for immunoprecipitation of the eALAS protein followed by gel separation. Lysates of membrane and matrix fractions contained 270 μg protein each.
Discussion
We describe a specific truncation of the erythroid isoform of ALAS that takes place in erythroid cells in the bone marrow. The function of the shorter form of eALAS has yet to be established, but one can assume that an altered function of the eALAS protein will serve to modulate heme synthesis.
Our data argue against a direct role of the cleaved N-terminus in membrane anchoring, although it is still possible it could serve as a binding site for other molecules. It is noteworthy that this sequence contains a typical heme binding site,16 suggesting a mechanism for end-product control of the eALAS protein. Such a contention may also be supported by the fact that this heme-binding site is lost with the alternative splicing of eALAS mRNA.24It could also be that cleavage of the N-terminus results in a loss of the ability of the eALAS protein to homodimerize. Yet other reasons for the shortening of eALAS may be the changes in eALAS protein stability or enzyme activity.
We believe there could be reasons why heme synthesis is controlled differently in bone marrow and peripheral blood. Excessive amounts of iron or heme easily result in the formation of aggressive free radicals via the Fenton reactions,32 which could lead to cell damage and cell transformation. Nucleated erythroblasts in the bone marrow could sustain an increased risk of erythroleukemic cell transformation. Thus a more stringent control of heme formation is needed. However, anucleate reticulocytes (which continue to synthesize proteins after entering the bloodstream) cannot transform, but they need a highly efficient hemoglobin synthesis and, as a result, might have less need of avoiding oxidative stress damage.
Some types of sideroblastic anemias are caused by point mutations in the eALAS gene,33 and 1 mutation evidently affects processing of the pre-eALAS protein.34 In other anemias, the reduced hemoglobin formation and the unclear molecular basis may also be associated with eALAS. For example, in the anemia of chronic disease (ACD), lower hemoglobin synthesis in erythroblasts was correlated to a reduced eALAS activity.35
This is the first description of an endoproteolytic site-specific processing activity in mitochondria, apart from proteases involved in cleavage of the import signal peptide.36-39 The protease activity is active in a strong ionic detergent, has no clear dependence on chelatable metals, and is not affected by most common protease inhibitors. Together with tissue specificity and the specific localization to the mitochondrial membrane fraction, we conclude that this protease activity must be different from known endoproteases in the mitochondria, such as mitochondrial processing peptidase (MPP) and mitochondrial intermediate peptidase (MIP), both of which are instrumental in cleavage of the signal peptide.40 An elastase-like protein (medullasin) from bone marrow was previously reported to inactivate aminolevulinate synthase.41Medullasin was later shown to be identical in sequence to neutrophil elastase.42,43 Unlike the present protease activity, it resides in lysosomes of granulocytes and is readily inhibited by elastatinal. Testing of purified medullasin and neutrophil elastase on the eALAS substrates did not reveal any similar cleavage (data not shown).
The protease activity seems to be tissue-specific, as we have not observed it in lysates from tissues other than bone marrow. We have further limited the activity to the erythroid cells in the marrow. This argues that there must be a limited number of targets for this protease activity. We also have not observed any changes in the protein profiles in total lysates under conditions that allow cleavage of the eALAS protein.
Many disorders affect hemoglobin function, and eALAS plays a pivotal role in the de novo formation of hemoglobin. A future challenge thus lies in identifying the specific function of the eALAS-short protein. Another issue will be the further characterization and regulation of the tissue-specific protease activity, as this might enable us to understand the switch between the long and short forms of eALAS.
Acknowledgments
We are grateful to Mike Timko for the pre–ALAS-E plasmids, Yosuke Aoki for medullasin, Susanne Widell for cell analysis, Cristina Szigyarto for discussions, and Matthias Hentze for initial support.
Supported by grant 06593-304 from the Swedish Natural Science Research Council, Sweden; the Swedish Association for Medical Research, Sweden (von Kantzow stipend); the Swedish Society of Medicine, Sweden; and the Lars Hierta and Magnus Bergvall foundations (Ö.M.), Sweden. While on sabbatical leave from the EMBL, Heidelberg, Germany, J.H.B was supported by CIBA-Geigy Switzerland and the European Union, and A.K. received a Wenner-Gren postdoctoral stipend. V.D. is on leave from the Vytautas Magnus University in Kaunas, Lithuania.
Submitted June 21, 1999; accepted March 3, 2000.
Reprints:Öjar Melefors, Microbiology and Tumor Biology Center, Karolinska Institutet, SE-171 77 Stockholm, Sweden; e-mail: ojar.melefors@mtc.ki.se.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



![Fig. 4. Inhibition of protease activity. / (A) Bone marrow cells were aliquoted into 6 different tubes containing RIPA and the indicated inhibitors (50 μg/mL elastatinal, 0.5 mg/mL alpha-1-antitrypsin, 5 mmol/L EDTA, and leupeptin/Pefabloc/pepstatin/aprotinin [LPPA]). A lysate of35S-methionine–labeled blood cells (300 μL each) was added, and the reaction was immunoprecipitated for 3 hours at 4°C before gel separation. (B) The experiment was repeated with varying amounts of alpha-1-antitrypsin (0.1, 0.25, 0.5, and 1 mg/mL, shown in lanes 3-6, respectively). 35S-methionine–labeled bone marrow cells were lysed in RIPA with or without 0.5 mg/mL alpha-1-antitrypsin followed by immunoprecipitation for 1 hour at 4°C without any preclearing (lanes 7 and 8).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/2/10.1182_blood.v96.2.740/5/m_bloo01442004w.jpeg?Expires=1764962661&Signature=ulqV17QbGpILlcekKL~H3OqTvM0EnE80G2sahckkVBdZPMo95bOepXTkpXZXSifVaCfVhva47HD932ReAGn8zpiELnurq9Rb2j-jTILo4AvzVt3f9rfaSW2LfbcI7dC5bnNbrjNeVsOGa8XEm0uM8j-XwoNghVn9YkwRr5-5IrI3GdMpZXGJSbd5ny6WWX5Fs4I-2M0CMtNEB3k0Zqm~f8KAMy6FDeEgSXBm95pPBRdYOxVZbj~Km-7bwtOFb92tXflnxvsAPbf-WKOlPrwFGkj9-QTQaruKc-UBClNH6zs1UpvmBL~Cx-ldCncCr7bDOvCooV1LFMXkKujMOwHwEA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal