Abstract
An acquired chromosomal translocation, t(8;13)(p11;q11-12), observed in a distinctive type of stem cell leukemia/lymphoma syndrome, leads to the fusion of the 5′ portion of ZNF198 and the 3′ portion of FGFR1. ZNF198–FGFR1 fusion transcripts encode 4 to 10 zinc fingers, a proline-rich region, and the intracellular portion of the FGFR1 (fibroblast growth factor receptor 1) receptor tyrosine kinase. We demonstrate that the ZNF198 proline-rich region constitutes a novel self-association domain. When fused to the intracellular domain of FGFR1, the ZNF198 proline-rich region is sufficient to cause oligomerization, FGFR1 tyrosine kinase activation, and transformation of Ba/F3 cells to IL-3 independent growth.
Various histologic subtypes of leukemia and lymphoma are associated with specific chromosome translocations,1-4and substantial strides have been made in determining the oncogenes targeted by those translocations. Many translocations in non-Hodgkin lymphomas, for example, those involving MYC in Burkitt lymphoma, cause up-regulation of the corresponding oncogenes through their juxtaposition with transcriptionally active chromosome regions. Other translocations, including many identified thus far in lymphoid and myeloid leukemias, produce fusion genes. The encoded chimeric oncoproteins may transform hematopoietic cells by gain-of-function or dominant loss-of- function mechanisms.5-12
Although most hematologic malignancies are monotypic, involving either myeloid or lymphoid lineages, others are associated with multilineage proliferation because of the transformation of a pluripotent hematopoietic stem cell. One example is chronic myelogenous leukemia, in which the characteristic bcr-abl fusion can be demonstrated in both myeloid and lymphoid lineages. Stem cell neoplasms have been of particular research interest because the underlying oncogenes are apt to function as differentiation and proliferation factors that regulate physiologic and pathologic hematopoiesis. In addition, the stem cell leukemias and lymphomas pose special challenges in clinical practice. Stem cell neoplasms are only curable using intensive chemotherapy, typically coupled with bone marrow or peripheral blood progenitor cell transplantation.13 14 Hence, new biologic insights—particularly those that can be translated into novel therapeutic approaches—may be particularly useful in these diseases.
A unique leukemia/lymphoma syndrome of acute myelogenous leukemia, eosinophilia, and lymphoblastic lymphoma has been described by several groups.15-23 Most patients have lymphoma and myeloid hyperplasia, and many have pronounced peripheral eosinophilia or prominent eosinophilic infiltrates in the involved bone marrow and lymph nodes. The myeloid hyperplasia usually progresses to acute myelogenous leukemia within a year of the original diagnosis. A specific chromosome translocation, t(8;13)(p11;q11-12), is found in lymphoma and myeloid leukemia cells from these patients, supporting bilineage differentiation from a transformed stem cell.16-18 The chromosomal translocation leads to the fusion of the 5′ portion of ZNF198 on chromosome 13 and the 3′ portion of FGFR1 on chromosome 8.24-27ZNF198 is a potential transcription factor that has 2 isoforms, containing either 4 or 10 atypical zinc fingers, a proline-rich region, and an acidic domain. FGFR1 is a transmembrane receptor tyrosine kinase (RTK) belonging to the fibroblast growth factor receptor family. All known FGFRs contain 2 or 3 immunoglobulin-like extracellular ligand-binding domains, a single-pass transmembrane domain, and an intracellular catalytic kinase domain.28-31 The ZNF198–FGFR1 fusion protein contains the intact tyrosine kinase domain of FGFR1 but lacks the N-terminal extracellular FGF binding domain and transmembrane domain, which are replaced by the ZNF198 atypical zinc finger and proline-rich domains.
Normal activation of RTKs involves ligand-dependent oligomerization. To determine the mechanism of ZNF198–FGFR1 oncogenicity, we evaluated the possibility of constitutive, ligand-independent oligomerization. We find that the proline-rich domain of ZNF198 functions as a novel oligomerization domain that promotes constitutive ZNF198–FGFR1 tyrosine kinase activity.
Materials and methods
DNA constructs
RNA was isolated from T-lymphoblastic lymphoma cells containing the t(8;13) translocation, and ZNF198–FGFR1 cDNA was amplified using Pyrococcus furiosus polymerase sequenced and subcloned in frame into derivatives of pcDNA3 (Invitrogen, Carlsbad, CA) that permit the expression of transcripts encoding C-terminal MYC or hemagglutinin (HA) epitopes. Truncated ZNF198–FGFR1 constructs were generated from polymerase chain reaction products encoding the appropriate ZNF198–FGFR1 domains. Retroviral constructs were produced by subcloning ZNF198–FGFR1 cDNA into forms of the vector MSCV containing G418 or green fluorescent protein markers.32
Transient transfection and in vitro transcription and translation
293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) and 10% fetal calf serum at 37°C under 5% CO2and were transfected with 10 μg of expression plasmid DNA using a standard calcium phosphate method.33 After 48 hours, the cells were lysed in ice-cold 1% NP-40, 50 mmol/L Tris pH 8.0, 100 mmol/L sodium fluoride, 30 mmol/L sodium pyrophosphate, 2 mmol/L sodium molybdate, 5 mmol/L EDTA, 2 mmol/L sodium vanadate, 5 μg/mL aprotinin, 5 μg/mL leupeptin, and 50 μg/mL phenylmethylsulfonyl fluoride (lysis buffer).
High-titer, helper-free recombinant retrovirus was produced through transient transfection of Bosc cells.33 In brief, 2.5 × 106 cells per 60-mm plate were cultured in DMEM with 10% fetal bovine serum to 80% confluence, re-fed with medium containing 25 μmol/L chloroquine, and transfected with 10 μg DNA. After 10 hours, the chloroquine-containing medium was replaced with fresh medium. The retroviral supernatant was harvested 48 hours after transfection.
In vitro expression of ZNF198–FGFR1 constructs was performed using the TNT Quick coupled transcription–translation system (Promega, Madison, WI) according to the manufacturer's instructions. In reactions programmed with more than 1 expression plasmid, the total input DNA was 0.5 μg/25μL reaction.
Immunoprecipitation and Western blot analysis
Protein lysates of in vitro expression polypeptides were diluted in 250 μL of lysis buffer, then incubated with various antibodies for 2 hours. After this 20 μL protein A–Sepharose (Pharmacia, Piscataway, NJ) was added for 1 hour at 4°C. After 3 washes in lysis buffer, proteins were eluted at 100°C in 40 μL of SDS–PAGE loading buffer and resolved by SDS–PAGE under reducing conditions (4%-12% gradient gels). To detect 35S-labeled proteins, the gels were dried and exposed to X-ray film for 0.5 to 12 hours. For Western blot analyses, the proteins were electrophoretically transferred to polyvinylidene difluoride membranes (Millipore, Waltham, MA), then blocked in PBS containing 0.1% Tween 20 (PBS-T) and 5% dried milk for 1 hour. Proteins were detected with anti-HA (Babco clone HA.11, Richmond, CA), anti-MYC (clone 9E10), anti-PY99 (Santa Cruz, CA), or anti-FGFR1 (Santa Cruz, CA) using a chemiluminescence method (ECL; Amersham, Piscataway, NJ).
Cell culture
Ba/F3 cells were grown in RPMI-1640 with 10% fetal calf serum and 15 ng/mL of IL-3. For viral infections, 2 mL of Ba/F3 cells at a concentration of 1.5 × 106 cells/mL were centrifuged, resuspended in 1 mL viral supernatant/polybrene mixture, and incubated at 37°C for 24 hours. The cells were then centrifuged and resuspended in cell culture medium. Cell selection was begun 24 hours later using Geneticin (800 μg/mL) and was continued for 10 days. Cell counts were determined daily using the trypan blue exclusion method.
Results
Ligand-independent ZNF198–FGFR1 oligomerization
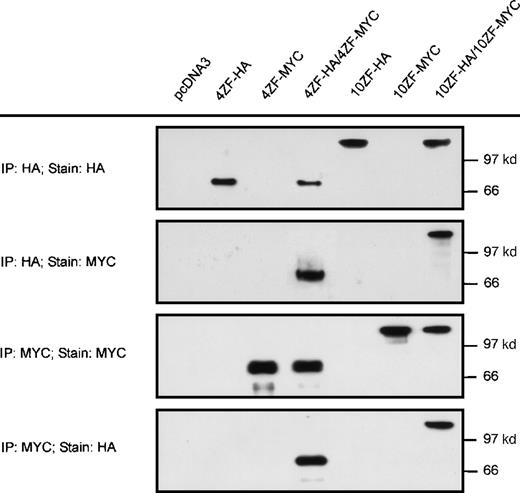
Based on precedence in other receptor tyrosine kinase oncoproteins,34-38 we hypothesized that ZNF198–FGFR1 transforming mechanisms involve aberrant activation of the FGFR1 kinase domain. Because FGFR1 kinase activation ordinarily requires dimerization,39 we evaluated the possibility that ZNF198–FGFR1 might self-associate. Two isoforms of ZNF198–FGFR1 identified in patients with the t(8;13) stem cell leukemia/lymphoma syndrome have either 4 or 10 zinc fingers, respectively, at the N-terminus. Derivative cDNA encoding each isoform was fused in frame with HA (10ZF-HA and 4ZF-HA) or MYC (10ZF-MYC and 4ZF-MYC) epitope tags (Figure 1) and expressed in 293T cells. The HA-tagged and MYC-tagged proteins were specifically immunoprecipitated by antibodies to the corresponding epitopes (Figure2). Immunoprecipitates prepared from cells coexpressing HA and MYC tagged ZNF198–FGFR1 demonstrated oligomerization of both the 4 and the 10 zinc finger ZNF198–FGFR1 isoforms (Figure 2).
Schematic of ZNF198/FGFR1 constructs. The full-length ZNF198–FGFR1 fusion proteins contain either 4 or 10 ZNF198 zinc fingers (depending on isoform), a ZNF198 proline-rich region, and the FGFR1 tyrosine kinase domain. One or more of these domains is deleted in each of the truncated fusion proteins. The fusion constructs were subcloned into the MSCV retrovirus vector or were tagged with HA or MYC and subcloned into the pcDNA 3.1 vector.
Schematic of ZNF198/FGFR1 constructs. The full-length ZNF198–FGFR1 fusion proteins contain either 4 or 10 ZNF198 zinc fingers (depending on isoform), a ZNF198 proline-rich region, and the FGFR1 tyrosine kinase domain. One or more of these domains is deleted in each of the truncated fusion proteins. The fusion constructs were subcloned into the MSCV retrovirus vector or were tagged with HA or MYC and subcloned into the pcDNA 3.1 vector.
Self-association of ZNF198–FGFR1.
The 4 and 10 zinc finger ZNF198–FGFR1 isoforms were tagged with HA or MYC, expressed individually (lanes 2, 3, 5, and 6) or coexpressed (lanes 4 and 7) in 293 T cells, then immunoprecipitated and detected with anti-HA or anti-MYC. Empty pcDNA 3 vector was also transfected as a negative control. Both 4 and 10 zinc finger ZNF198–FGFR1 isoforms self-associate (lanes 4 and 7).
Self-association of ZNF198–FGFR1.
The 4 and 10 zinc finger ZNF198–FGFR1 isoforms were tagged with HA or MYC, expressed individually (lanes 2, 3, 5, and 6) or coexpressed (lanes 4 and 7) in 293 T cells, then immunoprecipitated and detected with anti-HA or anti-MYC. Empty pcDNA 3 vector was also transfected as a negative control. Both 4 and 10 zinc finger ZNF198–FGFR1 isoforms self-associate (lanes 4 and 7).
Mapping an ZNF198 oligomerization interface
The primary structure of the ZNF198–FGFR1 fusion protein includes 3 major domains. The N-terminus is a ZNF198 potential DNA-binding domain, consisting of 4 to 10 atypical zinc fingers (Cys-X2-Cys-X19-20-Cys-X3-Cys; residues 1-175 or 1-796). The middle portion is also contributed by ZNF198 and contains 118 amino acids (residues 176-293 or 797-913 in the 4 and 10 zinc finger isoforms, respectively). This region has a high concentration (16%) of proline residues and contains 2 short proline–valine repeats. The C-terminus consists of part of the FGFR1 juxtamembrane domain and the entire tyrosine kinase domain (residues 294-687 or 914-1037 in the 4 and 10 zinc finger isoforms, respectively).
The ZNF198–FGFR1 oligomerization interface was mapped using a series of ZNF198–FGFR1 HA-tagged deletion constructs (Figure 1). Ability to oligomerize with full-length ZNF198–FGFR1 (tagged with an MYC epitope) was evaluated by expressing the constructs in reticulocyte lysates. A ZNF198 polypeptide consisting of the N-terminal 4 zinc fingers (4ZFΔPRΔTK-HA) did not oligomerize with the full-length ZNF198–FGFR1 oncoprotein (4ZF-MYC). By contrast, a ZNF198 polypeptide consisting of the same 4 zinc fingers with the immediately C-terminal proline-rich domain (4ZFΔTK-HA) did oligomerize with 4ZF-MYC (Figure3A), suggesting that the ZNF198 proline-rich region mediates self-association. Further evidence of a proline-rich region oligomerization domain was obtained from coexpression of full-length ZNF198–FGFR1 (4ZF-MYC) with a polypeptide lacking all zinc fingers but containing the ZNF198 proline-rich region and the FGFR1 tyrosine kinase domain (ΔZF-HA) (Figure 3A). Oligomerization function for the ZNF198 proline-rich region was then confirmed by expression of a 13-kd minipeptide containing only the proline-rich region (PR-HA). This minipeptide also coprecipitated full-length ZNF198–FGFR1 (4ZF-MYC) (Figure 3B).
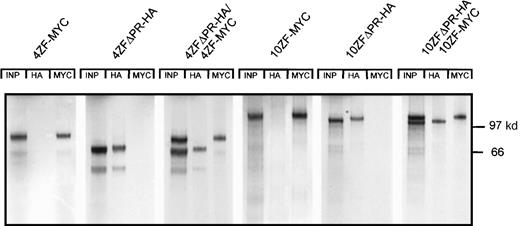
The ZNF198 proline-rich region is crucial for protein–protein interaction.
(A) Truncated ZNF198–FGFR1 fusion proteins with (4ZFΔTK-HA and ΔZF-HA) or without (4ZFΔPRΔTK-HA) the ZNF198 proline-rich region were expressed individually (columns 2, 4, and 6), or were coexpressed with the full-length 4 zinc finger ZNF198–FGFR1 isoform (4ZF-MYC) (columns 3, 5, and 7). In vitro expressions were performed using reticulocyte lysates (Promega; in vitro TNT system) in the presence of [35S] methionine. The proteins were analyzed directly (INP, input) or were immunoprecipitated with anti-HA or anti-MYC. Only fusion proteins containing the ZNF198 proline-rich region (columns 5 and 7) coprecipitated with full-length ZNF198–FGFR1. (B) A full-length 4 zinc finger ZNF198–FGFR1 isoform (4ZF-MYC) and a minipeptide containing only the ZNF198 proline-rich region (PR-HA) were expressed individually (columns 1 and 2) or coexpressed (column 3), then analyzed directly (INP) or immunoprecipitated with anti-HA or anti-MYC. PR-HA and 4ZF-MYC coprecipitated (column 3).
The ZNF198 proline-rich region is crucial for protein–protein interaction.
(A) Truncated ZNF198–FGFR1 fusion proteins with (4ZFΔTK-HA and ΔZF-HA) or without (4ZFΔPRΔTK-HA) the ZNF198 proline-rich region were expressed individually (columns 2, 4, and 6), or were coexpressed with the full-length 4 zinc finger ZNF198–FGFR1 isoform (4ZF-MYC) (columns 3, 5, and 7). In vitro expressions were performed using reticulocyte lysates (Promega; in vitro TNT system) in the presence of [35S] methionine. The proteins were analyzed directly (INP, input) or were immunoprecipitated with anti-HA or anti-MYC. Only fusion proteins containing the ZNF198 proline-rich region (columns 5 and 7) coprecipitated with full-length ZNF198–FGFR1. (B) A full-length 4 zinc finger ZNF198–FGFR1 isoform (4ZF-MYC) and a minipeptide containing only the ZNF198 proline-rich region (PR-HA) were expressed individually (columns 1 and 2) or coexpressed (column 3), then analyzed directly (INP) or immunoprecipitated with anti-HA or anti-MYC. PR-HA and 4ZF-MYC coprecipitated (column 3).
We next addressed the possibility that the ZNF198 proline-rich region might be requisite for ZNF198–FGFR1 oligomerization. Oligomerization potential was evaluated in 4 and 10 zinc finger forms of HA-tagged ZNF198–FGFR1 lacking the proline-rich region (4ZFΔPR-HA and 10ZFΔPR-HA, respectively). These proline-rich region deletion constructs were coexpressed with 4 and 10 zinc finger forms of MYC-tagged full-length ZNF198–FGFR1 (4ZF-MYC and 10ZF-MYC, respectively). Neither the 4 nor the 10 zinc finger isoforms of the proline-rich region deletion constructs interacted with full-length ZNF198–FGFR1 (Figure 4).
Deletion of the ZNF198 proline-rich region abolishes ZNF198–FGFR1 self-association.
4 and 10 zinc finger ZNF198–FGFR1 isoforms that were full-length (4ZF-MYC and 10ZF-MYC, respectively) or deleted for the ZNF198 proline-rich region (4ZFΔPR-HA and 10ZFΔPR-HA, respectively) were expressed individually (columns 1, 2, 4, and 5) or coexpressed (columns 3 and 6), and then analyzed directly (INP) or immunoprecipitated with anti-HA or anti-MYC.
Deletion of the ZNF198 proline-rich region abolishes ZNF198–FGFR1 self-association.
4 and 10 zinc finger ZNF198–FGFR1 isoforms that were full-length (4ZF-MYC and 10ZF-MYC, respectively) or deleted for the ZNF198 proline-rich region (4ZFΔPR-HA and 10ZFΔPR-HA, respectively) were expressed individually (columns 1, 2, 4, and 5) or coexpressed (columns 3 and 6), and then analyzed directly (INP) or immunoprecipitated with anti-HA or anti-MYC.
Transformation of hematopoietic cells
Transforming capabilities of various forms of ZNF198–FGFR1 were evaluated by retroviral transduction of cDNA into the murine IL-3–dependent Ba/F3 cell line. Three groups of MSCV-G418 ZNF198–FGFR1 constructs were assembled: 4 and 10 zinc finger versions of the full-length ZNF198–FGFR1 (4ZF/MSCV and 10ZF/MSCV, respectively); 4 and 10 zinc finger versions of ZNF198–FGFR1 deleted from the ZNF198 proline-rich region (4ZFΔPR/MSCV and 10ZFΔPR/MSCV, respectively); and a construct containing only the ZNF198 proline-rich region and the FGFR1 kinase domain (δZF/MSCV). High-titer recombinant retrovirus was obtained by transient transfection of Bosc cells, and Ba/F3 infections were then performed in the presence of polybrene.33 After the selection of retrovirally transduced cells with G418, cells were assessed for IL-3–independent growth. IL-3 independence was conferred by each construct encoding the proline-rich domain and FGFR1 tyrosine kinase domain, indicating that the zinc fingers are unnecessary for Ba/F3 cell transformation (Figure5A). By contrast, ZNF198–FGFR1 polypeptides lacking the ZNF198 proline-rich region (4ZFΔPR/MSCV and 10ZFΔPR/MSCV) did not transform Ba/F3 cells (Figure 5B). Taken together, these data indicate that the ZNF198 proline-rich region is essential for Ba/F3 cell transformation.
Ba/F3 transformation, by ZNF198–FGFR1, requires ZNF198 proline-rich region but not ZNF198 zinc fingers.
Ba/F3 cells were infected with various ZNF198–FGFR1 retroviral constructs. After G418 selection, the cells were cultured without IL-3, and viable cells were counted daily for 10 days. (A) Ba/F3 cells expressing an N-terminal truncated ZNF198–FGFR1 lacking all zinc fingers, but containing the ZNF198 proline-rich region (ΔZF/MSCV), proliferate as well in the absence of IL-3 as Ba/F3 expressing full-length ZNF198–FGFR1 isoforms (4ZF/MSCV and 10ZF/MSCV). Controls included Ba/F3 cells infected with empty vector and cultured in medium with or without IL-3, and uninfected Ba/F3 cells cultured without IL-3. (B) Ba/F3 cells expressing 4 and 10 zinc finger ZNF198–FGFR1 lacking the proline-rich region (4ZFΔPR/MSCV and 10ZFΔPR/MSCV) do not proliferate in the absence of IL-3. Controls are as in A.
Ba/F3 transformation, by ZNF198–FGFR1, requires ZNF198 proline-rich region but not ZNF198 zinc fingers.
Ba/F3 cells were infected with various ZNF198–FGFR1 retroviral constructs. After G418 selection, the cells were cultured without IL-3, and viable cells were counted daily for 10 days. (A) Ba/F3 cells expressing an N-terminal truncated ZNF198–FGFR1 lacking all zinc fingers, but containing the ZNF198 proline-rich region (ΔZF/MSCV), proliferate as well in the absence of IL-3 as Ba/F3 expressing full-length ZNF198–FGFR1 isoforms (4ZF/MSCV and 10ZF/MSCV). Controls included Ba/F3 cells infected with empty vector and cultured in medium with or without IL-3, and uninfected Ba/F3 cells cultured without IL-3. (B) Ba/F3 cells expressing 4 and 10 zinc finger ZNF198–FGFR1 lacking the proline-rich region (4ZFΔPR/MSCV and 10ZFΔPR/MSCV) do not proliferate in the absence of IL-3. Controls are as in A.
To exclude an autocrine mechanism of Ba/F3 cell transformation, noninfected Ba/F3 cells were cultured in medium conditioned by Ba/F3 cells transformed by the 4 zinc finger ZNF198–FGFR1 construct (4ZF/MSCV). The conditioned medium did not support IL-3 independent growth, indicating that the ZNF198–FGFR1 transformation is, at least in part, cell autonomous.
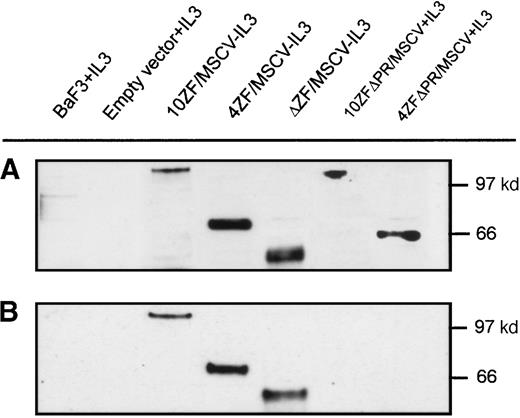
Tyrosine phosphorylation, resulting from cross-phosphorylation of oligomerized proteins, is a marker of FGFR1 activation. We performed Western blot analyses to determine whether Ba/F3 cell transformation correlates with ZNF198–FGFR1 tyrosine phosphorylation. Transforming ZNF198–FGFR1 fusion proteins containing the proline-rich region and either no zinc fingers, 4 zinc fingers, or 10 zinc fingers (ΔZF/MSCV, 4ZF/MSCV, and 10ZF/MSCV, respectively) were expressed in Ba/F3 cells (Figure 6A), and each of these polypeptides was tyrosyl phosphorylated (Figure 6B). By contrast, nontransforming ZNF198–FGFR1 fusion proteins lacking the ZNF198 proline-rich region and containing either 4 or 10 zinc fingers (4ZFΔPR/MSCV and 10ZFΔPR/MSCV, respectively) were expressed (Figure6A) but not tyrosyl phosphorylated (Figure 6B).
ZNF198–FGFR1 expression and tyrosine autophosphorylation.
(A) Verification of Ba/F3 cell ZNF198–FGFR1 expression by Western blot analysis using an anti-FGFR1 antibody. (B) Oligomeric ZNF198–FGFR1 fusion proteins (4ZF/MSCV, 10ZF/MSCV, and ΔZF/MSCV) are tyrosyl phosphorylated, whereas ZNF198–FGFR1 fusions lacking the ZNF198 proline-rich region (4ZFΔPR/MSCV and 10ZFΔPR/MSCV) did not self-associate and were not tyrosyl phosphorylated.
ZNF198–FGFR1 expression and tyrosine autophosphorylation.
(A) Verification of Ba/F3 cell ZNF198–FGFR1 expression by Western blot analysis using an anti-FGFR1 antibody. (B) Oligomeric ZNF198–FGFR1 fusion proteins (4ZF/MSCV, 10ZF/MSCV, and ΔZF/MSCV) are tyrosyl phosphorylated, whereas ZNF198–FGFR1 fusions lacking the ZNF198 proline-rich region (4ZFΔPR/MSCV and 10ZFΔPR/MSCV) did not self-associate and were not tyrosyl phosphorylated.
Discussion
The t(8;13) stem cell leukemia/lymphoma syndrome is a devastating disease in which both lymphoblastic lymphoma and acute myelogenous leukemia generally develop in affected patients. The chromosomal translocation t(8;13) results in the formation of aZNF198–FGFR1 fusion gene in which the tyrosine kinase domain is activated in a ligand-independent manner. In this study, we provide evidence that ZNF198–FGFR1 kinase activation results from constitutive oligomerization. We also show that ZNF198–FGFR1 oligomerization is mediated by a novel ZNF198 proline-rich region.
Ligand-independent oligomerization is a common mechanism of oncogenic activation for RTK oncogenes. Constitutive oligomerization can result from intragenic events, such as missense mutations, in-frame deletions, or in-frame insertions. Alternatively, oligomerization can result from gene fusions created by chromosomal rearrangements. Examples of intragenic mutations include those affecting the KIT juxtamembrane and phosphotransferase domains in gastrointestinal stromal tumors and mast cell tumors.40,41 These mutations result in constitutive KIT oligomerization and kinase activation.34-37 Similarly, germline mutations affecting 1 of 5 cysteines in the RET juxtamembrane domain, as found in most cases of multiple endocrine neoplasia type 2A (MEN 2A) and familial medullary thyroid carcinoma,42,43lead to cross-linking and activation through the formation of intermolecular disulfide bonds.44 Chromosomal mechanisms for RTK activation generally involve translocations or inversions, and the resultant fusion genes typically encode N-terminal oligomerization-inducing domains and C-terminal RTK domains. For example, t(5;12) translocation in patients with chronic myelomonocytic leukemia encodes a TEL–PDGFR β fusion protein,45 in which the TEL-pointed domain induces oligomerization and, consequently, constitutive activation of the PDGFR β tyrosine kinase.38Other fusion receptor tyrosine kinase oncogenes result from chromosomal rearrangements involving RET in thyroid carcinoma,46ALK in Ki-1 lymphoma,47ABL in chronic myeloid leukemia,48 andNTRK3 in fibrosarcoma.49-51 However, ZNF198–FGFR1, as reported herein, is the first fusion receptor tyrosine kinase oncoprotein in which constitutive oligomerization is mediated by a proline-rich region.
The ZNF198 oligomerizing proline-rich region has 2 short proline–valine repeats and contains 16% proline residues. Notably, proline-rich regions are often found within or adjacent to protein–protein interaction sites. Indeed, in a survey of more than 1600 protein–protein interaction sites, proline was the most common residue adjacent to the interaction sites.52 Proline has several unique properties believed to be of importance in protein–protein interaction. Proline disrupts α-helices and β-sheets, and its bulky pyrrolidine ring restricts the range of possible conformations and constrains the adjacent residue. Consequently, proline residues maintain the integrity of protein–protein interaction sites by establishing “brackets” that interrupt surrounding α-helix or β-sheet structures.52-55 The role of proline residues in protein–protein interactions is also indicated by the quaternary structures of oligomeric proteins anchored to each other by arm exchange.56 Prolines are often found at positions at which the exchanged arms take off from the protein cores, and mutation of these hinge prolines prevents oligomerization.56 In our studies, the ZNF198–FGFR1 oligomerization interface was localized to a 118-amino acid proline-rich region. Further studies are now needed to determine the minimal interaction site and the structural basis for self-association. It also remains to be determined whether ZNF198 oligomerizes in its native form and whether ZNF198 interacts with the family members ZNF261/DXS6673E,57ZNF262/KIAA0425,58 and ZNF258.59
Notably, Ollendorff et al60 have reported that an N-terminal truncated ZNF198–FGFR1 fusion protein, lacking the first 4 ZNF198 zinc fingers, did not associate with full-length ZNF198–FGFR1 in a yeast 2-hybrid assay. This finding suggested a contribution, for the first 4 zinc fingers, in ZNF198–FGFR1 oligomerization. The same report, on the other hand, provided evidence against such a role. Namely, the N-terminal truncated and full-length ZNF198–FGFR1 proteins oligomerized when coexpressed in Cos-1 cells. Similarly, our in vitro evaluations provide strong evidence that ZNF198 zinc fingers are not requisite for ZNF198–FGFR1-mediated cell transformation. Transforming potential, as assessed by the induction and kinetics of IL-3–independent growth in Ba/F3 cells, was comparable in ZNF198–FGFR1 proteins containing no zinc fingers, 4 zinc fingers, or 10 zinc fingers. Whereas the 4 and 10 zinc finger ZNF198–FGFR1 isoforms are coexpressed in human t(8;13) lymphoma cells, the “no zinc finger protein” ZNF198–FGFR1 does not occur naturally. However, the studies reported herein suggest that the ZNF198 zinc fingers are nonessential in ZNF198–FGFR1 oligomerization and oncogenic transformation. In contrast, the novel proline-rich ZNF198 oligomerization domain is critical for self-association, kinase activation, and cellular transformation and thus is a potential target for drug development.
RTK signaling pathways play an important role in cell mitosis, differentiation, migration, and metabolism, and their abnormal activation has been observed in an increasing number of hematologic and solid tumors. RTKs have distinct ligands, and they use complex signaling networks in transducing extracellular signals to the nucleus. However, a recent study revealed that the activation of various RTKs is accompanied by broadly overlapping effects on the expression of immediate early genes (IEG).61 In particular, the sets of IEGs induced by PDGFR β and FGFR1 were virtually identical. The similar IEG expression profiles for PDGFR β and FGFR1 are notable because constitutively activated forms of PDGFR β and FGFR1 are found in phenotypically distinctive hematopoietic neoplasms. PDGFR β fusion proteins have been reported only in a subset of patients with juvenile chronic myelomonocytic leukemia45 and, unlike ZNF198–FGFR1, have not been described in lymphoid neoplasms. Further studies are needed to determine whether the different clinicopathologic associations for PDGFR β and FGFR1 oncoproteins result from subtle differences in aberrant PDGFR β versus FGFR1 signaling or from unique interactions involving the N-terminal, non-RTK, components of the fusion proteins.
Supported by National Institutes of Health grant R01 CA72791 and American Cancer Society grant RPG-00-108-01-MGO.
Reprints:Sheng Xiao/Jonathan A. Fletcher, Department of Pathology, Brigham and Women's Hospital, 75 Francis Street, Boston, MA 02115; e-mail: sxiao@rics.bwh.harvard.edu orjfletcher@rics.bwh.harvard.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


![Fig. 3. The ZNF198 proline-rich region is crucial for protein–protein interaction. / (A) Truncated ZNF198–FGFR1 fusion proteins with (4ZFΔTK-HA and ΔZF-HA) or without (4ZFΔPRΔTK-HA) the ZNF198 proline-rich region were expressed individually (columns 2, 4, and 6), or were coexpressed with the full-length 4 zinc finger ZNF198–FGFR1 isoform (4ZF-MYC) (columns 3, 5, and 7). In vitro expressions were performed using reticulocyte lysates (Promega; in vitro TNT system) in the presence of [35S] methionine. The proteins were analyzed directly (INP, input) or were immunoprecipitated with anti-HA or anti-MYC. Only fusion proteins containing the ZNF198 proline-rich region (columns 5 and 7) coprecipitated with full-length ZNF198–FGFR1. (B) A full-length 4 zinc finger ZNF198–FGFR1 isoform (4ZF-MYC) and a minipeptide containing only the ZNF198 proline-rich region (PR-HA) were expressed individually (columns 1 and 2) or coexpressed (column 3), then analyzed directly (INP) or immunoprecipitated with anti-HA or anti-MYC. PR-HA and 4ZF-MYC coprecipitated (column 3).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/2/10.1182_blood.v96.2.699/5/m_bloo01453003w.jpeg?Expires=1769154399&Signature=CzME3Amz~HTjQi-qqwqumUuzB7uuxt4wYNMp~-4rG9~stkSIDuAXXVoXWmLH8zGqIDgVCCmAq2~UtRji2kh75xI3JadBXePluj~hLb25~IajaYX64v-wfYIiMPQh8jOJj-9mhB~ajvcrwM1-iwxEYoTvUweQznE12Dj1lyNShJkBN9JEyvV10bc6nwmepEcBymK4UShdjUjsVvRMj~zRYJaEU1-EZ~1zkPUa2AOqUwzJoGZQ2-kKt4zAv9l4ol3DiA2~JHE0TEILdpdGtdhOZZInb09mW5Qw0HMeWxWGVmw7lP6xtJngRrelAVabu0fLVeJ731inCt8QXyIyCSGQow__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal