Abstract
Megaloblastic anemia 1 (MGA1) is an autosomal recessive disorder caused by the selective intestinal malabsorption of intrinsic factor (IF) and vitamin B12/cobalamin (Cbl) in complex. Most Finnish patients with MGA1 carry the disease-specific P1297L mutation (FM1) in the IF-B12 receptor, cubilin. By site-directed mutagenesis, mammalian expression, and functional comparison of the purified wild-type and FM1 mutant forms of the IF–Cbl-binding cubilin region (CUB domains 5-8, amino acid 928-1386), we have investigated the functional implications of the P1297L mutation. Surface plasmon resonance analysis revealed that the P1297L substitution specifically increases the Kd for IF–Cbl binding several-fold, largely by decreasing the association rate constant. In agreement with the binding data, the wild-type protein, but not the FM1 mutant protein, potently inhibits 37°C uptake of iodine 125–IF–Cbl in cubilin-expressing epithelial cells. In conclusion, the data presented show a substantial loss in affinity of the FM1 mutant form of the IF–Cbl binding region of cubilin. This now explains the malabsorption of Cbl and Cbl-dependent anemia in MGA1 patients with the FM1 mutation.
Autosomal recessive megaloblastic anemia 1 (MGA1), alias, Imerslund-Gräsbeck syndrome, is a serious juvenile disorder1,2 caused by the malabsorption of vitamin B12–cobalamin (Cbl), the coenzyme for methionine synthase and methylmalonyl CoA mutase. More than 200 patients globally have been diagnosed,3 with clusters of cases reported in Norway,1 Finland,2 and several Middle Eastern countries.3 In contrast to patients with classical pernicious anemia, patients with MGA1 have normal production of intrinsic factor (IF), the Cbl-binding protein facilitating the uptake of the vitamin in the terminal portion of the ileum. Furthermore, many patients with MGA1 have significant Cbl-resistant proteinuria, a symptom not typical of pernicious anemia.
The IF–Cbl complex is removed from the intestinal lumen by means of a high-affinity receptor, the 460-kd epithelial protein cubilin.4-8 Cubilin-mediated endocytosis resembles classical endocytosis mediated by the low-density lipoprotein (LDL) receptor family of proteins.9 In intestinal cells, IF undergoes lysosomal degradation, and, by still unknown mechanisms, Cbl is transported out of the lysosomes. Later, Cbl complexes with transcobalamin (transcobalamin II), which is secreted into circulation from the basolateral side of the intestinal cells.10
Cubilin has a unique structural organization of extracellular protein modules comprising 8 epidermal growth factor repeats followed by 27 contiguous CUB domains.4,6 (CUB is an abbreviation of C1r/s, Uegf, and bone morphogenic protein-1, the first proteins known to contain the CUB domain.) The cubilin-encoding human gene CUBN is localized on the short arm of chromosome 10,6 the region in which the disease locus is identified in Norwegian and Finnish MGA1 patients.11Recently, 2 disease-specific CUBN mutations were identified in Finnish MGA1 patients.11 The most prevalent mutation, designated Finnish mutation 1 (FM1), accounts for 31 of 34 disease chromosomes from this group of patients. FM1 is a missense mutation causing substitution of a proline to a leucine at residue position 1297 of cubilin. Impaired function of the binding site is a possible consequence of the FM1 mutation because molecular analysis of recombinant fragments of rat cubilin has disclosed CUB domain 5-8 (amino acid 912-1369 of rat cubilin) as the region harboring the IF-Cbl–binding site.12 The other mutation, a single nucleotide mutation (designated FM2) identified in 1 patient and 1 carrier, apparently activates a cryptic splice site, which results in the in-frame insertion of multiple stop codons in the CUB domain 6.11
The current study was undertaken to define the functional implications of the disease-causing FM1 mutation in the IF-Cbl–binding region. Using site-directed mutagenesis of mammalian-expressed CUB 5-8 fragments, we performed a comparative functional analysis of the ligand-binding region. These data now provide the molecular information linking the disease-causing malabsorption of Cbl and the underlying genetic mutation in patients with FM1-type MGA1.
Materials and methods
Ligands
Human and porcine IF–Cbl were kindly donated by Dr Ebba Nexø, Aarhus University Hospital, Denmark.13
Expression of wild-type and mutant ligand-binding domain of human cubilin
Human cubilin cDNA fragments encoding CUB domains 5-8 were amplified by polymerase chain reaction (PCR) with the cloned Pfu DNA polymerase (Stratagene, La Jolla, CA). The wild-type fragment was amplified in a 1-step reaction using primers matching human cubilin bp 2808-2825 and bp 4167-4184 and extending the fragment with enzyme restriction sites (BamHI, EcoRV). For amplification of the mutated fragment, a mega-primer of 277 bp was amplified by PCR using a forward primer 5′-tagggtatctgaatccttattctga-3′, including the mutation causing the P1297L substitution and a reverse primer matching human cubilin bp 4167-4184 and extending the fragment with theEcoRV restriction site. This mega-primer was used for amplification of the mutated fragment, together with a forward primer matching human cubilin bp 2808-2825 and extending the fragment with the enzyme-restriction site BamHI. All PCR products were purified with the QIAEX II gel extraction kit (Qiagen, Chatsworth, CA). Upon purification, both fragments were subcloned into the pSecTag2B expression vector (Invitrogen, Groningen, The Netherlands) using the restriction enzymes BamHI and EcoRV (New England Biolabs, Beverly, MA) and the T4 DNA ligase (Amersham Pharmacia Biotech AB, Uppsala, Sweden). The pSecTag2B vector encodes the murine immunoglobulin κ chain leader sequence, which was used as leader peptide for protein secretion of both fragments. This vector also encodes the ampicillin and Zeocin (Invitrogen) resistance genes, allowing selection in Escherichia coli and mammalian cells. Plasmids were transformed using DH5α-competent cells (Clontech, Palo Alto, CA), and plasmid DNA was isolated by the Qiagen Maxiprep method (Qiagen) and sequenced before transfection as described previously.4 Chinese hamster ovary (CHO) K-1 cells were transfected using DOSPER liposomal transfection reagent (Roche Diagnostics, Mannheim, Germany), and stable transfected CHO clones were established by limited dilution and selection with 500 μg/mL Zeocin (Invitrogen). Clones were grown in serum-free medium for CHO cells (HyQ-CCM 5 from HyClone Logan, UT) with 300 μg/mL Zeocin.
Detection of expression products
Secretion of the recombinant human cubilin fragments was determined by Western blotting. Twenty μL growth medium was loaded onto 8% to 16% polyacrylamide gels and subjected to nonreducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) in 25 mmol/L Tris-base, 192 mmol/L glycine, and 0.1% SDS. After electrophoresis, proteins were transferred to a polyvinyl membrane (Sequi-Blot PVDF Membrane; Bio-Rad, Hercules, CA) in 25 mmol/L Tris-base and 192 mmol/L glycine. The membrane was blocked in 10 mmol/L Tris-base, 100 mmol/L NaCl, 2% polyoxyethylene–sorbitan monolaurate (Tween 20; Sigma, St. Louis, MO), and washed in 2 mmol/L CaCl2, 1 mmol/L MgCl2, 10 mmol/L HEPES (Sigma), 140 mmol/L NaCl, and 0.05% Tween 20. The membrane was incubated with a polyclonal antibody against human cubilin14 diluted 1:2000 in 2 mmol/L CaCl2, 1 mmol/L MgCl2, 10 mmol/L HEPES, 140 mmol/L NaCl, 0.05% Tween 20, and 2% nonfat dried milk (MD Foods, Aarhus, Denmark) for 2 hours at room temperature. After the membrane was washed, it was incubated with goat alkaline phosphatase-conjugated antirabbit immunoglobulins (DAKO A/S, Copenhagen, Denmark) diluted 1:1000 in 2 mmol/L CaCl2, 1 mmol/L MgCl2, 10 mmol/L HEPES, 140 mmol/L NaCl, 0.05% Tween 20, and 2% nonfat dried milk for 1 hour at room temperature. Antigen–antibody complexes were detected with the use of 5-bromo-4-chloro-3-indolyl-phosphate–nitro blue tetrazolium (Promega, Madison, WI) as a chromogenic substrate.
Ligand-affinity precipitation and purification of wild-type and mutated ligand-binding domain of human cubilin
The conditioned medium of the transfected cells secreting recombinant human cubilin fragments into the medium was precipitated with CNBr-activated Sepharose 4B beads (Amersham Pharmacia Biotech AB) coupled with porcine IF–Cbl (1 mg/mL gel) as described previously.12 Larger-scale purification of the wild-type and the mutated ligand-binding domains of human cubilin was performed by IF–Cbl affinity chromatography.7
Surface plasmon resonance
Affinity measurements of the binding of human IF–Cbl to purified human wild-type cubilin CUB 5-8 and human FM1 mutant cubilin CUB 5-8 were performed by surface plasmon resonance analyses on a BIAcore 2000 instrument (Biacore AB, Uppsala, Sweden) as described.7,12The BIAcore sensor chips (type CM5; Biacore AB) were activated with a 1:1 mixture of 0.2 mol/LN-ethyl-N′-(3-dimethylaminopropyl) carbodiimide and 0.05 mol/L N-hydroxysuccimide in water. Human cubilin was immobilized at a concentration of 40 μg/mL in 10 mmol/L sodium acetate, pH 4.5,6 and purified human cubilin fragments at concentrations of 25 to 40 μg/mL in 10 mmol/L sodium acetate, pH 4.0. The remaining binding sites were blocked with 1 mol/L ethanolamine, pH 8.5. The surface plasmon resonance signals generated from immobilized human full-length cubilin, human wild-type CUB 5-8, and human FM1 mutant CUB 5-8 corresponded to 33, 58, and 54 fmol of receptor or receptor fragment/mm2, respectively. The on and off rates for binding of human IF–Cbl were recorded by the flow of 10, 20, 50, and 100 nmol/L human IF–Cbl. Flow cells were regenerated with 6 mol/L guanidine-HCl6 or 1.6 mol/L glycine-HCl, pH 3, and the binding data were analyzed using the BIA evaluation program version 3.0 (Biacore AB).
Uptake of IF–Cbl in cultured yolk sac cells
Cubilin-expressing Brown Norway rat yolk sac epithelial cells immortalized by mouse sarcoma virus15 16 were grown to confluence in 24-well plates (Nunc; Life Technologies A/S, Taastrup, Denmark) in Dulbecco's modified Eagle's medium (DMEM; Life Technologies A/S) containing 10% fetal calf serum. Incubation with iodine 125–IF–Cbl was carried out in serum-free DMEM supplemented with 0.2% bovine serum albumin. Degradation of labeled proteins was measured by precipitation of the incubation medium with 12.5% trichloroacetic acid. Cell-associated radioactivity was measured by radioactivity determination of the washed cell layer in 0.5 mol/L NaOH. Chloroquine and leupeptin from Sigma were applied for the time course.
Proteolytic digestion of wild-type and mutated ligand-binding domain of human cubilin
Purified wild-type and mutant human cubilin fragments (approximately 0.6 μg) were digested with 0.2, 1, 5, and 25 μg/mL trypsin (Sigma) or chymotrypsin (Roche Diagnostics) for 3 hours at 37°C (total volume, 20 μL) in 10 mmol/L NaH2PO4, 150 mmol/L NaCl, 0.6 mmol/L CaCl2, pH 7.4, with and without 2 mmol/L EDTA.
Results
Figure 1, panel A, shows the known structural organization of cubilin, including its 27 contiguous CUB domains, and outlines the IF–Cbl-binding CUB 5-8 region—the subject of mutagenesis and functional analyses in this study. The CUB 5-8 region was expressed in stably transfected CHO cells in 2 variants, the wild-type form and the FM1 mutant form having the P1297L mutation in CUB domain 8. Figure 1, panel B, shows a Western blot of the 2 analogues, both of which were secreted into the cell medium.
Mammalian expression of the IF–Cbl-binding region of wild-type and FM1 mutant cubilin.
(A) The modular protein organization of human cubilin (460 kd) and the IF–Cbl-binding region (cubilin CUB domains 5-8, amino acid 928-1386) expressed in CHO cells stably transfected with miniconstructs of human cubilin cDNA. The region was expressed in 2 variants, the wild-type CUB 5-8 region and the corresponding FM1 mutant form containing the P1297L mutation. (B) Western blotting of the conditioned CHO cell medium containing the 2 analogues of the CUB 5-8 region. The estimated size (approximately 95 kd) of the proteins indicates that glycosylation accounts for approximately 40% of the mass.
Mammalian expression of the IF–Cbl-binding region of wild-type and FM1 mutant cubilin.
(A) The modular protein organization of human cubilin (460 kd) and the IF–Cbl-binding region (cubilin CUB domains 5-8, amino acid 928-1386) expressed in CHO cells stably transfected with miniconstructs of human cubilin cDNA. The region was expressed in 2 variants, the wild-type CUB 5-8 region and the corresponding FM1 mutant form containing the P1297L mutation. (B) Western blotting of the conditioned CHO cell medium containing the 2 analogues of the CUB 5-8 region. The estimated size (approximately 95 kd) of the proteins indicates that glycosylation accounts for approximately 40% of the mass.
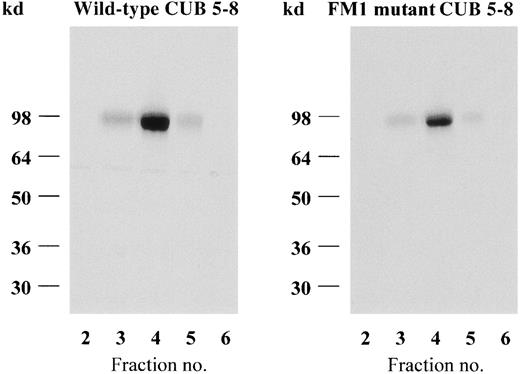
The wild-type CUB 5-8 protein and the FM1 mutant analogue were purified from the conditioned CHO cell media by IF–Cbl affinity chromatography (Figure 2). This single-step procedure isolated the 2 recombinant proteins in a pure homogeneous form. However, though similar amounts and concentrations of the 2 expression products were loaded on identical IF–Cbl–Sepharose 4B columns, the yield of the FM1 mutant protein was generally lower than that of the wild-type protein (Figure 2). Analysis of the flow-through from the IF–Cbl columns showed that affinity chromatography virtually depleted the medium of wild-type CUB 5-8 protein, whereas it retained less of the FM1 mutant form. The possibility that the expressed FM1 mutant CUB 5-8 protein contained 2 fractions, representing a ligand-binding and an inactive form, was excluded by repeated IF–Cbl affinity chromatography recovering more than 90% of the FM1 mutant CUB 5-8 protein in a purified form (not shown). The structural stability of the 2 purified protein analogues was analyzed by proteolytic digestion with trypsin and chymotrypsin. This analysis revealed that both CUB 5-8 analogues were resistant to proteolysis, indicating a compact folding. Interestingly, this resistance to proteolysis required calcium in the buffer, as seen in Figure3, which shows the digestion with trypsin in the presence and absence of EDTA. SDS–PAGE of the digested CUB 5-8 analogues did not reveal any major changes in the fragmentation patterns in the absence or the presence of calcium (Figure 3).
Elution profiles of IF–Cbl-purified wild-type and FM1 mutant CUB 5-8 proteins.
Nonreducing SDS–PAGE (silver staining) of the eluted fractions.
Elution profiles of IF–Cbl-purified wild-type and FM1 mutant CUB 5-8 proteins.
Nonreducing SDS–PAGE (silver staining) of the eluted fractions.
Proteolytic digestion of the wild-type and FM1 mutant CUB 5-8 proteins.
Analysis by reducing SDS–PAGE and silver staining. The left panel shows the degradation of the wild-type CUB 5-8 protein with various concentrations of trypsin in the presence (−Ca++) and absence (+Ca++) of EDTA. The weak band at the position of the asterisk is not a proteolytic degradation product, as seen in the first lane. The right panel shows the similar effect of trypsin on the FM1 mutant CUB 5-8 protein.
Proteolytic digestion of the wild-type and FM1 mutant CUB 5-8 proteins.
Analysis by reducing SDS–PAGE and silver staining. The left panel shows the degradation of the wild-type CUB 5-8 protein with various concentrations of trypsin in the presence (−Ca++) and absence (+Ca++) of EDTA. The weak band at the position of the asterisk is not a proteolytic degradation product, as seen in the first lane. The right panel shows the similar effect of trypsin on the FM1 mutant CUB 5-8 protein.
The IF–Cbl binding affinity of the 2 purified expression products was further characterized by surface plasmon resonance analysis (Figure4) on a BIAcore 2000 instrument (Biacore AB). The 2 CUB 5-8 proteins were immobilized on similar CM5 sensor chips in flow cells in which the binding of flowing IF–Cbl was recorded. Measurements of the binding at various IF–Cbl concentrations showed, in agreement with the affinity chromatography data, that the 2 CUB 5-8 protein sensor chips had a similar capacity for binding IF–Cbl (measured at higher than 1 μmol/L concentrations of ligand; data not shown) but a substantial difference in affinity (measured at 10-50 nmol/L ligand; Figure 4) for the ligand. The wild-type CUB 5-8 protein exhibited a high IF–Cbl binding affinity (Kd = 2 nmol/L), virtually identical to that of the binding of IF–Cbl to native, full-length cubilin,6 whereas the FM1 mutant CUB 5-8 protein bound less efficiently (Kd = 10 nmol/L). Examination of the rate constants, which determine the overallKd (=koff/kon), showed that the association rate (kon) was more affected (legend to Figure 4) than the dissociation rate constant (koff). To exclude that a difference in immobilization on the flow sensor chip might account for an estimated difference in affinity of the 2 CUB 5-8 analogues, we performed the reverse experiment, in which IF–Cbl was the immobilized component and the CUB 5-8 proteins represented the analyte in the flow buffer. This experiment confirmed a substantial decrease in affinity of the FM1 mutant CUB 5-8 protein due to an approximately 3-fold decrease in the association rate (not shown).
Surface plasmon resonance analysis of the binding of IF–Cbl flowing along BIAcore CM5 sensor chips with immobilized wild-type or FM1 mutant form of CUB 5-8.
(A) The IF-Cbl binding to the wild-type CUB 5-8 protein with a flow concentration of 10, 20, and 50 nmol/L ligand. (B) The IF–Cbl binding to the FM1 mutant CUB 5-8 protein with a flow concentration of 10, 20, and 50 nmol/L ligand. The BIA evaluation 3.0 software showed the best fit to a 1-site model with the following binding parameters: wild-type CUB 5-8: Kd = 2 nmol/L; kon = 1.3 × 105mol/L−1s−1; koff= 3.0 × 10−4s−1; FM1 mutant CUB 5-8: Kd= 10 nmol/L; kon = 4.2 × 10.4 mol/L−1 s−1;koff = 4.1 × 10−4s−1.
Surface plasmon resonance analysis of the binding of IF–Cbl flowing along BIAcore CM5 sensor chips with immobilized wild-type or FM1 mutant form of CUB 5-8.
(A) The IF-Cbl binding to the wild-type CUB 5-8 protein with a flow concentration of 10, 20, and 50 nmol/L ligand. (B) The IF–Cbl binding to the FM1 mutant CUB 5-8 protein with a flow concentration of 10, 20, and 50 nmol/L ligand. The BIA evaluation 3.0 software showed the best fit to a 1-site model with the following binding parameters: wild-type CUB 5-8: Kd = 2 nmol/L; kon = 1.3 × 105mol/L−1s−1; koff= 3.0 × 10−4s−1; FM1 mutant CUB 5-8: Kd= 10 nmol/L; kon = 4.2 × 10.4 mol/L−1 s−1;koff = 4.1 × 10−4s−1.
Using an established sarcoma virus-immortalized cubilin-expressing yolk sac cell line,15,16 we investigated the potential effect of the FM1 mutation on the level of cubilin-mediated uptake of iodine 125–IF–Cbl. Previous data on this cell line5 demonstrated a high iodine 125–IF–Cbl uptake more than 80% inhibitable with polyclonal anticubilin antibodies. Figure5A shows the time course for cellular association and degradation of IF–Cbl in the yolk sac cell line and a strong inhibitory effect on degradation by leupeptin and chloroquine, leading to the cellular accumulation of IF–Cbl (Figure 5B). This indicates that the receptor-mediated uptake in these cells is followed by lysosomal degradation, and the total uptake can, therefore, be estimated as the sum of cell-associated and degraded radioactivity. Figure 6 shows the inhibitory effect of the wild-type and FM1 mutant CUB 5-8 proteins on the total uptake of iodine 125–IF–Cbl after 2 hours. Concentrations of 10 μg/mL or higher of the wild-type CUB 5-8 protein inhibited uptake by 75% to 85% (n = 3), the same level of inhibition as previously seen with polyclonal anticubilin antibodies.5 The half-maximal inhibition (IC50) was approximately 1 μg/mL (20 nmol/L) for the wild-type CUB 5-8 protein. An approximately 20-fold higher concentration of the FM1 mutant CUB 5-8 protein was needed to reach the same level of inhibition (IC50 = 20 μmol/L).
Uptake of 125I–IF–Cbl in cubilin-expressing epithelial cells and inhibition with the wild-type and FM1 mutant CUB 5-8 proteins.
(A) Time-course for cell association (•) and degradation (○) of iodine 125–IF–Cbl. (B) Time-course for uptake and degradation of iodine 125–IF–Cbl in the presence of chloroquine and leupeptin. Values are the mean ± 1 SD of triplicate experiments.
Uptake of 125I–IF–Cbl in cubilin-expressing epithelial cells and inhibition with the wild-type and FM1 mutant CUB 5-8 proteins.
(A) Time-course for cell association (•) and degradation (○) of iodine 125–IF–Cbl. (B) Time-course for uptake and degradation of iodine 125–IF–Cbl in the presence of chloroquine and leupeptin. Values are the mean ± 1 SD of triplicate experiments.
Effect of wild-type and FM1 mutant CUB 5-8 proteins on the total uptake of 125I–IF–Cbl in epithelial cells.
Values are the mean ± 1 SD of triplicate experiments and are shown as the percentage of uptake in relation to the control uptake of iodine 125–IF–Cbl without any added CUB 5-8 protein.
Effect of wild-type and FM1 mutant CUB 5-8 proteins on the total uptake of 125I–IF–Cbl in epithelial cells.
Values are the mean ± 1 SD of triplicate experiments and are shown as the percentage of uptake in relation to the control uptake of iodine 125–IF–Cbl without any added CUB 5-8 protein.
Discussion
The data presented here show that the FM1 mutation inCUBN11 of Finnish patients with MGA1 has a substantial negative effect on IF–Cbl binding affinity of the CUB 5-8 cubilin region. The strong decrease in affinity implies that less IF–Cbl may be bound and internalized in the short segment of the terminal ileum taking up the vitamin-carrier complex. This implication of the FM1 mutation in patients with MGA1 is illustrated by the BIAcore data (Figure 4), which, analogous to the conditions in the intestine, show receptor binding of IF–Cbl flowing unidirectionally in a tube-like compartment. With a flow of 10 to 50 nmol/L ligand, the FM1 mutant receptor picks up 6- to 8-fold less IF–Cbl exposed to the surface. However, the exact negative effect in vivo of the decreased cubilin affinity may vary because different physiological factors—such as ligand concentration, composition and volume of the intestinal fluid, flow rate, cubilin density, and length and area of the cubilin-expressing intestinal surface—all contribute to the final fraction of IF–Cbl taken up. Furthermore, the BIAcore data were recorded at 25°C, which is below the physiological temperature. The rate constants for protein interactions are usually temperature sensitive, and the affinities measured by surface plasmon resonance may differ from the affinities at 37°C. In fact, the data on the inhibitory effect of the cellular uptake of the CUB 5-8 proteins at 37°C indicated an approximately 20- to 30-fold stronger inhibitory potency of the wild-type CUB 5-8 proteins compared to the FM1 mutant protein.
The BIAcore data further indicated that the association rate constant is the main binding parameter affected in the FM1 mutant protein, whereas the dissociation rate constant appears only slightly altered. It is tempting to speculate that the mutation may cause a structural change that affects the routing of the ligand to the binding site. The P1297L mutation may cause such a structural change because of the distinctive 65° angle in between the N and Cα atoms in the proline residue. The fact that the proline in this position is conserved in all species investigated support the crucial importance of this residue.12
Limited proteolysis of the CUB 5-8 analogous suggested no major differences in structure and calcium-protective effect. The effect of calcium is an interesting observation because it may represent a mechanism whereby the receptor protects itself against digestion in the proteolytically active milieu of the small intestine. Furthermore, it indicates that calcium binds to cubilin. This is also indicated by the fact that calcium is a prerequisite for ligand binding to cubilin, and CUB domains of other proteins (C1r/s) are known to bind calcium.
Disease-causing mutations with functional consequences on endocytic receptors have, to our knowledge, only been reported in the gene encoding the LDL receptor. Close to 250 different mutations are known in this gene, and many of these cause critical amino acid changes in the ligand-binding type A repeats, leading to decreased affinity for the LDL particle.17 A part of this group of mutations affects primarily the binding of calcium,18 which is also essential for ligand binding to the LDL receptor. Other mutations affect the stability, processing, or trafficking/endocytosis of the LDL receptor. Similar functional implications of mutations in CUBNmay explain the appearance of MGA1 in ethnic populations other than the Finnish. However, defects in other genes encoding accessory activity important for cubilin function might also cause MGA1. This possibility has, in particular, become evident from a recent genetic analysis19 of a canine model20 resembling human MGA1. These dogs present absent brush border expression of cubilin, but the functional defect and disease show no linkage with the canineCUBN locus.19
It is worth noting that since the early 1970s, the number of newly identified MGA1 cases in Finland has declined, both the general type and the FM1 type. Therefore, speculation11 has risen about whether this decline results from changes in the population structure or from drastic changes in Finnish dietary habits, such as the increase in meat consumption during the last 50 years, may have influenced the penetrance of the mutant gene. An interesting perspective concerning the treatment for FM1 type MGA1, and perhaps other forms of MGA1, arises if the latter holds true. A controlled Cbl-rich diet instead of the present, regular Cbl injections may then be an optional alternative to avoid relapse of megaloblastic anemia.
In conclusion, our data provide molecular and functional evidence explaining the effect of the disease-specific FM1 mutation in patients with MGA1. Future studies may disclose new MGA1-causing mutations inCUBN or in other genes affecting the function of cubilin.
Acknowledgments
We thank Dr Ebba Nexø for providing purified IF–Cbl and Kirsten Lassen for excellent technical assistance.
Supported by the Novo Nordisk Foundation, the Danish Medical Research Council, the Danish Biomembrane Center, Aarhus University, and the Velux Foundation.
Reprints:Søren K. Moestrup, Department of Medical Biochemistry, University of Aarhus, Ole Worms Allé, blgn. 170, Aarhus, Denmark; e-mail: skm@biobase.dk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal