Abstract
WNT proteins compose a family of secreted signaling molecules that regulate cell fate and behavior. The possible influence of WNTs on hematopoietic cell fate was examined. Both hematopoietic progenitor cell (HPC)–enriched embryonic avian bone marrow cells and the quail mesodermal stem cell line QCE6 were used for these studies. Under optimized conditions, the bone marrow and QCE6 cells behaved identically and developed into red blood cells (RBCs), monocytes, macrophages, granulocytes, and thrombocytes. This broad range of blood cell phenotypes exhibited by QCE6 cells was dependent on their active expression of WNT11. However, when QCE6 cells were prevented from producing WNT11—by expression of a stably transfected WNT11 antisense transgene—the cultures were dominated by highly vacuolated macrophages. RBCs were absent from these cultures, and the presence of monocytes was greatly diminished. Exposure of these WNT11 antisense cells to soluble WNT11 or WNT5a restored the broad range of blood cell phenotypes exhibited by parental QCE6 cells. Overexpression of WNT protein in QCE6 cells further increased the prevalence of RBCs and monocytes and greatly diminished the appearance of macrophages. Accordingly, treatment of HPC-enriched bone marrow cultures with soluble WNT11 or WNT5a inhibited macrophage formation. Instead, monocytes and RBCs were the prevalent cells displayed by WNT-treated bone marrow cultures. Together, these data indicate that WNTs may play a major role in regulating hematopoietic cell fate.
Introduction
The various cell types that make up the blood are thought to emanate from a common multipotent stem cell of mesodermal origin. During embryogenesis, blood cell formation results from at least 2 independent episodes. Hematopoietic cells are first observed within the blood islands of early somite stage embryos. These cellular aggregates arise from ventral mesodermal cells that have migrated to extra-embryonic regions during gastrulation. These “primitive” hematopoietic progenitor cells (HPCs) will yield primarily erythrocytes, which provide circulating blood cells for the developing embryo. Shortly thereafter, intra-embryonic dorsal mesoderm contributes to a second wave of blood cell formation—referred to as definitive hematopoiesis. These HPCs, which originate within the embryo, give rise to all the myeloid and lymphoid cell lineages, supplying the starting material for adult hematopoiesis. As embryogenesis proceeds, hematopoietic cells home to various sites, such as the spleen, thymus, and liver. Hematopoiesis becomes established in the bone marrow, the principal site of HPC regeneration and maturation in the adult, during the later stages of embryonic development.1-4
Although avian hematopoiesis is very similar to mammalian hematopoiesis, there are several noticeable differences. First, red blood cells (RBCs) are nucleated.5 Second, heterophils, which along with basophils and eosinophils compose the granulocytic cell types in the bird, are not exhibited in mammals. Third, thrombocytes, which are equivalent to the nonnucleated mammalian platelet, are the progeny of mononuclear thromboblasts.6In comparison with mouse and human studies, the field of avian hematopoiesis has been hindered by the absence of suitable cell identification markers. Very few clusters-of-differentiation antibodies exist that react to avian leukocytes.7 Hence, cytological examination with azure stains (eg, May-Grünwald-Giemsa and Wright-Giesma) has remained the predominant means of identifying specific blood cell types in the bird. Additionally, the detection of leukocyte-associated enzymatic activities, such as acid phosphatase (AP), tartaric acid–resistant phosphatase (TARP), naphthol AS-D chloroacetate esterase (CAE), and α-naphthyl acetate esterase (ANAE), has been used to further verify the phenotype of avian blood cells.8
A large number of individual extracellular signaling factors that regulate hematopoietic cell differentiation have been described. Many of these factors—such as granulocyte-macrophage colony-stimulating factor, erythropoietin, and the various interleukins—were initially identified and studied primarily because of their involvement with hematopoiesis.9-11 Additionally, other molecules, whose importance were initially determined for nonblood lineages, were shown to play major roles in blood cell formation—including bFGF, transforming growth factor β, BMP4, and stem cell factor (SCF).12,13 Among these latter factors are WNT proteins, whose importance for patterning emerging embryonic tissues has been well established.14,15 WNTs comprise a large and highly conserved group of secreted signaling proteins that are encoded by at least 18 distinct genes in the vertebrate genome. In embryonic mice, WNT gene expression has been observed in hematopoietic tissue, with both WNT5a and WNT10b exhibited in yolk sac and fetal liver.16 WNT5a has also been shown to be expressed by CD34+Lin− progenitor cells in human bone marrow.17 Culture experimentation with either mouse or human hematopoietic cells indicated that WNTs may act as growth factors for progenitor cell populations.16 17
In prior investigations, we examined the ability of WNTs to promote the differentiation of mesoderm during early embryogenesis.18,19 To assist in these studies, we used the mesodermal stem cell line QCE6 as a cell culture model for studying the regulated diversification of mesoderm. QCE6 cells were derived from the mesodermal layer of early gastrula stage Japanese quail (Coturnix coturnix japonica) embryos20,21 and possess the potential to differentiate into cardiomyocytes, endothelial cells, and RBCs.22 For the present study, we examined whether QCE6 cells have the capability to give rise to other blood lineages in addition to erythrocytes. Here, we report that QCE6 cells will exhibit as diverse a hematopoietic potential as HPCs taken directly from quail bone marrow. Since the ability of QCE6 cells to undergo cardiomyocyte differentiation was WNT dependent,18we additionally asked whether WNT expression would influence the blood cell differentiation of these cells. We found that WNTs profoundly affect the diversity of hematopoietic cell phenotypes that are exhibited by both QCE6 and bone marrow–derived cells. Specifically, we find that WNT signals inhibit the formation of macrophages and increase the prevalence of monocyte and erythrocyte cell phenotypes.
Materials and methods
Isolation of avian bone marrow cells
Femurs were collected from embryonic day 14 (ED14) quail embryos. Primary cells were obtained by flushing bone marrow cavities with Dulbecco's modified Eagle's medium (DMEM) (Sigma, St. Louis, MO) plus 100 IU/mL penicillin–100 μg/mL streptomycin (pen/strep) (Gibco BRL, Grand Island, NY). Bone marrow cords were dissociated via repetitive passage through a 20-gauge needle followed by filtration through a 40-μm nylon cell sieve (Falcon Labware, Franklin Lakes, NJ). The mononuclear cell fraction was isolated from samples, pooled from 4 dozen embryos per experiment, by centrifugation over a Ficoll-Paque (Pharmacia, Inc, Piscataway, NJ) gradient as described.23 Cells were resuspended at a concentration of 1 × 106 cells per milliliter in DMEM supplemented with 10% fetal bovine serum plus pen/strep, and subjected to overnight adherence selection on tissue-culture plastic to separate adherent cells.
Methylcellulose cultures
Purified avian bone marrow or QCE6 cells were seeded at 5 × 104 cells per milliliter into dishes (Nalge Nunc, Rochester, NY) containing semisolid medium that was composed of the following: DMEM, 1.2% of 1500 centipoise methylcellulose (Sigma), 20% fetal bovine serum (Sigma), 10% chick serum (Sigma; heat inactivated for 1 hour at 56°C), 5% WeHi-3–conditioned medium,24100 μmol/L N-acetyl-D-glucosamine (Sigma), 1% deionized bovine serum albumin (Intergen, Purchase, NY), 10% ConA-stimulated spleen cell–conditioned medium,25 200 μg/mL iron-saturated transferrin (Sigma), 1 IU/mL erythropoietin (R & D Systems, Minneapolis, MN), 100 ng/mL SCF (PeproTech, Rocky Hill, NJ), 10 ng/mL insulinlike growth factor I (PeproTech), 500 ng/mL hydrocortisone (Sigma), and pen/strep. The medium was supplemented with 10% NaHCO3 to sustain a pH concentration of 8.0, which proved optimal for blood cell differentiation.26 Duplicate cultures were incubated for up to 7 days at 37°C in a 5% CO2 atmosphere.
Fibrin gel cultures
Purified avian bone marrow or QCE6 cells were plated at 5 × 104 cells per milliliter culture in medium containing DMEM, 1 mg/mL human fibrinogen (Sigma), 20% fetal bovine serum, 10% chick serum (heat inactivated for 1 hour at 56°C), 5% WeHi-3–conditioned medium, 1% deionized bovine serum albumin, 1 IU/mL erythropoietin, 100 ng/mL SCF, 500 ng/mL hydrocortisone, 27.4 mmol/L NaHCO3, 200 μg/mL iron-saturated transferrin, and pen/strep. The mixture was allowed to clot at 37°C for 15 minutes following the addition of 1 IU/mL bovine thrombin (Sigma). Gels were overlaid with an equal volume of medium without fibrinogen to inhibit dehydration. Cultures were plated in duplicate and incubated for up to 7 days at 37°C in a 5% CO2 atmosphere.
Stably transfected QCE6 cell sublines
The production of stable transfectants of QCE6 cells with altered levels of WNT11 expression has been previously described.18 Briefly, full-length WNT11 complementary DNA (cDNA), in either sense or antisense orientations, was inserted immediately downstream of the cytomegalovirus promoter in the eukaryotic expression vector pcDNA3 (Invitrogen, Carlsbad, CA) and then introduced into QCE6 cells by means of LipofectAMINE (Gibco BRL). Cells that stably incorporated the transgenes were selected by their resistance to neomycin and subsequently cloned. Multiple subclones that contained either the sense or the antisense WNT11 cDNA were generated and referred to as WNT11ox (ox stands for overexpress) and WNT11αs cells, respectively. For the present study, we exclusively used a single WNT11 sense clone, WNT11ox/3, and 2 individual antisense subclones, WNT11αs/4 and WNT11αs/6. To maintain expression of the transgenes, these stably transfected cells were continuously passaged in the presence of 200 μg/mL G418 (Gibco BRL). To determine relative levels of WNT11 protein produced by WNT11ox/3, QCE6, WNT11αs/6, and WNT11αs/4 cells were grown in 60-mm tissue-culture dishes until confluency was reached. The medium was removed from each dish and replaced with 5 mL minimal essential medium (MEM) containing 5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL selenium (Sigma) plus 50 μg/mL heparin (which solubilizes WNT protein bound to cell surfaces and matrix). Following overnight incubation, the heparin-treated media were collected and assayed for WNT11 protein expression by immunoblot analysis as described.19 To ensure that the volumes of secreted protein used for blotting represented equal numbers of cells, total cell protein was prepared from these cultures following collection of the culture fluids. Culture plates were scraped after the addition of hypotonic buffer (5 mmol/L Tris pH 7.5, 5 mmol/L egtazic acid, 5 mmol/L EDTA, 2 mmol/L phenylmethylsulfonyl fluoride, 2 μg/mL Leupeptin, 1 μg/mL Pepstatin A); the resulting cell lysates were passed several times through 25-gauge needles, incubated on ice for 2 hours, and brought to isotonicity with 10 × phosphate-buffered saline (PBS). These protein samples were electrophoretically separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels, transferred to polyvinylidene fluoride (PVDF) membrane, and probed with streptavidin-conjugated alkaline phosphatase (Tropix, Bedford, MA) to detect the mitochondrial protein pyruvate carboxylase.27 28
For producing WNT5a-expressing QCE6 cells, we followed a similar protocol except that the expression vector employed was pcDNA3.1/myc-6xHis A (Invitrogen). This allowed for the production of a WNT5a fusion protein with a myc-6xHis tag at the carboxy terminus. To accomplish this, Xenopus WNT5a cDNA (a gift from Randall Moon) was amplified by polymerase chain reaction (PCR) to produce full coding length sequence that included 31 base pairs of the 5′-untranslated region up to the last amino-acid codon. The PCR fragment, which contained terminal EcoRI and XbaI sites from the PCR primers, was subsequently ligated in-frame withmyc-6xHis tag of pcDNA3.1A. Following LipofectAMINE-mediated transfection and G418 selection, individual clones were isolated by means of cloning rings. After expansion, culture medium was collected from these clones and immunoblotted for myc-tagged fusion protein—by means of epitope-specific antibody (Santa Cruz Biotechnology, Santa Cruz, CA)—to identify high-secreting sublines. For the present studies, we used a single WNT5a-expressing clonal subline, which is referred to as WNT5a/QCE6.
Generation of WNT-containing conditioned media and purification of WNT5a protein
Conditioned media were harvested from confluent cultures of either WNT11ox/3 or WNT5a/QCE6 cells grown in serum-free media containing 5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL selenium, and 0.4% albumin in vitamin-enriched MEM (Sigma). The presence of WNT11 and WNT5a protein within these conditioned media was verified by immunoblotting by means of WNT11-specific18 19 ormyc-tag–reactive (Santa Cruz Biotechnology) antibodies, respectively. Purification of WNT5a protein from conditioned media was accomplished by means of nickel–nitrilotriacetic acid (Ni-NTA) spin columns (Qiagen Inc, Valencia, CA), which binds 6xHis-tagged proteins with high affinity. Conditioned media from WNT5a/QCE6 cells (WNT5a CM) was supplemented with imidazole (Qiagen) at a final concentration of 5 mmol/L. Individual Ni-NTA columns were pre-equilibrated with NaH2PO4 pH8, 300 mmol/L NaCl, and 5 mmol/L imidazole and then loaded with 500 μL aliquots of WNT5a CM per 5 mmol/L imidazole. After samples were passed through the columns by low-speed centrifugation (at 700g), columns were washed 4 times with NaH2PO4 pH8, 300 mmol/L NaCl, and 20 mmol/L imidazole. Metal-binding protein was collected by eluting twice from each column with 200 μL volumes of NaH2PO4 pH8, 300 mmol/L NaCl, and 250 mmol/L imidazole. Afterward, pooled samples were dialyzed 4 times against 500 volumes of PBS and stored at −70°C. Protein concentrations of WNT5a-containing conditioned media and Ni-NTA column–purified WNT5a–containing fraction were determined by means of the Micro BCA protein assay reagent kit (Pierce, Rockford, IL), according to the manufacturer's instructions.
To assess the influence of ectopic WNT protein on blood cell diversification, embryonic bone marrow cells, QCE6 cells, or QCE6 stably transfectants were plated in either methylcellulose (MeC) or fibrin gel (FB) cultures as described above, except that conditioned medium collected from either WNT11ox/3 or WNT5a/QCE6 cells was substituted for DMEM. Alternatively, Ni-NTA column–purified WNT5a protein was added to the cultures at a final concentration of 5% vol/vol.
Assays for cell differentiation
Both MeC and FB cultures were monitored microscopically each day for cell growth and differentiation. Cultures were scored in situ according to number and cell phenotype of colonies, with medium (20 to 50 cells) and large (more than 50 cells) colonies tabulated independently. Prior to staining, live cultures were documented with an Olympus IMT-2 inverted microscope with the use of Ektachrome ASA 100 film (Kodak, Rochester, NY). Both MeC and FB cultures were processed for May-Grünwald-Giemsa,6 esterase, and phosphatase stains. For MeC cultures, similar colonies were picked and cytospun onto electrostatically charged microscope slides (Fisher Scientific, Suwanee, GA). For FB cultures, gels were transferred onto microscope slides and partially dehydrated according to Lanotte's technique.29 Designation of blood cell phenotypes within these cultures, following May-Grünwald-Giemsa staining, was according to previous documentation of avian blood cells.6Naphthol AS-D CAE, ANAE, AP, and TARP stains were performed by means of Sigma kits 90-A1, 90-C2, and 387-A.
Results
Hematopoietic potential of avian embryonic bone marrow
To examine the molecular regulation of HPC diversification, we first optimized conditions for eliciting a full range of blood cell phenotypes from embryonic avian bone marrow. Hematopoietic cell progenitors were obtained from the bone marrow by separation of cell suspensions on Ficoll-Paque gradients and subsequent removal of adherent cells by overnight incubation on tissue-culture plastic (see “Materials and methods”). Staining of bone marrow cells before (Figure 1A) or after (Figure 1B) Ficoll-Paque purification with May-Grünwald-Giemsa (MGG) solution verified that these purification procedures removed most differentiated cells and left primarily nondifferentiated cells having a blast-like cell morphology (Figure 1B).
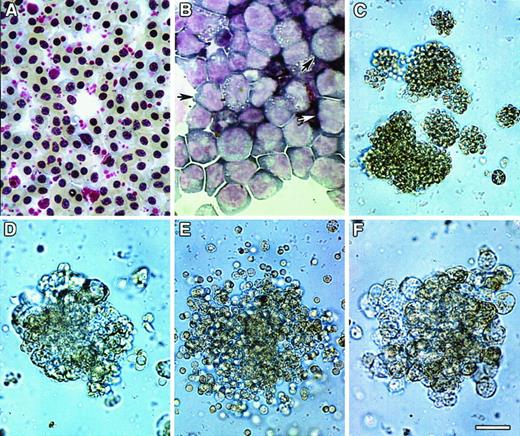
Colony formation of bone marrow cultures.
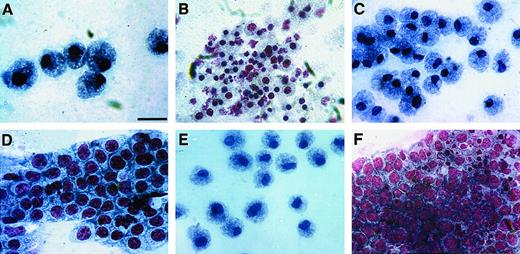
HPCs were obtained from ED14 quail embryonic bone marrow by separation on Ficoll-Paque density gradients and subsequent removal of cells adherent to tissue-culture plastic. The enrichment of progenitor cells by these procedures was confirmed by staining of bone marrow cellular isolates with May-Grünwald-Giemsa (MGG) solution either before (A) or following (B) Ficoll-Paque separation. Note that prior to separation, fully differentiated RBCs were the predominant cell type obtained from the bone marrow (A), whereas the Ficoll-Paque gradients allowed for the isolation of cellular populations that were primarily nondifferentiated blasts (arrows) (B). (C-F) HPC-enriched bone marrow cells were incubated in MeC cultures as described in “Materials and methods.” Cultures were incubated for 6 days prior to microscopic imaging. Representative examples of the types of colonies that formed in these cultures are erythroid (C), granulocyte/macrophage (D), granulocyte/erythroid/macrophage (E), and macrophage (F). Scale bar: A, 15 μm; B, 17 μm; C, 40 μm; D, F, 36 μm; E, 48 μm.
Colony formation of bone marrow cultures.
HPCs were obtained from ED14 quail embryonic bone marrow by separation on Ficoll-Paque density gradients and subsequent removal of cells adherent to tissue-culture plastic. The enrichment of progenitor cells by these procedures was confirmed by staining of bone marrow cellular isolates with May-Grünwald-Giemsa (MGG) solution either before (A) or following (B) Ficoll-Paque separation. Note that prior to separation, fully differentiated RBCs were the predominant cell type obtained from the bone marrow (A), whereas the Ficoll-Paque gradients allowed for the isolation of cellular populations that were primarily nondifferentiated blasts (arrows) (B). (C-F) HPC-enriched bone marrow cells were incubated in MeC cultures as described in “Materials and methods.” Cultures were incubated for 6 days prior to microscopic imaging. Representative examples of the types of colonies that formed in these cultures are erythroid (C), granulocyte/macrophage (D), granulocyte/erythroid/macrophage (E), and macrophage (F). Scale bar: A, 15 μm; B, 17 μm; C, 40 μm; D, F, 36 μm; E, 48 μm.
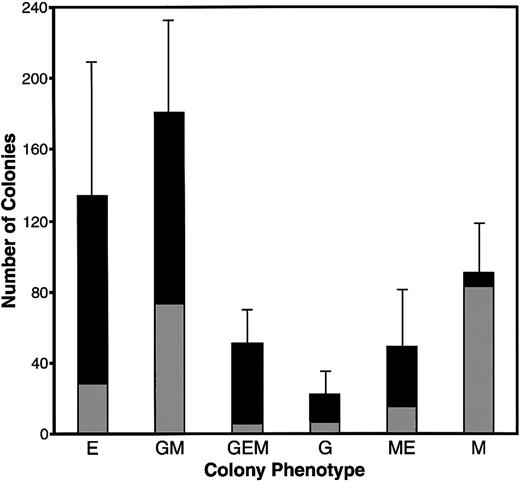
Extensive experimentation led to the development of culture conditions for promoting complete hematopoiesis in both MeC suspension cultures and fibrin (FB) gels (or plasma clot gels), the latter being the most common method used for avian hematopoiesis.26,30-32Similar to our previous experience with culturing avian mesodermal cells,22 we found that a mixture of chicken and bovine sera, in addition to cytokines, provided optimal culture conditions for supporting cell growth and differentiation. Under these conditions, in both MeC and FB gels, 6 morphologically distinct colonies were observed: erythroid (E) (Figure 1C), granulocyte/macrophage (GM) (Figure 1D), granulocyte/erythroid/macrophage (GEM) (Figure 1E), granulocyte (G), macrophage/erythroid (ME) and macrophage (M) (Figure1F). Quantitation of the MeC cultures (Figure2) showed that the most frequently occurring colony types were erythroid and granulocyte/macrophage, which yielded 134 and 181 total colonies per 5 × 104 bone marrow cells. Macrophage colonies also occurred at a high rate; however, most of the macrophage-only clusters were of smaller (20 to 50 cell) size. The bone marrow culture data shown in this report used ED14 quail embryos as the tissue source. Additionally, these culture conditions were replicated for analogously staged ED16 chick bone marrow, which produced identical results.
Colony formation of quail bone marrow cells in MeC.
HPC-enriched bone marrow from ED14 quail embryos was incubated in MeC for 6 days, prior to colony counting. The numbers of colonies per 5 × 104 seeded HPCs per culture are indicated on the y-axis. These colonies numbers were tabulated according to both cell phenotype and size, with gray and black bars denoting colonies of 20 to 50 cells and more than 50 cells, respectively. The results are presented as mean values for 5 separate experiments, with the error bars corresponding to the SD of total numbers for each colony category. The categories indicated on the x-axis are for colonies of erythroid (E), granulocyte/macrophage (GM), granulocyte/erythroid/macrophage (GEM), granulocyte (G), macrophage/erythroid (ME), and macrophage (M) lineage cells.
Colony formation of quail bone marrow cells in MeC.
HPC-enriched bone marrow from ED14 quail embryos was incubated in MeC for 6 days, prior to colony counting. The numbers of colonies per 5 × 104 seeded HPCs per culture are indicated on the y-axis. These colonies numbers were tabulated according to both cell phenotype and size, with gray and black bars denoting colonies of 20 to 50 cells and more than 50 cells, respectively. The results are presented as mean values for 5 separate experiments, with the error bars corresponding to the SD of total numbers for each colony category. The categories indicated on the x-axis are for colonies of erythroid (E), granulocyte/macrophage (GM), granulocyte/erythroid/macrophage (GEM), granulocyte (G), macrophage/erythroid (ME), and macrophage (M) lineage cells.
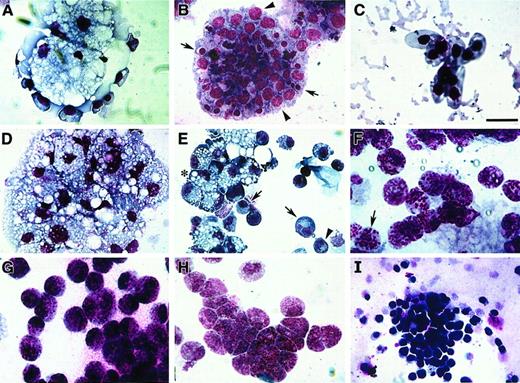
Avian bone marrow cells cultured in either MeC or FB gels displayed maximal colony growth and cell differentiation by day 6 of incubation. At this time, cell phenotype was analyzed further by harvesting the cultures and staining them with MGG solution. Cytological examination indicated that bone marrow–derived cells were able to produce colonies containing RBCs, granulocytes, monocytes, macrophages and thrombocytes—in both MeC and FB gels (Figure3). As with human hematopoiesis,33 developing erythroblasts were found often in close association with macrophages (Figure 3A)—although pure colonies of erythroblasts (Figure 3B) and mature RBCs were also highly prevalent. Macrophages were typified by their highly vacuolated cell phenotype and dark blue cytoplasm (Figure 3A,D,E). Also contained within macrophage-positive colonies were cells exhibiting a monocyte or promonocyte phenotype, which are characterized as macrophage-lineage cells.34 These premacrophage cells, which we will collectively refer to as monocytes, display a blue-gray cytoplasm with or without azurophilic granules (Figure 3E). Granular leukocytes in these cultures consisted of heterophil myelocytes with characteristic magenta granules and rings (Figure 3F), basophil myelocytes with abundant dark blue and magenta granules (Figure 3G), and eosinophil myelocytes with smaller, evenly sized dark pink granules (Figure 3H). The smallest cell, the thrombocyte, was easily detected by its large, dark purple nucleus and clear gray cytoplasm (Figure 3I). The relative occurrence of these various blood cell types within MeC and FB cultures appeared to be similar.
Blood cell phenotypes exhibited by avian bone marrow.
Bone marrow–derived HPCs were cultured for 6 days in either MeC (A, C, D, E, I) or FB (B, F-H) gels and subsequently stained with MGG solution. Representative cell types that developed from the bone marrow cultures were as follows: clustered vacuolated macrophages surrounded by erythroblasts (A), colony-containing erythroblasts (arrowheads) and developing erythrocytes (arrows) (B), mature RBCs (C), aggregated macrophages (D), mixed areas containing macrophages (asterisk), monocytes (arrows) and heterophils (arrowhead) (E), heterophils (note granules; arrow) (F), basophils (G), eosinophils (H), and thrombocytes (I). All the cell phenotypes shown were equally prevalent in both MeC and FB cultures. Scale bar: A, B, 13 μm; C, H, 8 μm; D, G, I, 14 μm; E, 20 μm; F, 10 μm.
Blood cell phenotypes exhibited by avian bone marrow.
Bone marrow–derived HPCs were cultured for 6 days in either MeC (A, C, D, E, I) or FB (B, F-H) gels and subsequently stained with MGG solution. Representative cell types that developed from the bone marrow cultures were as follows: clustered vacuolated macrophages surrounded by erythroblasts (A), colony-containing erythroblasts (arrowheads) and developing erythrocytes (arrows) (B), mature RBCs (C), aggregated macrophages (D), mixed areas containing macrophages (asterisk), monocytes (arrows) and heterophils (arrowhead) (E), heterophils (note granules; arrow) (F), basophils (G), eosinophils (H), and thrombocytes (I). All the cell phenotypes shown were equally prevalent in both MeC and FB cultures. Scale bar: A, B, 13 μm; C, H, 8 μm; D, G, I, 14 μm; E, 20 μm; F, 10 μm.
Hematopoietic potential of QCE6 cells
Having ascertained optimal protocols for embryonic avian bone marrow hematopoiesis, we next examined the diversification of the mesodermal stem cell line QCE6 under the same conditions. As reported previously,22 QCE6 cells have the capacity to differentiate to RBCs. To investigate their full hematopoietic potential, we plated QCE6 cells in conditions devised for bone marrow–derived HPCs. As was observed with avian embryonic HPCs (Figure3), QCE6 cells cultured in either MeC or FB gels gave rise to a broad range of blood cell phenotypes (Figure4). Specifically, as indicated by the pattern of MGG staining, QCE6 cells differentiated into RBCs, granulocytes, monocytes, macrophages, and thrombocytes (Figure4).
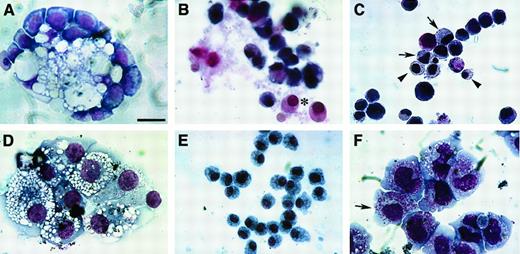
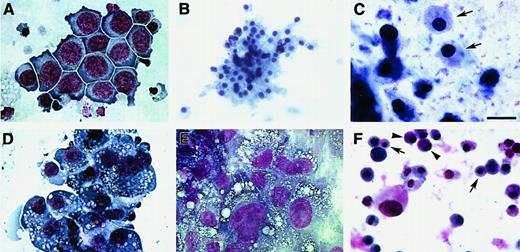
Blood cell phenotypes exhibited by QCE6 cells.
QCE6 cells were cultured for 6 days in either MeC (A, D, F) or FB (B, C, E) gels and subsequently stained with MGG solution. Representative cell types that developed from the QCE6 cultures were as follows: clustered vacuolated macrophages surrounded by erythroblasts (A), RBCs (asterisk) (B), mixed group of RBCs (arrowheads) and monocytes (arrows) (C), aggregated macrophages (D), monocytes (E), and early developing heterophils (note granules) (arrow) (F). All the cell phenotypes shown were equally prevalent in both MeC and FB cultures. Scale bar: A, 10 μm; B, F, 11 μm; C, E, 13 μm; D, 12 μm.
Blood cell phenotypes exhibited by QCE6 cells.
QCE6 cells were cultured for 6 days in either MeC (A, D, F) or FB (B, C, E) gels and subsequently stained with MGG solution. Representative cell types that developed from the QCE6 cultures were as follows: clustered vacuolated macrophages surrounded by erythroblasts (A), RBCs (asterisk) (B), mixed group of RBCs (arrowheads) and monocytes (arrows) (C), aggregated macrophages (D), monocytes (E), and early developing heterophils (note granules) (arrow) (F). All the cell phenotypes shown were equally prevalent in both MeC and FB cultures. Scale bar: A, 10 μm; B, F, 11 μm; C, E, 13 μm; D, 12 μm.
As an additional means to compare the cell phenotypes that developed from bone marrow and QCE6 cells, we examined MeC and FB gel cultures for the distribution of specific enzyme activities associated with leukocytes: AP, TARP, naphthol AS-D CAE, and ANAE. The results we obtained with avian bone marrow cells were consistent with those reported by other investigators.35 All leukocytes that developed from avian bone marrow exhibited positive AP activity (Figure5A). Addition of tartaric acid limited this AP activity (ie, TARP) to macrophage subpopulations. Both CAE and ANAE activity were detected in granulocytes, macrophages, and monocytes. Similarly to mouse and human, the ANAE enzyme activity was fluoride sensitive in monocytes8 and therefore was greatly reduced in these cells following NaF treatment (Figure 5B). These patterns of enzyme activity appeared to be similarly exhibited by QCE6 cells. For example, all leukocytes that developed from QCE6 cells exhibited AP activity (Figure 5C), with TARP activity restricted to macrophages (Figure 5D). Also, as with bone marrow–derived cells, QCE6 cells exhibiting granulocyte, macrophage, or monocyte phenotypes were both CAE-positive (Figure 5E) and ANAE-positive. Again, as observed with bone marrow–derived cells, monocytes were the only QCE6-derived cell type whose ANAE activity was inhibited by treatment with NaF (Figure 5F). Together, these data indicate that QCE6 cells will exhibit a differentiation potential similar to bone marrow HPCs when placed in a culture environment that promotes hematopoiesis.
Enzyme histochemistry of avian bone marrow and QCE6 cells.
MeC cultures of either quail bone marrow HPCs (A-B) or QCE6 cells (C-F) were cultured for 6 days, cytospun onto microscope slides, and assayed for leukocyte-associated enzyme activities. Representative staining for quail bone marrow cultures is shown for both AP (A) and NaF-resistant ANAE (B) activities. (A) All bone marrow–derived leukocytes were AP positive (reddish brown stain). (B) NaF pretreatment of bone marrow cultures distinguished macrophages, which remained ANAE positive (arrow), and the now ANAE-negative monocytes (arrowhead). (C) Similarly to bone marrow cells, all leukocytes that developed from QCE6 cells exhibited positive AP activity (reddish brown stain). (D) High levels of TARP activity were limited to QCE6-derived macrophage subpopulations (arrows). In contrast, tartaric acid treatment significantly reduced staining of monocytes (asterisk). (E) Naphthol AS-D CAE activity (brown stain) was exhibited by leukocytes of QCE6-cell origin. (F) As was shown in panel B with bone marrow cells, NaF pretreatment of QCE6 cells selectively inhibited ANAE activity to allow macrophages (brown stain; arrows) to be distinguished from unstained monocytes (arrowheads). For all these enzyme activities, the patterns of enzyme distribution were identical within cultures derived from either bone marrow or QCE6 cells. Scale bar: A, C, F, 10 μm; B, 12 μm; D, 5 μm; E, 11 μm.
Enzyme histochemistry of avian bone marrow and QCE6 cells.
MeC cultures of either quail bone marrow HPCs (A-B) or QCE6 cells (C-F) were cultured for 6 days, cytospun onto microscope slides, and assayed for leukocyte-associated enzyme activities. Representative staining for quail bone marrow cultures is shown for both AP (A) and NaF-resistant ANAE (B) activities. (A) All bone marrow–derived leukocytes were AP positive (reddish brown stain). (B) NaF pretreatment of bone marrow cultures distinguished macrophages, which remained ANAE positive (arrow), and the now ANAE-negative monocytes (arrowhead). (C) Similarly to bone marrow cells, all leukocytes that developed from QCE6 cells exhibited positive AP activity (reddish brown stain). (D) High levels of TARP activity were limited to QCE6-derived macrophage subpopulations (arrows). In contrast, tartaric acid treatment significantly reduced staining of monocytes (asterisk). (E) Naphthol AS-D CAE activity (brown stain) was exhibited by leukocytes of QCE6-cell origin. (F) As was shown in panel B with bone marrow cells, NaF pretreatment of QCE6 cells selectively inhibited ANAE activity to allow macrophages (brown stain; arrows) to be distinguished from unstained monocytes (arrowheads). For all these enzyme activities, the patterns of enzyme distribution were identical within cultures derived from either bone marrow or QCE6 cells. Scale bar: A, C, F, 10 μm; B, 12 μm; D, 5 μm; E, 11 μm.
WNT11 expression alters hematopoietic diversification of QCE6 cells
The findings on the hematopoietic potential of QCE6 cells suggest that they may have utility as a cell line model for examining the regulated diversification of blood cell progenitors. In prior studies,18,19 we examined the function of WNT-secreted signaling proteins in diverting mesodermal progenitors to cardiac phenotypes. Since blood cell progenitors have been shown to respond to WNT signals,16,17 we investigated whether WNTs may similarly influence hematopoietic cell diversification. As an initial inquiry into this possibility, we examined the blood cell potential of QCE6 cells as a function of WNT expression. QCE6 cells express WNT11 messenger RNA and protein, a characteristic they share with a subset of nondifferentiated mesodermal cells in the early gastrula.18 Previously, we described the generation of QCE6 subclones with altered levels of WNT11 expression.18This was accomplished by stably transfecting QCE6 cells with transgenes that expressed either WNT11 sense or antisense RNA, yielding variant subclones that produce either constitutively higher or diminished levels of WNT11 protein, respectively. For the present study, we have used an individual WNT11 sense QCE6 stable transfectant cell line, referred to as WNT11ox/3, and 2 individually derived WNT11 antisense sublines of QCE6 cells: WNT11αs/4 and WNT11αs/6. The relative levels of WNT11 produced by these cell lines were assayed by immunoblot analyses (Figure 6).
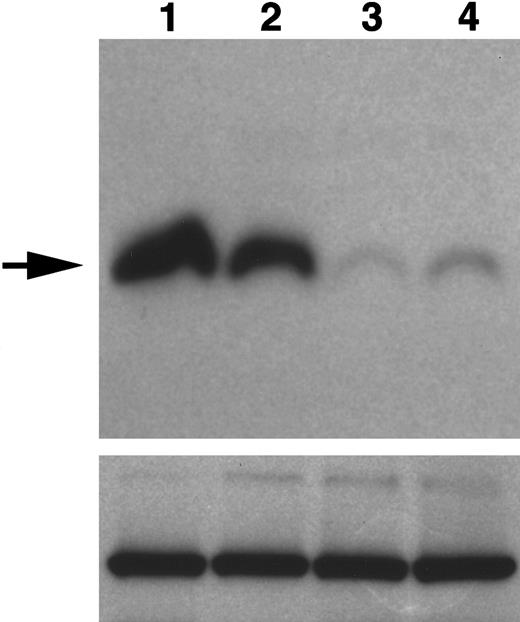
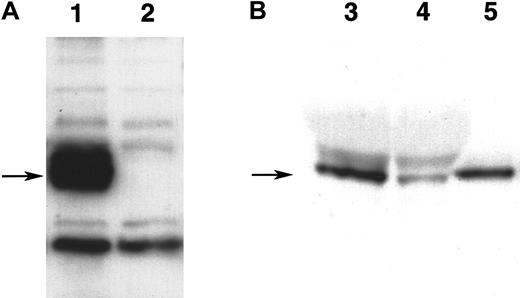
Expression of WNT11 protein by QCE6 cells and WNT11 stable transfectants.
Each lane contains protein isolated from cultures of WNT11ox/3 (lane 1), QCE6 (lane 2), WNT11αs/6 (lane 3), or WNT11αs/4 cells (lane 4). Confluent cultures of each cell line were treated overnight with 50 μg/mL heparin (which solubilizes WNT protein bound to cell surfaces and matrix). After culture fluids were collected, total cell protein was isolated following cell lysis in hypotonic buffer. The top panel shows cell supernatants blotted for WNT11, as indicated by an individual band of approximately 48 kd (arrow). The greatest amount of WNT11 protein was exhibited by WNT11ox/3 cells (lane 1), with QCE6 cells showing more moderate levels of this protein (lane 2). In comparison, both WNT11αs/6 (lane 3) and WNT11αs/4 cells (lane 4) produced very low amounts of WNT11. The bottom panel shows total cell protein (5 μg/lane) blotted for pyruvate carboxylase to ensure that the volumes of culture supernatants added to the WNT11 gel were representative of similar cell numbers.
Expression of WNT11 protein by QCE6 cells and WNT11 stable transfectants.
Each lane contains protein isolated from cultures of WNT11ox/3 (lane 1), QCE6 (lane 2), WNT11αs/6 (lane 3), or WNT11αs/4 cells (lane 4). Confluent cultures of each cell line were treated overnight with 50 μg/mL heparin (which solubilizes WNT protein bound to cell surfaces and matrix). After culture fluids were collected, total cell protein was isolated following cell lysis in hypotonic buffer. The top panel shows cell supernatants blotted for WNT11, as indicated by an individual band of approximately 48 kd (arrow). The greatest amount of WNT11 protein was exhibited by WNT11ox/3 cells (lane 1), with QCE6 cells showing more moderate levels of this protein (lane 2). In comparison, both WNT11αs/6 (lane 3) and WNT11αs/4 cells (lane 4) produced very low amounts of WNT11. The bottom panel shows total cell protein (5 μg/lane) blotted for pyruvate carboxylase to ensure that the volumes of culture supernatants added to the WNT11 gel were representative of similar cell numbers.
In parallel with the parental QCE6 cells, these various QCE6 stably transfected sublines were cultured in conditions that support hematopoiesis (Figure 7). In common with nontransfected QCE6 cells, WNTox/3 cells plated in MeC or FB gels produced a broad range of blood cell phenotypes, which included RBCs, granulocytes, monocytes, and thrombocytes. However, WNT11ox/3 cells displayed a shift toward the erythrocyte lineage, with an increase in both the frequency of erythrocyte-containing colonies (Figure 7) and the proportion of erythroblasts (Figure8A) and mature RBCs (Figure 8B-C) within these colonies. Another feature of WNT11ox/3 hematopoiesis was a dramatic decrease in macrophage formation, with a corresponding increase in the numbers of cells exhibiting a monocyte or promonocyte morphology (Figure 7). In contrast, highly vacuolated macrophages dominated the MeC and FB gel cultures of WNT11αs/4 cells (Figure8D-E). Cells exhibiting a monocyte morphology were much less frequent, as compared with the parental QCE6 or WNT11ox cells. Most striking was the complete absence of erythroblasts and RBCs in cultures of WNT11αs/4 cells (Figure 7). Likewise, WNT11αs/6 cells also demonstrated a phenotypic shift from RBCs/monocytes to macrophages.
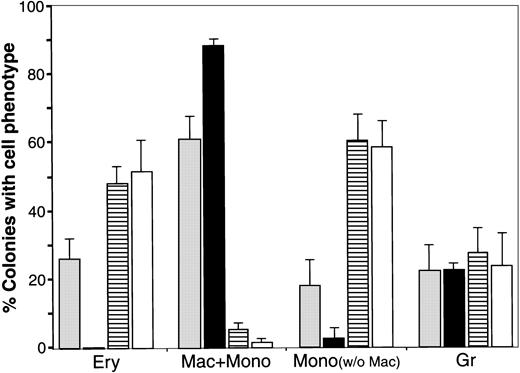
Effect of WNT expression on the distribution of blood cell phenotypes exhibited by QCE6 cells.
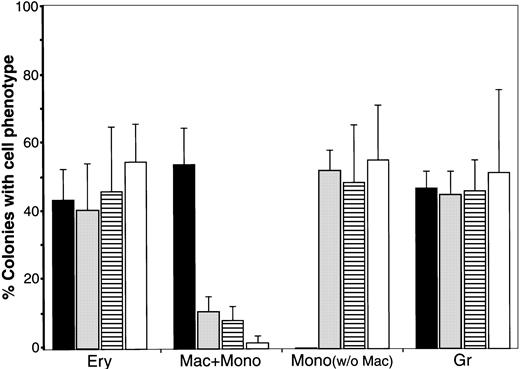
The level of WNT expression alters the distribution of blood cell phenotypes exhibited by QCE6 cells. Parental QCE6 cells and stably transfected QCE6 sublines were incubated in MeC (5 × 104cells per culture) for 6 days prior to colony counting. Colonies were scored as described in the legend to Figure 2, except that an additional colony category was tallied. For the bone marrow cultures reported in Figure 2, monocyte phenotypes were detected only in cultures that contained macrophages. Here, colonies that contained monocytes but no macrophages were observed. After tabulation of the different blood colony types for each culture, mixed and individual lineage colonies were added to determine the proportion of colonies that contained a given blood cell phenotype. The results are presented as the mean values per culture, with the error bars corresponding to the SD. Total numbers of MeC colonies counted from duplicate cultures in 2 independent experiments were 344 colonies for the parental QCE6 (░), 271 colonies for WNT11αs (▪), 405 colonies for WNT11ox/3 (▤), and 401 colonies for WNT5a-transfected QCE6 (WNT5a/QCE6, ■). The phenotypic groups indicated on the x-axis are colonies that contained RBCs (Ery), macrophages plus monocytes (Mac + Mono), monocytes without macrophages (Mono), and granulocytes (Gr). Note that WNT11αs cells did not yield any erythroid lineage cells, and both WNT11ox/3 and WNT5a/QCE6 cells displayed very few macrophages. It should also be noted that, although not indicated in this graph, WNT11αs Mac + Mono colonies consisted primarily of macrophages, and the WNT11ox/3 and WNT5a/QCE6 Mac + Mono colonies contained far higher numbers of monocytes than macrophages.
Effect of WNT expression on the distribution of blood cell phenotypes exhibited by QCE6 cells.
The level of WNT expression alters the distribution of blood cell phenotypes exhibited by QCE6 cells. Parental QCE6 cells and stably transfected QCE6 sublines were incubated in MeC (5 × 104cells per culture) for 6 days prior to colony counting. Colonies were scored as described in the legend to Figure 2, except that an additional colony category was tallied. For the bone marrow cultures reported in Figure 2, monocyte phenotypes were detected only in cultures that contained macrophages. Here, colonies that contained monocytes but no macrophages were observed. After tabulation of the different blood colony types for each culture, mixed and individual lineage colonies were added to determine the proportion of colonies that contained a given blood cell phenotype. The results are presented as the mean values per culture, with the error bars corresponding to the SD. Total numbers of MeC colonies counted from duplicate cultures in 2 independent experiments were 344 colonies for the parental QCE6 (░), 271 colonies for WNT11αs (▪), 405 colonies for WNT11ox/3 (▤), and 401 colonies for WNT5a-transfected QCE6 (WNT5a/QCE6, ■). The phenotypic groups indicated on the x-axis are colonies that contained RBCs (Ery), macrophages plus monocytes (Mac + Mono), monocytes without macrophages (Mono), and granulocytes (Gr). Note that WNT11αs cells did not yield any erythroid lineage cells, and both WNT11ox/3 and WNT5a/QCE6 cells displayed very few macrophages. It should also be noted that, although not indicated in this graph, WNT11αs Mac + Mono colonies consisted primarily of macrophages, and the WNT11ox/3 and WNT5a/QCE6 Mac + Mono colonies contained far higher numbers of monocytes than macrophages.
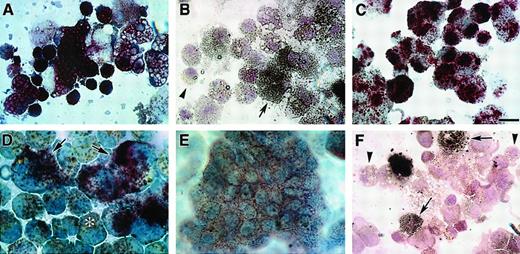
Blood cell phenotypes exhibited by QCE6 cells in response to altered levels of WNT protein.
WNT11ox/3 (A-C), WNT11αs/4 (D-E), or WNT5a/QCE6 cells (F) were cultured for 6 days in hematopoietic-promoting conditions prior to phenotypic analysis with MGG stain. Panels A and D show cells grown in MeC, and panels B, C, E, and F display cells cultured in FB gels. The prevalent cell phenotypes exhibited by WNT11ox/3 cells were erythroid or monocyte phenotypes, although granulocytes, thrombocytes, and a few macrophages were also observed. An erythroblast cluster (A), an RBC cluster (B), and a high magnification view of RBCs (arrows) (C), which developed from WNT11ox/3 cells. (D-E) In contrast, macrophages were the predominant cell type derived from WNT11αs/4 cells. Monocytes were very rare and RBCs were never observed in cultures of WNT11αs/4 cells. (F) Similarly to WNT11ox/3 cells, stably transfected WNT5a/QCE6 cells contained primarily RBCs (arrows) and monocytes (arrowheads). Scale bar: A, 10 μm; B, 25 μm; C, 6.5 μm; D-F, 12 μm.
Blood cell phenotypes exhibited by QCE6 cells in response to altered levels of WNT protein.
WNT11ox/3 (A-C), WNT11αs/4 (D-E), or WNT5a/QCE6 cells (F) were cultured for 6 days in hematopoietic-promoting conditions prior to phenotypic analysis with MGG stain. Panels A and D show cells grown in MeC, and panels B, C, E, and F display cells cultured in FB gels. The prevalent cell phenotypes exhibited by WNT11ox/3 cells were erythroid or monocyte phenotypes, although granulocytes, thrombocytes, and a few macrophages were also observed. An erythroblast cluster (A), an RBC cluster (B), and a high magnification view of RBCs (arrows) (C), which developed from WNT11ox/3 cells. (D-E) In contrast, macrophages were the predominant cell type derived from WNT11αs/4 cells. Monocytes were very rare and RBCs were never observed in cultures of WNT11αs/4 cells. (F) Similarly to WNT11ox/3 cells, stably transfected WNT5a/QCE6 cells contained primarily RBCs (arrows) and monocytes (arrowheads). Scale bar: A, 10 μm; B, 25 μm; C, 6.5 μm; D-F, 12 μm.
As a further test on the ability of WNTs to control blood cell diversification, we stably introduced into QCE6 cells a transgene encoding for WNT5a. We chose WNT5a for these studies since its functional properties closely resemble those of WNT11,36,37 and it is expressed within both embryonic and adult hematopoietic tissue.16 17 To facilitate detection of WNT5a protein by QCE6 stably transfected sublines, this molecule was expressed as a fusion protein with a myc-6xHis tag at the carboxy end. Accordingly, expression of WNT5a protein was demonstrated by immunoblotting for the myc epitope tag (Figure9A). As was observed for the overexpression of WNT11 (Figure 8A-C), cells that expressed WNT5a showed a phenotypic shift toward the RBC lineage (Figure 8F), with macrophage formation occurring very infrequently (Figure 7).
Detection of WNT5a protein.
(A) Expression of WNT5a protein in stably transfected QCE6 cells. Both lanes contain 25 μg of protein from culture lysates of either stably transfected WNT5a/QCE6 (lane 1) or parental QCE6 cells (lane 2), which were subsequently blotted with antibody reactive to themyc-fusion tag. Arrow indicates the position of an approximately 50-kd band that is present only in the stably transfected cells, which corresponds to the WNT5a fusion protein. (B) Partial purification of WNT5a protein from WNT5a-conditioned media. This shows equal volumes of either nonpurified WNT5a CM (lane 3), the effluent fraction of WNT5a CM run over an Ni-NTA column (lane 4), or the nickel-binding fraction eluted from the same column (lane 5). An arrow indicates the position of an approximately 50-kd WNT5a band that specifically bound to the Ni-NTA column owing to the multiple histidine residues of the fusion protein tag. The protein concentrations of WNT5a CM and Ni-NTA column–purified WNT5a–containing fraction were 14 μg/μL and 52 ng/μL, respectively.
Detection of WNT5a protein.
(A) Expression of WNT5a protein in stably transfected QCE6 cells. Both lanes contain 25 μg of protein from culture lysates of either stably transfected WNT5a/QCE6 (lane 1) or parental QCE6 cells (lane 2), which were subsequently blotted with antibody reactive to themyc-fusion tag. Arrow indicates the position of an approximately 50-kd band that is present only in the stably transfected cells, which corresponds to the WNT5a fusion protein. (B) Partial purification of WNT5a protein from WNT5a-conditioned media. This shows equal volumes of either nonpurified WNT5a CM (lane 3), the effluent fraction of WNT5a CM run over an Ni-NTA column (lane 4), or the nickel-binding fraction eluted from the same column (lane 5). An arrow indicates the position of an approximately 50-kd WNT5a band that specifically bound to the Ni-NTA column owing to the multiple histidine residues of the fusion protein tag. The protein concentrations of WNT5a CM and Ni-NTA column–purified WNT5a–containing fraction were 14 μg/μL and 52 ng/μL, respectively.
Exposure to WNT protein shifts the blood cell phenotypes displayed by WNT11αs cells
QCE6 cells exhibit a hematopoietic potential that is characteristic of bone marrow HPCs. In contrast, WNT11αs cells exhibit a shift in cell fate toward macrophage formation. To verify that the inhibition of WNT11 production is principally responsible for the cell demographic profile exhibited by WNT11αs cells, we investigated the response of these cells to exogenously added WNT11 or WNT5a. In a previous study, we demonstrated that conditioned media from WNT11ox cells contains functionally active WNT11 protein.19 Similarly, WNT5a/QCE6 cells produce high amounts of soluble secreted WNT5a protein (Figure 9B). Thus, WNT11αs/4 cells were plated in FB gels in the absence or presence of either WNT11ox- or WNT5a/QCE6-conditioned media (WNT11 CM or WNT5a CM). In contrast to the outcome in the absence of WNT-containing media (Figure 7), WNT11αs cells treated with either WNT11 CM or WNT5a CM generated significant numbers of RBCs. Also, the proportion of monocytes was increased in response to these treatments. To verify that this phenotypic shift was in fact due to WNT protein, we purified WNT5a from conditioned media. Taking advantage of the carboxy-terminal 6xHis fusion tag, we separated WNT5a protein from contaminating proteins on the basis of its affinity for Ni+ (Figure 9B). As observed with the conditioned media treatments, purified WNT5a protein provoked WNT11αs cells to give rise predominantly to monocytes (Figure10A) and RBCs (Figure 10B).
Effect of WNT11 and WNT5a on expression of blood cell phenotypes.
WNT11 and WNT5a inhibit macrophage formation and promote RBC and monocyte differentiation. WNT11αs/4 cells cultured in the presence of purified WNT5a protein gave rise to predominantly monocytes (A) and RBCs (B). Few macrophages were observed in these cultures. Identical results were obtained when WNT11αs/4 cells were treated with WNT11 CM or WNT5a CM. These results are in complete contrast to the phenotypic profile of WNT11αs/4 cells cultured in the absence of WNT, where macrophages were the predominant cell type, monocytes were infrequently observed, and RBCs were totally absent (see Figure 7, 8). (C-F) Quail bone marrow HPCs also showed increased prevalence of monocytes and RBCs in response to these treatments. Shown here are bone marrow cultures treated with either WNT11 CM (C-D) or purified WNT5a protein (E-F) that produced monocytes (C-E), erythroblasts (D), and developing RBCs (F). Macrophages were infrequently observed in bone marrow cultures in response to either WNT11 or WNT5a. Scale bar: A, D, 10 μm; B-C, 12 μm; E, 13 μm; F, 14 μm.
Effect of WNT11 and WNT5a on expression of blood cell phenotypes.
WNT11 and WNT5a inhibit macrophage formation and promote RBC and monocyte differentiation. WNT11αs/4 cells cultured in the presence of purified WNT5a protein gave rise to predominantly monocytes (A) and RBCs (B). Few macrophages were observed in these cultures. Identical results were obtained when WNT11αs/4 cells were treated with WNT11 CM or WNT5a CM. These results are in complete contrast to the phenotypic profile of WNT11αs/4 cells cultured in the absence of WNT, where macrophages were the predominant cell type, monocytes were infrequently observed, and RBCs were totally absent (see Figure 7, 8). (C-F) Quail bone marrow HPCs also showed increased prevalence of monocytes and RBCs in response to these treatments. Shown here are bone marrow cultures treated with either WNT11 CM (C-D) or purified WNT5a protein (E-F) that produced monocytes (C-E), erythroblasts (D), and developing RBCs (F). Macrophages were infrequently observed in bone marrow cultures in response to either WNT11 or WNT5a. Scale bar: A, D, 10 μm; B-C, 12 μm; E, 13 μm; F, 14 μm.
Exposure to WNT protein shifts the blood cell phenotypes displayed by bone marrow–derived HPCs
The above results demonstrated that WNTs are able to regulate the diversification of blood phenotypes exhibited by QCE6 cells. To extend these findings, we went back to studying blood cell progenitors obtained from avian bone marrow. Quail HPCs were plated in FB gels, in the presence or absence of conditioned media containing WNT11 or WNT5a. WNT11 CM (Figure 10C-D) and WNT5a CM treatment yielded similar outcomes, with cultures containing monocytes (Figure 10C), RBCs (Figure10D), granulocytes, and thrombocytes. More importantly, in comparison with the control cultures, macrophage formation was markedly reduced. Instead, the conditioned media greatly increased the occurrence of monocytes (and to a lesser extent RBCs) as compared with cells cultured in the absence of ectopic WNT protein (Figure11). When we repeated these experiments with quail bone marrow cells using purified WNT5a protein, the same result was produced: greater numbers of monocytes (Figure 10E) and RBCs (Figure 10F), with macrophages appearing very infrequently in these cultures (Figure 11).
Effect of WNT protein on the distribution of blood cell phenotypes in quail bone marrow cells.
Exposure to WNT protein alters the distribution of blood cell phenotypes displayed by quail bone marrow cells. HPC-enriched bone marrow from ED14 quail embryos was incubated in MeC for 6 days in the absence (▪) or presence of either WNT11 CM (░), WNT5a CM (▤), or purified WNT5a protein (■). Colonies were scored and tabulated as described in the legend to Figure 7. After tabulation of the different blood colony types for each culture, mixed and individual lineage colonies were added to determine the proportion of colonies that contained a given blood cell phenotype. The results are presented as the mean values per culture, with the error bars corresponding to the SD. Total number of MeC colonies counted from multiple experiments were 2875 total colonies for the control, 301 colonies for WNT11 CM–treated, 374 colonies WNT5a CM–treated, and 214 colonies WNT5a protein–treated HPC cultures. The phenotypic groups indicated on the x-axis are for colonies that contained RBCs (Ery), macrophages plus monocytes (Mac + Mono), monocytes without macrophages (Mono), and granulocytes (Gr). Note that the control cultures never displayed colonies that contained monocytes without macrophages. Yet treatment with the WNT preparations provoked large numbers of monocyte-positive, macrophage-negative colonies. It should also be noted that, although not indicated in this graph, in response to the WNT treatments, the proportion of RBCs within mixed phenotypic colonies (ie, GEM and ME colonies) appeared to be enhanced.
Effect of WNT protein on the distribution of blood cell phenotypes in quail bone marrow cells.
Exposure to WNT protein alters the distribution of blood cell phenotypes displayed by quail bone marrow cells. HPC-enriched bone marrow from ED14 quail embryos was incubated in MeC for 6 days in the absence (▪) or presence of either WNT11 CM (░), WNT5a CM (▤), or purified WNT5a protein (■). Colonies were scored and tabulated as described in the legend to Figure 7. After tabulation of the different blood colony types for each culture, mixed and individual lineage colonies were added to determine the proportion of colonies that contained a given blood cell phenotype. The results are presented as the mean values per culture, with the error bars corresponding to the SD. Total number of MeC colonies counted from multiple experiments were 2875 total colonies for the control, 301 colonies for WNT11 CM–treated, 374 colonies WNT5a CM–treated, and 214 colonies WNT5a protein–treated HPC cultures. The phenotypic groups indicated on the x-axis are for colonies that contained RBCs (Ery), macrophages plus monocytes (Mac + Mono), monocytes without macrophages (Mono), and granulocytes (Gr). Note that the control cultures never displayed colonies that contained monocytes without macrophages. Yet treatment with the WNT preparations provoked large numbers of monocyte-positive, macrophage-negative colonies. It should also be noted that, although not indicated in this graph, in response to the WNT treatments, the proportion of RBCs within mixed phenotypic colonies (ie, GEM and ME colonies) appeared to be enhanced.
Discussion
Previous investigations have shown that WNTs may act as determiners of cell lineage,14,15 either by pushing nondifferentiated cells toward more specialized phenotypes,38,39 shifting cells between alternative cell fates,40 41 or both. Here, we report that WNT proteins can significantly influence the phenotypic distribution of blood cells—obtained from data with both bone marrow HPCs and a stem cell line that exhibits a broad hematopoietic cell potential.
QCE6 cells serve as a cell line model of hematopoiesis
The QCE6 cell line was derived from anterior mesodermal cells of an early gastrula stage quail embryo,20,21 analogous to Hamburger and Hamilton42 stage 4. Although QCE6 cells were initially characterized for their cardiac and endothelial cell potential, it was subsequently demonstrated that these cells can develop into RBCs.22 Intrigued by this observation, we began to examine whether QCE6 cells might have an even broader cell potential. To investigate the capability of this cell line to give rise to nonerythroid blood cells, we first optimized culture conditions for blood cell diversification of hematopoietic progenitor cells. The HPC source used for these experiments was bone marrow from late-stage embryonic quail or chick embryos. Under optimized conditions, bone marrow–derived cells were able to develop into RBCs, granulocytes, monocytes, macrophages, and thrombocytes. When QCE6 cells were then cultured under the identical conditions, they also gave rise to the same broad range of blood cell types. The phenotypes of these cells were verified by cytological examination with May-Grünwald-Giemsa stain and the pattern of leukocyte-associated enzyme reactivity. Side-by-side comparison with bone marrow–derived cells clearly indicated that the cell types generated from QCE6 cells are differentiated RBCs, granulocytes, monocytes, macrophages, and thrombocytes.
The differentiation of QCE6 cells into multiple hematopoietic cell types raises the issue of why these cells display this phenotypic potential. Although QCE6 cells were obtained from a mesodermal area that does not normally give rise to blood cells, other studies have shown that this region is capable of producing RBCs when cultured in the presence of yolk sac endoderm.43 44 The potential of early embryonic mesoderm to give rise to additional blood cell types has not been shown. Whether the multipotentiality of QCE6 cells, as evidenced by the data presented here, is totally reflective of the tissue from which it was derived is an issue for a later study. What is shown is that QCE6 cells will behave in a manner nearly identical to bone marrow HPCs when exposed to hematopoietic-promoting conditions. Thus, this cell line has utility as a tool to examine mechanisms that regulate hematopoiesis. This was dramatically illustrated when we examined the influence of WNT expression on the phenotypic distribution of blood cells, as information obtained from QCE6 cells appeared to be directly applicable for the regulated diversification of bone marrow HPCs.
WNTs influence the diversification of blood cell progenitors
QCE6 cells express WNT11—a property they share with a subset of anterior mesodermal cells from the early gastrula.18 Under culture conditions optimized for hematopoiesis of bone marrow HPCs, QCE6 cells will give rise to a similar broad range of differentiated blood cell types. However, alteration of WNT expression by these cells greatly influenced the formation of specific blood cell phenotypes by these cells. Inhibition of WNT11 production by QCE6 cells, via stable transfection of a WNT11 antisense transgene, produced cultures that were dominated by macrophages, produced few if any monocytes, and were devoid of RBCs. Accordingly, the formation of RBCs and monocytes by these WNT11αs cells was restored when the cells were exposed to ectopic WNT protein—either in the form of WNT11- or WNT5a-containing conditioned media or as purified WNT5a protein. Moreover, when WNT levels were increased in QCE6 cells by stable insertion of WNT11 or WNT5a transgenes, the prevalence of RBCs and monocytes was significantly increased, with the appearance of macrophages markedly reduced in these cultures. When bone marrow HPCs were exposed to WNT11 or WNT5a, macrophages appeared very infrequently within the cultures. Instead, monocytes were now the prevalent blood cell that arose from the bone marrow–derived cells, with increased numbers of RBCs also apparent.
The expression of WNT5a in the yolk sac and by hematopoietic progenitors in the bone marrow16,17 implicates this molecule as a candidate regulator of blood cell formation in situ. That WNT5a and WNT11 similarly influenced the distribution of blood cell phenotypes in our cultures is consistent with previous observations that these 2 proteins share functional activities.36,37Although WNTs are a highly conserved group of molecules, they do exhibit dramatic differences in their functional activities. Thus, it has been useful to subdivide WNTs into 2 broad classes—referred to as WNT1 and WNT5a classes—on the basis of their functional properties.14 It is therefore likely that any influence WNT1-class proteins (eg, WNT1, WNT3a, and WNT8) have on blood cell development would be very different than exhibited by WNT5a-class molecules (eg, WNT5a and WNT11). In this regard, it is interesting to compare the activities of WNT5a with WNT10b, a molecule that has also been shown to be expressed in hematopoietic tissue.16,17In the studies of Van Den Berg et al,17 CD34+cells from human bone marrow were cocultured with feeder cells that were infected with either WNT5a- or WNT10b-expressing virus. Although both WNTs increased the incidence of blood colony formation in MeC cultures, only WNT5a was able to stimulate increased burst-forming unit–erythrocytic numbers. Thus, there is precedence that WNT5a-class proteins uniquely stimulate RBC development.
Diversification of blood cells
In the culture conditions used for our experiments, both bone marrow HPCs and QCE6 cells gave rise to RBCs, granulocytes, monocytes, macrophages, and thrombocytes. When QCE6 cells were prevented from producing WNT11 (ie, WNT11αs cells), they no longer generated RBCs, and the appearance of monocytes was infrequent. Instead, cultures of WNT11αs cells were dominated by macrophages. Accordingly, WNT11αs cells behaved like normal QCE6 cells when ectopic WNT11 or WNT5a was added to the cultures. Overexpressing WNT protein in QCE6 cells, with either WNT11 or WNT5a transgenes, inhibited macrophage differentiation, but increased the prevalence of RBCs and monocytes. Exposure of bone marrow HPCs to either WNT11 or WNT5a similarly inhibited macrophage formation, as macrophages were infrequently observed in these latter cultures. In this instance, RBC numbers were moderately enhanced, with a much greater enhancement of monocytes produced from the bone marrow cells in response to the WNT signal. This relative enhancement by WNTs on RBC and monocyte development was the principal difference we observed in our results with bone marrow and QCE6 cells. This variance may reflect that QCE6 cells, being a cloned cell line, present a less complex cellular environment than does the relatively heterogeneous population of bone marrow HPCs. An additional consideration is that expression of WNT5a (or other WNTs) by quail bone marrow, as has been shown for human bone marrow,17 may moderate the relative enhancement of RBC formation by ectopically added WNT protein. Overall, the QCE6 cells behaved remarkably similar to the bone marrow cells in the MeC and FB gel cultures. Not only did this cell line generate the same range of blood cell phenotypes as did bone marrow–derived hematopoietic progenitors, but also that macrophage development was identically affected in response to altered WNT expression.
Together, the data derived from the bone marrow and QCE6 cells suggest that WNT5a or a similar signal (ie, WNT11) influences blood cell formation by shifting cell fate from a macrophage to an RBC phenotype. Interestingly, the same WNT signals also inhibit the differentiation of monocytes to macrophages. Thus, both WNT11 and WNT5a promote the expression of RBCs and monocytes at the expense of macrophages. These results raise the question of whether similar WNT responsiveness indicates linkage between 2 regulatory events that both have an impact on macrophage lineage. In other words, are RBCs and monocytes more closely linked than previously suspected?
Acknowledgment
We thank Randall Moon for his generosity in providing us with the Xenopus WNT5a cDNA.
Supported by National Institutes of Health grant HL55923 (C.A.E.), American Heart Association Grant 9808097U (C.A.E.), American Heart Association Grant-in-Aid 9950638N (L.M.E.), and South Carolina Research Initiative Grant (C.A.E., L.M.E.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Carol A. Eisenberg, Department of Cell Biology and Anatomy, Medical University of South Carolina, 173 Ashley Ave, Suite 652, PO Box 250508, Charleston, SC 29425; e-mail: eisenbec@musc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal