Abstract
Gene transfer in human hematopoietic stem cells (HSCs) has great potential for both gene therapy and the understanding of hematopoiesis. As HSCs have extensive proliferative capacities, stable gene transfer should include genomic integration of the transgene. Lentiviral vectors are now preferred to oncoretroviral vectors especially because they integrate in nondividing cells such as HSCs, thereby avoiding the use of prolonged cytokine stimulation. Human immunodeficiency virus type-1 (HIV-1) has evolved a complex reverse transcription strategy including a central strand displacement event controlled in cis by the central polypurine tract (cPPT) and the central termination sequence (CTS). This creates, at the center of HIV-1 linear DNA molecules, a 99-nucleotide-long plus-strand overlap, the DNA flap, which acts as a cis-determinant of HIV-1 genome nuclear import. The reinsertion of the DNA flap sequence in an HIV-derived lentiviral vector promotes a striking increase of gene transduction efficiency in human CD34+ hematopoietic cells, and the complementation of the nuclear import defect present in the parental vector accounts for this result. In a short ex vivo protocol, the flap-containing vector allows efficient transduction of the whole hierarchy of human HSCs including both slow-dividing or nondividing HSCs that have multiple lymphoid and myeloid potentials and primitive cells with long-term engraftment ability in nonobese diabetic/severe combined immunodeficiency mice (NOD/SCID).
Introduction
Human hematopoietic stem cells (HSCs) have the potential to regenerate the entire hematopoietic system, and as such, they are the target cells both for gene therapy and the understanding of mechanisms regulating normal and pathological hematopoiesis. As HSCs have extensive proliferative capacities, stable gene transfer should include genomic integration of the transgene. Retroviral vectors based on Moloney murine leukemia virus (MoMuLV) have been very popular because they systematically integrate their genome into the host cell chromatin. However, like the wild-type virus, MoMuLV-derived vectors do not integrate in nondividing cells including human HSCs that are almost all quiescent. Many prestimulation protocols using different cytokine combinations have been developed to trigger cell cycling of HSCs, and thus these protocols induce HSC susceptibility to transduction by the oncoretrovirus-derived, mitosis-dependent gene-transfer vectors. However, prolonged cytokine stimulation can induce differentiation together with proliferation, thereby leading to the loss of fundamental properties of HSCs during the transduction process. The use of prolonged cytokine stimulation to trigger cell cycling can be overcome by using lentivirus-derived vectors because lentiviruses have evolved a mitosis-independent nuclear import strategy that accounts for their unique capacity, among retroviruses, to replicate efficiently in nondividing target cells.2 3
The search for the viral determinants responsible for the active nuclear import of the human immunodeficiency virus type-1 (HIV-1) complementary DNA (cDNA) genome is an active but controversial field of investigation.4-12 HIV-1 has evolved a complex reverse transcription strategy including a central strand displacement event controlled in cis by the central polypurine tract (cPPT) and the central termination sequence (CTS).13-16 Reverse transcription results in a linear DNA molecule bearing in its center a stable 99-nucleotide-long plus-strand overlap, here referred to as the central DNA flap. We have previously established that the central DNA flap of HIV-1 is involved in a late step of HIV-1 genome nuclear import, immediately prior to or during viral DNA translocation through the nuclear pore.1 This result has important implications for the design of efficient lentiviral vectors because it is essential to maintain the lentiviral nuclear import determinants within vector constructs. Classical retroviral vector constructs are replacement vectors in which the entire viral coding sequences between the 2 long terminal repeats (LTRs) are replaced by the sequences of interest.17 In the case of lentiviral vectors, this leads to the deletion of the central cis-active cPPT and CTS sequences, which suggests that such replacement vector constructs are not optimal.
We show here that introduction of the DNA flap sequence in an HIV-derived lentiviral vector,18 the resulting vector being called TRIP vector, dramatically increased gene transduction efficiency in human CD34+ hematopoietic cells by strongly stimulating nuclear import of the vector pre-integration complex. Most importantly, we show that during a 24-hour ex vivo transduction protocol, an efficient and equivalent transduction of the whole hierarchy of human HSCs is indeed achieved. The transduced cells include slow and nondividing HSCs, which have multiple lymphoid and myeloid potential, and primitive cells, which are able to establish long-term human hematopoiesis in nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice.
Materials and methods
Collection and fractionation of cells
Adult bone marrow (BM), peripheral blood mobilized cell (PBMC), and cord blood (CB) samples were collected with the informed consent of the patients and mothers according to approved institutional guidelines. CD34+ and CD34+CD38− cells (representing 20% of total CD34+ cells) were purified as previously described.19 CD34+ cells were used immediately or after storage in liquid nitrogen.
Lentiviral vector supernatants
The TRIP vector plasmid was constructed as previously described.1 Briefly, a 178–base pair (bp) polymerase chain reaction (PCR) fragment encompassing cPPT and CTS was inserted in the unique ClaI site of HR, the HIV-1–derived vector described by Naldini et al,18 in which theLacZ reporter gene was replaced by the enhanced green fluorescent protein (EGFP) coding sequence (Clontech Laboratories, Heidelberg, Germany), as shown in Figure1A. Vector particles were produced by transient calcium-phosphate cotransfection of 293 T cells with the vector plasmid, an encapsidation plasmid lacking Vif-, Vpr-, Vpu-, and Nef-accessory HIV-1 proteins (p8.91),20,21 and a vesicular stomatitis virus (VSV) envelope-expression plasmid (pHCMV-G),22 as previously described.18Because some variation is observed in the efficiency of transduction between different batches of vector, all comparative experiments between HR and TRIP vectors were performed with stocks of vector particles produced in parallel. The concentration of vector particles was normalized by measuring the P24 (HIV-1 capsid protein) content of supernatants. Both vectors encode EGFP under the transcriptional control of an internal cytomegalovirus (CMV) promoter.
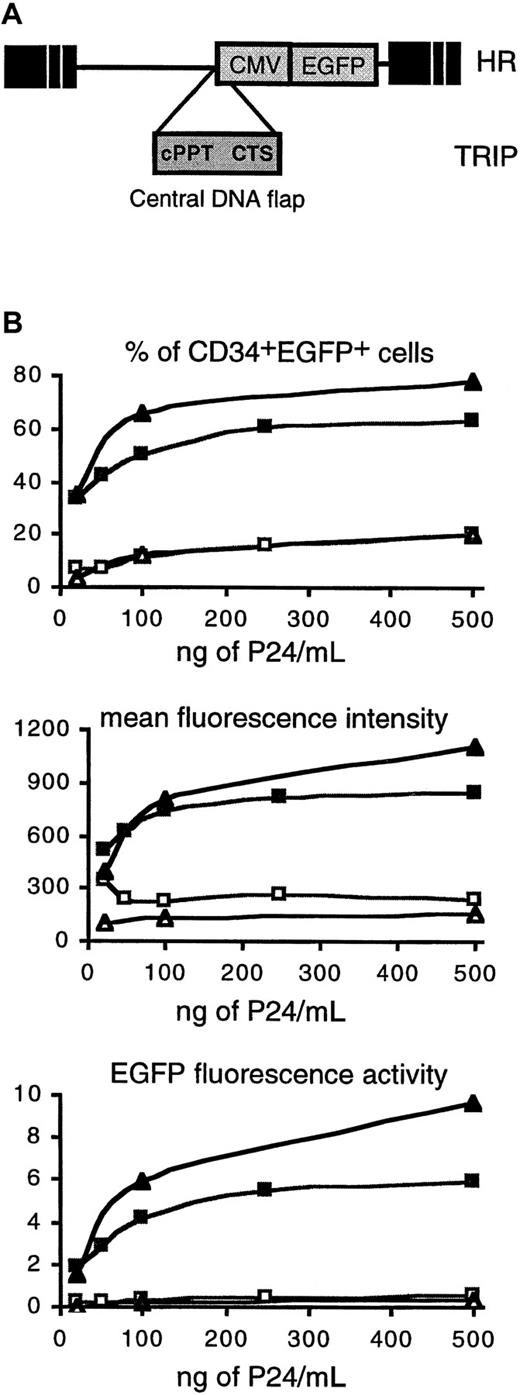
The central DNA flap increases the transduction efficiency of human CD34+ and CD34+CD38− CB cells by lentiviral vectors.
(A) Vector constructs. The TRIP vector was derived from the previously described HR vector18 by the insertion, immediately upstream of the internal CMV promoter, of a central HIV-1 DNA fragment encompassing the cPPT and CTS cis-active sequences. cPPT and CTS cis-active sequences are responsible for the formation of the DNA flap during HIV-1 reverse transcription.13 15 Both vectors encode EGFP. (B) Dose-response transduction experiments with TRIP (▪, ▴) and HR (■, ▵) vectors. CD34+ CB cells (▪, ■) or CD34+CD38− cells (▴, ▵) from the same CB were exposed to vector particles for 24 hours in serum-free medium in the presence of cytokines, and after an additional 48 hours of culture, EGFP expression was analyzed by flow cytometry (“Materials and methods”). The upper panel shows the percentage of CD34+EGFP+ cells, and the middle panel shows the MFI of the CD34+EGFP+ cell population. The lower panel shows the EGFP fluorescence activity of the CD34+EGFP+ cell population (percentage of CD34+EGFP+ cells multiplied by the MFI, arbitrary units). All are plotted as a function of vector concentration normalized according to P24 levels. The data shown are the results of one representative experiment out of 3 independent experiments.
The central DNA flap increases the transduction efficiency of human CD34+ and CD34+CD38− CB cells by lentiviral vectors.
(A) Vector constructs. The TRIP vector was derived from the previously described HR vector18 by the insertion, immediately upstream of the internal CMV promoter, of a central HIV-1 DNA fragment encompassing the cPPT and CTS cis-active sequences. cPPT and CTS cis-active sequences are responsible for the formation of the DNA flap during HIV-1 reverse transcription.13 15 Both vectors encode EGFP. (B) Dose-response transduction experiments with TRIP (▪, ▴) and HR (■, ▵) vectors. CD34+ CB cells (▪, ■) or CD34+CD38− cells (▴, ▵) from the same CB were exposed to vector particles for 24 hours in serum-free medium in the presence of cytokines, and after an additional 48 hours of culture, EGFP expression was analyzed by flow cytometry (“Materials and methods”). The upper panel shows the percentage of CD34+EGFP+ cells, and the middle panel shows the MFI of the CD34+EGFP+ cell population. The lower panel shows the EGFP fluorescence activity of the CD34+EGFP+ cell population (percentage of CD34+EGFP+ cells multiplied by the MFI, arbitrary units). All are plotted as a function of vector concentration normalized according to P24 levels. The data shown are the results of one representative experiment out of 3 independent experiments.
Transduction protocol
Tissue culture plates were coated with fibronectin (Bio-Whittaker France, Emerainville, France) according to the manufacturer's instructions. Human CD34+ populations were plated at 2-3 × 105 cells per mL in serum-free medium23 in the presence of 4 μg/mL polybrene (Sigma-Aldrich, Saint Quentin Fallavier, France) and the following recombinant human (rh) cytokines: 100 ng/mL stem cell factor (SCF) (Amgen, Neuilly sur Seine, France); 100 ng/mL Flt-3 ligand (FL) (Immunex Corporation, Seattle, WA); 60 ng/mL interleukin-3 (IL-3) (Novartis France, Rueil-Malmaison, France); and 10 ng/mL pegylated–megacaryocyte growth and differentiation factor (PEG-MGDF) (Kirin Brewery, Tokyo, Japan).
Lentiviral vector particles were added at a concentration corresponding to 100 ng viral P24/mL (multiplicity of infection, ≈ 100) during 24 hours. Cells were then washed and cultured in vitro in conditions that support lymphoid and myeloid differentiation19for 48 hours to ensure maximal transgene expression. EGFP expression was analyzed in the CD34+ cell fraction by fluorescence-activated cell sorter (FACS) flow cytometry (FACScan, Becton Dickinson, Pont de Claix, France). The 48-hour time point was chosen to minimize EGFP fluorescence due to pseudotransduction; less than 1% of the cells expressed EGFP in the presence of 2 μM Nevirapin (Roche, Meylan, France), an inhibitor of reverse transcriptase.
Hematopoietic cell cultures
Colony-forming cells (CFCs) and long-term culture-initiating cells (LTC-ICs) were assayed as described.24,25 Lymphoid (B and natural killer [NK]) and myeloid (granulomonocytic and erythrocytic) potentials were assessed as described19 on MS5 stromal cells in the presence of rhSCF; rhFL; rhPEG-MGDF; and rhIL-2, rhIL-7, and rhIL-15. The cytokine cocktail also contained 10 ng/mL rhIL-3 and, in some experiments, 2 U/mL rhEPO (erythropoietin) (CILAG, Issy les Moulineaux, France). In all cases, cells were analyzed for EGFP expression by flow cytometry and phenotyped using the following mouse monoclonal antibodies (mAbs): CD19-PE (phycoerythrin) (Becton Dickinson), CD15, CD14, GPA-PE (PharMingen, Pont de Claix, France), CD11b-PE, CD56, and CD34-PE-Cy5 (Immunotech, Villepinte-Roissy CDG, France). Unspecific staining was detected using irrelevant mouse immunoglobulin G1 (IgG1) and IgM mAbs.
PCR analysis
Integration of the EGFP transgene was analyzed by PCR analysis on DNA extracted from CFC-derived colonies and from cells in lymphomyeloid cultures. DNA was prepared as previously described.26 Amplification of genomic DNA was performed on the entire extract with primers that amplify part of the vector-encoded EGFP sequence (5′-CCCTCGAGCTAGAGTCGCGGCCG-3′ sense primer and 5′-CCGGATCCCCACCGGTCGCCACC-3′ antisense primer). The amplification was performed for 35 cycles at an annealing temperature of 62°C, resulting in an 800-bp PCR product.
Assessment of the number of cell divisions
CD34+ cells were stained with PKH26 (Sigma-Aldrich) according to the manufacturer's instructions. Immediately after labeling, a CD34+ cell population corresponding to a narrow band of PKH26bright cells was isolated by cell sorting27 and transduced as described above. An aliquot of these cells was cultured in parallel in the presence of 0.1 μg/mL colcemid (Demecolcemid, Sigma-Aldrich) to serve as an internal control of nondivided cells. Fluorescence of PKH26-labeled transduced cells was analyzed by flow cytometry at the end of the 24-hour transduction protocol as well as after an additional 48 hours of culture in lymphomyeloid conditions. At that time cells were analyzed for both EGFP expression and PKH26 staining. In 2 experiments cells having undergone 0 or 1 division were sorted, and single cell clones were cultured in LTC and lymphomyeloid conditions. Sorting gates were defined in comparison with colcemid-treated cells.
Transplantation of CD34+ cells into NOD/SCID mice
Immediately after transduction, 7-10 × 104 cells were intravenously injected into sublethally irradiated NOD-LtSz-scid/scid (NOD/SCID) mice (3-3.5 Gy at 0.43 Gy/mn in an x-ray Phillips RT250 irradiator). Fifteen weeks later, BM cells were harvested from recipient mice, and the presence of human cells was assessed in individual mice by flow cytometry using the following mouse antihuman mAbs: CD38-PE, CD19-PE, CD34-PE-Cy5, and CD14-PE. EGFP expression was also analyzed in each differentiated population. Human CD34+CD38− cells were then sorted on a FACS Vantage (Becton Dickinson) from the mouse BM and cultured in bulk or as single cells in LTC and lymphomyeloid conditions. Ten control mice were injected with the same number of mock-transduced cells.
Statistical analyses
Statistical analyses were done using the Studentt test.
Results
Lentiviral vector–mediated gene transfer into CD34+cells is enhanced by the central DNA flap
First, using a 24-hour transduction protocol in the presence of cytokines, we compared the transduction efficiencies of CD34+ and CD34+CD38− CB cells with HIV-1–derived vectors that lacked (HR18 vector) or harbored (TRIP vector), the triple-stranded DNA flap structure (Figure1A). In both vectors, EGFP transgene expression was driven from an internal CMV promoter. The presence of the HIV-1 DNA flap in the lentiviral vector strikingly increased transduction efficiencies in CD34+ CB cells, as measured by the percentage of CD34+EGFP+ cells (Figure 1B, upper panel, indicated by squares). This result was independent of the quantity of vector used. In a total of 8 experiments, using a concentration of vector particles corresponding to 500 ng P24/mL, 36% ± 16% (mean ± SD; range, 17%-63%) and 11% ± 5% (range, 5%-20%) of CD34+ cells expressed EGFP after exposure to the TRIP and HR vectors, respectively (P = .0002). This higher transduction efficiency of TRIP versus HR vector was also true for the more immature CD34+CD38− cells because in 3 experiments using the same concentration of vector particles, 46% ± 28% (mean ± SD; range, 30%-78%) and only 11% ± 8% (range, 5%-20%) of cells were EGFP+ after exposure to the TRIP and HR vectors, respectively (Figure 1B, upper panel, indicated by triangles). It is interesting to note that at high concentrations of vector particles, the mean fluorescence intensity (MFI) was higher in cells transduced with the TRIP vector compared to the HR vector (Figure1B, middle panel), reflecting an increased level of EGFP protein in TRIP-transduced cells. Combining both the percentage of EGFP+ cells and the mean fluorescence intensity, the presence of the DNA flap sequence could enhance EGFP synthesis in CD34+ cells by more than a 10-fold factor (Figure 1B, lower panel).
The central DNA flap promotes nuclear import of a lentiviral vector in CD34+ cells
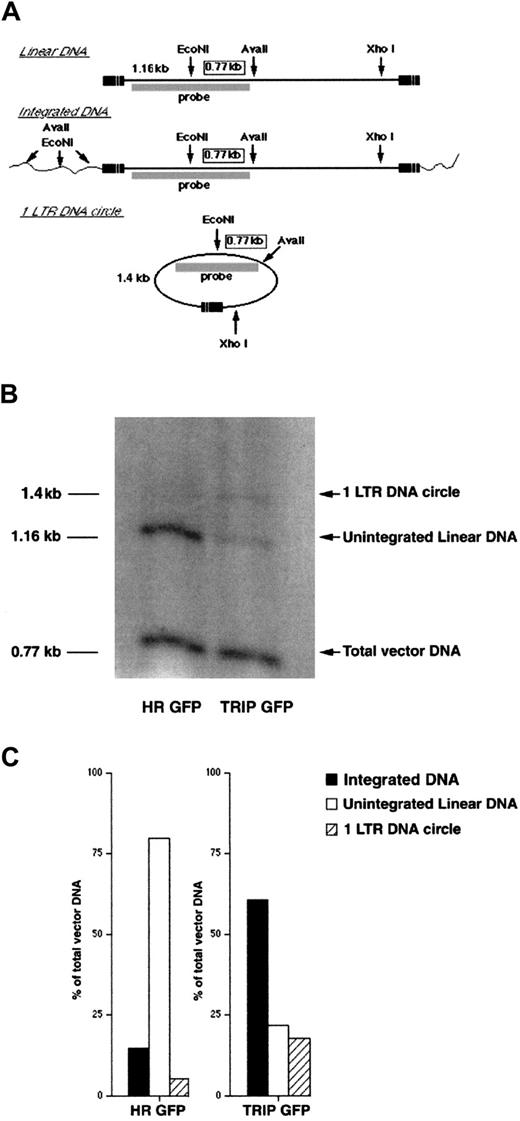
To determine whether the effect of the flap sequence on nuclear import was responsible for the observed increase in transduction efficiency, we precisely quantified the different forms of the intracellular vector DNA present within human CD34+ CB cells 48 hours after exposure to the TRIP or HR vectors (Figure2). Once reverse-transcribed in the cytoplasm, the linear vector DNA is imported into the nucleus, where it either integrates or circularizes, generally into one LTR DNA circle. Using a specific strategy of digestion and probing (Figure 2A), these different DNA forms were accurately quantified by a Southern blot assay in CD34+-transduced cells. In cells exposed to the TRIP vector, 60% of the total vector DNA had integrated into the host cell DNA, 18% had circularized into one LTR circle, and 22% remained as stable unintegrated linear DNA. As previously described,282 LTR DNA circles were present only in trace amounts. Of note, this intracellular vector DNA profile was similar to the profile observed in cells infected with wild-type HIV-1,1 indicating an efficient nuclear import process. In cells exposed to the HR vector, a similar amount of vector DNA was synthesized, indicating that the presence of the DNA flap sequence did not influence the rate of reverse transcription. However, a marked alteration in the pattern of intracellular HR vector DNA was evident. In cells exposed to the HR vector, there was a striking accumulation of untranslocated linear DNA forms (representing up to 80% of total vector DNA), and only low amounts of nuclear forms (Figure 2B,C).
Quantitative analysis of vector DNA nuclear import in human CD34+ cells.
(A) Strategy for the determination of vector DNA status in human CD34+ cells. DNA from human CD34+-transduced cells was extracted 24 hours after the end of transduction and digested with EcoNI and AvaII. DNA was further digested with XhoI to minimize transfer bias due to the large size of circular DNA fragments. Southern blot was then performed using a PCR-generated DNA fragment exactly overlapping the EcoNI site as a probe. The internal 0.77-kb EcoNI-AvaII DNA fragment, common to all vector DNA species irrespective of their integrated or unintegrated state, indicates the total amount of reverse-transcribed vector DNA in transduced cells. The 1.16-kb and 1.4-kb signals correspond to unintegrated linear DNA and one LTR circular DNA, respectively. Due to the diversity of vector DNA integration sites, the sizes of digested genomic fragments containing the 5′-part of the vector DNA were very heterogeneous, and thus, the integrated vector DNA could not be revealed on the blot. Because the PCR-generated probe exactly overlaps the EcoNI site, the intensity of each signal is directly proportional to the amount of the corresponding vector DNA species. Thus, the amount of integrated proviral DNA can be calculated by subtracting the signals of unintegrated linear and circular vector DNA from the total amount of vector DNA. (B) Southern blot analysis of the vector DNA forms in CD34+-transduced cells. (C) Phosphorimage quantitation of intracellular vector DNA profiles in CD34+ cells transduced by TRIP or HR vectors. The results are expressed as percentages of total vector DNA. The data shown in panels B and C are the results of 1 representative experiment out of 3 independent experiments.
Quantitative analysis of vector DNA nuclear import in human CD34+ cells.
(A) Strategy for the determination of vector DNA status in human CD34+ cells. DNA from human CD34+-transduced cells was extracted 24 hours after the end of transduction and digested with EcoNI and AvaII. DNA was further digested with XhoI to minimize transfer bias due to the large size of circular DNA fragments. Southern blot was then performed using a PCR-generated DNA fragment exactly overlapping the EcoNI site as a probe. The internal 0.77-kb EcoNI-AvaII DNA fragment, common to all vector DNA species irrespective of their integrated or unintegrated state, indicates the total amount of reverse-transcribed vector DNA in transduced cells. The 1.16-kb and 1.4-kb signals correspond to unintegrated linear DNA and one LTR circular DNA, respectively. Due to the diversity of vector DNA integration sites, the sizes of digested genomic fragments containing the 5′-part of the vector DNA were very heterogeneous, and thus, the integrated vector DNA could not be revealed on the blot. Because the PCR-generated probe exactly overlaps the EcoNI site, the intensity of each signal is directly proportional to the amount of the corresponding vector DNA species. Thus, the amount of integrated proviral DNA can be calculated by subtracting the signals of unintegrated linear and circular vector DNA from the total amount of vector DNA. (B) Southern blot analysis of the vector DNA forms in CD34+-transduced cells. (C) Phosphorimage quantitation of intracellular vector DNA profiles in CD34+ cells transduced by TRIP or HR vectors. The results are expressed as percentages of total vector DNA. The data shown in panels B and C are the results of 1 representative experiment out of 3 independent experiments.
This intracellular DNA profile strongly suggests a defect in the nuclear import of HR DNA, which leads to a decreased proportion of nuclear viral DNA species and a concomitant increase in the proportion of untranslocated linear DNA molecules. Thus, the strong defect in the nuclear import of the HR vector DNA in CD34+ cells was complemented by the insertion of the DNA flap sequence. Upon establishing the benefit of the central DNA flap on gene transduction in CD34+ and in CD34+CD38− cells, a subpopulation enriched in primitive hematopoietic cells,29 30 we performed all further experiments with the TRIP vector.
The TRIP vector efficiently transduces CD34+ cells isolated from various sources
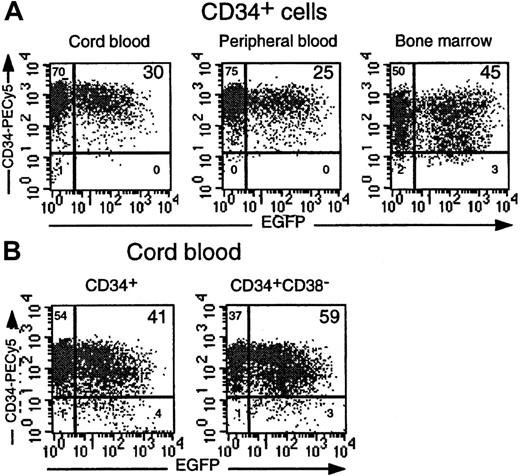
We next tested the ability of the TRIP vector to transduce CD34+ cells from CB, PBMC, and BM cells. A TRIP vector inoculum corresponding to 100 ng P24/mL was used in all experiments. In 40 experiments, 40% ± 16% (range, 17%-78%) of CD34+CB cells were transduced. Interestingly, omitting cytokines led to a drop in the transduction efficiency (9% ± 5%; range, 2%-19%; n = 14) as previously observed.27 31 We also successfully transduced CD34+ cells from adult sources such as cytokine-mobilized PBMC and BM cells (29% ± 10% and 41% ± 5%, respectively; n = 4) (Figure3A). PCR analysis on CFC-derived colonies confirmed the proportion of progenitors that had been transduced (data not shown).
CD34+ cells from different hematopoietic origins are efficiently transduced by the TRIP vector.
(A) Comparison of EGFP expression in purified CD34+ cells from CB, adult PBMC, and BM cells transduced in parallel using a TRIP vector concentration of 100 ng P24/mL. Dot-plot analyses demonstrating EGFP and CD34 expression are shown. (B) EGFP expression in identically transduced CD34+ and CD34+CD38−cells isolated from the same CB sample. Percentages of gated cells are indicated in each quadrant.
CD34+ cells from different hematopoietic origins are efficiently transduced by the TRIP vector.
(A) Comparison of EGFP expression in purified CD34+ cells from CB, adult PBMC, and BM cells transduced in parallel using a TRIP vector concentration of 100 ng P24/mL. Dot-plot analyses demonstrating EGFP and CD34 expression are shown. (B) EGFP expression in identically transduced CD34+ and CD34+CD38−cells isolated from the same CB sample. Percentages of gated cells are indicated in each quadrant.
High-efficiency transduction of slow-dividing immature hematopoietic cells
The main goal of gene transfer in human HSCs is to target immature stem cells with extensive proliferating and differentiating capacities. To demonstrate that the TRIP vector was able to transduce such cells, different approaches were used. First, we observed that both CD34+ and CD34+CD38− cells from the same CB samples were susceptible to transduction by the TRIP vector (mean transduction efficiencies of 41% ± 5% and 57% ± 6%, respectively; n = 5) (Figures 1B and 3B). Second, as HSCs exhibit a low mitotic activity,32 it was important to demonstrate that slow-dividing or nondividing cells were transduced in our protocol. To this end, CD34+ CB cells were labeled with PKH26, a fluorescent dye that intercalates into the cell membrane and is diluted as the cell divides. A homogeneous population of PKH26bright CD34+ cells was sorted and transduced. Immediately after exposure to the vector particles, more than 95% of the CD34+ cells exhibited the same fluorescent intensity as colcemid-treated cells, which indicated that they had not undergone mitosis during the transduction period (Figure4A, day 1). Indeed, 80% of these cells had not yet begun to replicate their DNA (data not shown).
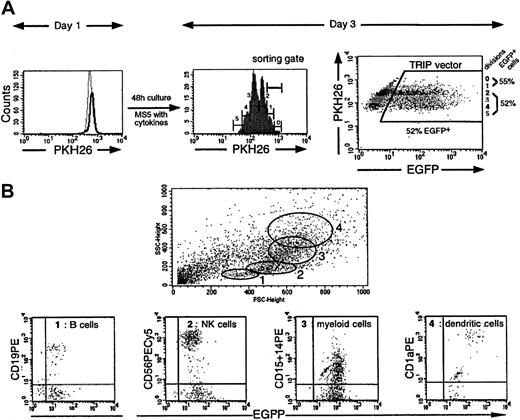
The TRIP vector allows efficient transduction of slow-dividing multipotent CD34+ CB cells.
(A) EGFP expression was studied as a function of cell divisions. The results of one representative experiment (Table 1, experiment 1) out of 2 experiments are illustrated here. CD34+PKH26bright cells were sorted on day 0 and transduced during 24 hours with 500 ng P24/mL TRIP viral stock. On day 1 the PKH26 fluorescence profile of the transduced CD34+cells (indicated by a solid line) was compared to that of the nondivided colcemid-treated CD34+ cells (indicated by a broken line), demonstrating that at the end of the vector exposure, more than 95% of the cells had not undergone division. A histogram profile of the PHK26 fluorescence in the cells exposed to the vector after an additional 48 hours of culture in lymphomyeloid conditions is shown (day 3). At this time point, cells have undergone between 0 and 5 divisions (gates 0 to 5). The dot-plot on the right shows EGFP expression in each of the division peaks indicated in the histogram. (B) Flow cytometry analysis of the progeny of a single TRIP-transduced CD34+ cell, sorted using the gate corresponding to the slow-dividing population. Briefly, slow-dividing CD34+cells were sorted (the sorting gate corresponds to gates 0 and 1 in the histogram in panel A) and cultured at one cell per well in culture conditions allowing the differentiation of both lymphoid (B and NK) and myeloid cells. Three weeks later, cells from clones containing at least 500 cells were labeled with mAbs directed against CD19 (B cells), CD56 (NK cells), CD15 (granulocytes), CD14 (myelomonocytic cells), and CD1a (dendritic cells). Each clone was analyzed individually, and the forward scatter and side scatter criteria (FSC/SSC) profiles were carefully monitored. Expressions of markers in cells present in the specific FSC/SSC morphological gates (gates 1 to 4) are shown.
The TRIP vector allows efficient transduction of slow-dividing multipotent CD34+ CB cells.
(A) EGFP expression was studied as a function of cell divisions. The results of one representative experiment (Table 1, experiment 1) out of 2 experiments are illustrated here. CD34+PKH26bright cells were sorted on day 0 and transduced during 24 hours with 500 ng P24/mL TRIP viral stock. On day 1 the PKH26 fluorescence profile of the transduced CD34+cells (indicated by a solid line) was compared to that of the nondivided colcemid-treated CD34+ cells (indicated by a broken line), demonstrating that at the end of the vector exposure, more than 95% of the cells had not undergone division. A histogram profile of the PHK26 fluorescence in the cells exposed to the vector after an additional 48 hours of culture in lymphomyeloid conditions is shown (day 3). At this time point, cells have undergone between 0 and 5 divisions (gates 0 to 5). The dot-plot on the right shows EGFP expression in each of the division peaks indicated in the histogram. (B) Flow cytometry analysis of the progeny of a single TRIP-transduced CD34+ cell, sorted using the gate corresponding to the slow-dividing population. Briefly, slow-dividing CD34+cells were sorted (the sorting gate corresponds to gates 0 and 1 in the histogram in panel A) and cultured at one cell per well in culture conditions allowing the differentiation of both lymphoid (B and NK) and myeloid cells. Three weeks later, cells from clones containing at least 500 cells were labeled with mAbs directed against CD19 (B cells), CD56 (NK cells), CD15 (granulocytes), CD14 (myelomonocytic cells), and CD1a (dendritic cells). Each clone was analyzed individually, and the forward scatter and side scatter criteria (FSC/SSC) profiles were carefully monitored. Expressions of markers in cells present in the specific FSC/SSC morphological gates (gates 1 to 4) are shown.
We then analyzed EGFP expression as a function of cell division 48 hours after transduction as a function of cell division. As it is generally believed that cells which are more immature divide slowly in vitro in the presence of cytokines, we arbitrarily distinguished a “slow-dividing” (0 and 1 division illustrated in Figure 4A, middle panel) cell population and a “rapid-dividing” (more than 2 divisions) cell population. Interestingly, 40.5% ± 17% of the slow-dividing cells and 36% ± 12% of the rapid-dividing cells in these populations (n = 3) expressed EGFP, respectively, indicating that they had been transduced at equivalent levels (Figure 4A, right panel). To confirm the immaturity of the slow-dividing cells, they were sorted (sorting gate shown in Figure 4A) and tested for the presence of LTC-ICs and cells able to generate both lymphoid (B and NK cells) and myeloid (granulomacrophagic cells) progenies. The proportions of slow-dividing cells that were LTC-ICs were 15% and 27% in experiments 1 and 2; of these LTC-ICs 33% and 50%, respectively, were EGFP+ by PCR analysis (Table 1). In these same slow-dividing cells, lymphomyeloid progenitors were identified. In experiment 1, 29.5% and in experiment 2, 24% of the progenitors were multipotent, ie, capable of differentiation in B, NK, and myeloid (M) cells. Of note, 40% of these clones also differentiated into dendritic (D) cells (Figure 4B, experiment 1). Importantly, 61% (experiment 1) and 37.5% (experiment 2) of these multipotent lymphomyeloid progenitors were transduced (as assessed by EGFP expression) compared to 22% to 45% of more mature CD34+ cells that were committed to only one or 2 lineages (Table 1).
Transduced long-term culture-initiating cells and multipotent lymphomyeloid progenitors are detected in slow-dividing CD34+ cord blood cells
| . | . | No. transduced/total progenitors (% of transduced) . | ||||
|---|---|---|---|---|---|---|
| Exp. . | Transduced per total cells, %‡ . | Long-term culture* . | Lymphomyeloid culture† . | |||
| LTC-ICs . | Proliferating clones . | Monopotent . | Bipotent . | Tripotent . | ||
| 1 | 55 | 3/9 (33) | 28/61 (46) | 13/29 (45) | 4/14 (28.5) | 11/18 (61) |
| 2 | 21 | 8/16 (50) | 18/67 (27) | 8/37 (22) | 4/14 (28.5) | 6/16 (37.5) |
| . | . | No. transduced/total progenitors (% of transduced) . | ||||
|---|---|---|---|---|---|---|
| Exp. . | Transduced per total cells, %‡ . | Long-term culture* . | Lymphomyeloid culture† . | |||
| LTC-ICs . | Proliferating clones . | Monopotent . | Bipotent . | Tripotent . | ||
| 1 | 55 | 3/9 (33) | 28/61 (46) | 13/29 (45) | 4/14 (28.5) | 11/18 (61) |
| 2 | 21 | 8/16 (50) | 18/67 (27) | 8/37 (22) | 4/14 (28.5) | 6/16 (37.5) |
LTC-ICs indicate long-term culture-initiating cells; CB, cord blood; EGFP, enhanced green fluorescent protein.
In each experiment, 60 slow-dividing cells were sorted and cultured at one cell per well. Cultures were terminated after 5 weeks, and the contents of each well were individually plated in clonogenic assays. Out of 60 input cells, 9 cells (15%) in experiment 1 and 16 cells (27%) in experiment 2 were retrospectively identified as LTC-ICs.
One hundred twenty slow-dividing cells were sorted and cultured at one cell per well. In experiments 1 and 2, 61 and 67 clones, respectively, contained enough cells (more than 500 cells) after 2 to 4 weeks to allow phenotyping by flow cytometry. In experiments 1 and 2, 29 of 61 clones and 37 of 67 clones, respectively, contained only cells from one lineage defining monopotent progenitors (CD19+ B, CD56+ NK, or CD14+ CD15+ myeloid cells). Similarly, 14 of 61 input cells in experiment 1 and 14 of 67 input cells in experiment 2 were bipotent progenitors (B+NK, B+M, and NK+M), and 18 (29.5%) of 61 input cells and 16 (24%) of 67 input cells, respectively, were tripotent cells (B+NK+M). These proportions are similar to those we have previously observed with unmanipulated CD34+ CB cells.19
Transduction efficiency was evaluated as the percentage of slow-dividing CD34+ cells expressing EGFP 48 hours after exposure to TRIP vector particles (Figure 4).
Altogether these results indicate that the TRIP vector has the ability to transduce slow-dividing progenitors, which include primitive LTC-IC and multipotential (NK+B+M+D) progenitors. The lymphomyeloid potential of the slow-dividing sorted cells was similar to that observed with fresh CD34+ CB cells,19 indicating that contact with and/or transduction by the TRIP vector had no detrimental effect on the hematopoietic potential of CD34+ CB cells.
Long-term NOD/SCID repopulating cells are transduced by the TRIP vector
Finally, because long-term in vivo transplantability of human cells in NOD/SCID mice is considered a hallmark of cell immaturity,33 we injected CD34+ CB cells immediately after transduction into NOD/SCID mice (3 experiments). Fifteen weeks after transplantation, human hematopoietic cells (CD34+ progenitors, CD19+ B-lymphoid, and CD14/CD15+ granulocyte and myelomonocytic cells) were present in the BM of 10 of 11 mice. (The proportion of human cells varied between 0.5% and 62%, which is comparable to controls injected with untransduced cells). Surprisingly, in 2 experiments (6 mice), it was noted that whereas 50% of the injected cells expressed EGFP, more than 80% of the human cells detected 15 weeks later were EGFP+. Analysis of a representative mouse is shown in Figure 5A. To demonstrate that CD34+CD38− cells present in the BM of NOD/SCID mice had been transduced and were functional, in one experiment these cells were sorted and grown in lymphomyeloid bulk culture. After 2 weeks these cells produced CD19+ B, CD56+ NK, and CD14+ monocytic cells and GPA+erythroblasts in the presence of rhEPO, and all these cells expressed EGFP (Figure 5B). Moreover, analysis of the progeny of single-cell cultures indicated that multipotent EGFP+ B + NK + M progenitors were present among CD34+CD38−human cells engrafted in NOD/SCID mice (Figure 5C) at a frequency of 20%, which is identical to that observed in comparable experiments with fresh CD34+ CB cells.19 In addition, this cell population contained transduced LTC-ICs (data not shown). These results definitively demonstrate that the TRIP vector efficiently transduces functional primitive and multipotent CD34+CD38− human CB cells with the ability to sustain long-term hematopoiesis in NOD/SCID mice.
The TRIP vector efficiently transduces NOD/SCID mice long-term reconstituting human cells.
(A) Phenotype of cells harvested from the BM of NOD/SCID mice grafted 15 weeks earlier with 7 × 104CD34+ CB cells transduced with the TRIP vector. At that time, human CD34+ cells were isolated by immunomagnetic selection. CD34+CD38− primitive cells were further sorted on a FACS Vantage. (B) Phenotype of cells differentiated from 104 CD34+CD38− cells, purified from reconstituted mice, following bulk culture in lymphomyeloid conditions. rhEPO was added to the bulk culture to allow terminal erythroid differentiation (glycophorin A [GPA+] cells). (C) The experiment was performed as described in panel B, except that the cells were cultured at 1 cell per well in the absence of rhEPO. Progeny of 39 of 120 cultures proliferated sufficiently to be phenotyped by FACS and were assessed 2 to 3 weeks later using flow cytometry, as illustrated in Figure 4B. Eight of 39 (20%) clones produced CD19+ B, CD56+ NK, and CD11b+ myeloid cells defining multipotential progenitors. Shown in panel C is a representative analysis of the progeny of an input multipotent progenitor.
The TRIP vector efficiently transduces NOD/SCID mice long-term reconstituting human cells.
(A) Phenotype of cells harvested from the BM of NOD/SCID mice grafted 15 weeks earlier with 7 × 104CD34+ CB cells transduced with the TRIP vector. At that time, human CD34+ cells were isolated by immunomagnetic selection. CD34+CD38− primitive cells were further sorted on a FACS Vantage. (B) Phenotype of cells differentiated from 104 CD34+CD38− cells, purified from reconstituted mice, following bulk culture in lymphomyeloid conditions. rhEPO was added to the bulk culture to allow terminal erythroid differentiation (glycophorin A [GPA+] cells). (C) The experiment was performed as described in panel B, except that the cells were cultured at 1 cell per well in the absence of rhEPO. Progeny of 39 of 120 cultures proliferated sufficiently to be phenotyped by FACS and were assessed 2 to 3 weeks later using flow cytometry, as illustrated in Figure 4B. Eight of 39 (20%) clones produced CD19+ B, CD56+ NK, and CD11b+ myeloid cells defining multipotential progenitors. Shown in panel C is a representative analysis of the progeny of an input multipotent progenitor.
Discussion
In an attempt to transduce very primitive human HSCs, we tested the efficiency of lentiviral vectors that in contrast to murine oncoretrovirus-derived vectors, can integrate in the genome of nondividing cells. Based on our recent demonstration that inclusion of the central DNA flap of HIV-1 dramatically enhances viral genome nuclear import,1 we derived a new construct, designated the TRIP vector, by inserting this central DNA flap in the HR vector. Here we show that this TRIP lentiviral vector, which encodes the EGFP transgene, can efficiently transduce very primitive human progenitors during a very short culture period (24 hours) in the presence of cytokines. This conclusion is based on 2 sets of data: (1) Approximately 40% of CD34+ cells could be transduced and could express the EGFP transgene during a sustained period of time. With selected batches of vector, it is of note that more than 60% of the CD34+ cells could reproducibly be transduced. (2) Similar high proportions of cells derived from myeloid LTC-ICs, multipotent lymphomyeloid progenitors, and long-term repopulating NOD/SCID mouse cells expressed EGFP, indicating the efficient transduction of progenitors functionally close to stem cells.
A major determinant was the use of the modified vector including the central DNA flap. Indeed, we first compared the transduction efficiencies of HR and TRIP vectors on CD34+and CD34+CD38− cells from human CB. The amount of both vectors used in transduction experiments was normalized by the P24 capsid protein content of vector supernatants. This method proved to be reliable. First, the amounts of P24 antigen in HR and TRIP vector stocks produced in parallel were very comparable. Second, transduction with the P24-normalized amounts of HR and TRIP vectors resulted in the approximately equivalent quantities of reverse-transcribed vector DNA in the vector-exposed cells (Figure 2), indicating the presence of similar levels of functional vector particles in normalized vector stocks. Regardless of the dose of vector used and based on the percentage of EGFP+ fluorescent cells, the flap-containing vector was more efficient in transducing CD34+ and CD34+CD38− CB cells than the HR vector.
When a high concentration of TRIP vector particles was used for transduction, an increase in the fluorescence intensity of transduced cells was also observed. Because the insertion of the DNA flap sequence does not influence the transcriptional activity of the internal CMV promoter,1 the higher level of EGFP expression observed with the TRIP vector is likely to be due to multiple integration events of the vector genome into target cells. However, increasing the vector dose did not permit the CD34+ cell population to be transduced to homogeneity. This could be explained if a fraction of the CD34+ cell population was refractory to transduction by lentiviral vectors that, for instance, might be related to the variable proportion of cells being in the G0 or early G1 phase.34,35 In the case of HIV-1, it has been shown that in naive lymphocytes, which are in G0, the HIV replicative cycle aborts due to an incomplete reverse transcription process.36,37 A second possibility explaining the refractoriness of certain CD34+ cells to lentiviral vector transduction could be the lack of nuclear import of the retrotranscribed HIV-1 genome.38 Finally, toxicity is possibly induced at a high concentration of vector particles.
The cPPT and CTS cis-active sequences, which are responsible for the formation of the DNA flap during reverse transcription, are located at the center of lentiviral genomes and overlap the 3′-region of the integrase coding sequence (POL). The crucial role of the DNA flap in vector DNA nuclear import most probably accounts for the previously reported heterogeneous efficiency of transduction of CD34+ cells by lentiviral vectors. Indeed, an efficient transduction is described using vectors in which the transgene replaces the envelope and/or Nef coding sequences of the HIV-1 genome.29,39,40 However, the HIV-1 GAG and POL proteins are encoded by such vector constructs, thereby representing a major disadvantage because deficient HIV particles are produced by the transduced cells. A much lower efficiency of transduction has been reported with classical “gutless” nonimmunogenic lentiviral vectors deleted for the entire coding sequences and thus for the central DNA flap sequence.27 41
In this study the HR vector appeared significantly less efficient than the TRIP vector in transducing CD34+ and CD34+CD38− CB cells. Thus, in all experiments assessing the potential of transduced cells, the TRIP vector was used. Despite the optimized nuclear import observed with the TRIP vector, we could not transduce more than 20% of the CD34+ CB cells in the absence of cytokine during the transduction protocol. This rather low efficiency might be due to the inability of metabolically inactive cells to efficiently reverse-transcribe the vector, as was previously described for the wild-type HIV-1.35 However, we clearly demonstrate here that the TRIP lentiviral vector integrated into cytokine-activated CD34+ cells that did not complete mitosis (Figure 4A). Indeed, 55% of the cells that divided slowly, ie, completing 0 or 1 division in 3 days, expressed EGFP. Among the slow-dividing cells, up to 50% and 60% of LTC-IC and lymphomyeloid progenitors, respectively, identified in single-cell experiments were transduced. This demonstrates that the TRIP vector can integrate into slowly proliferating multipotent progenitors, an important result because it is usually admitted that the most primitive multipotent cells are not actively cycling.31,32 42
Previous experiments have shown that NOD/SCID-repopulating cells could be transduced by the HR vector, with transduction of as many as 27% of human CD45+ cells.41 As we expected, the TRIP vector resulted in a better transduction efficiency in NOD/SCID-repopulating cells. Surprisingly, whereas only 50% of TRIP-transduced CD34+ CB cells expressed EGFP 48 hours after transduction in 2 experiments, at least 80% of their progeny harvested from BM of engrafted NOD/SCID mice were transduced (6 of 7 recipient mice). Similarly, Miyoshi et al41 detected a transduction efficiency of 35% in CFCs recovered from NOD/SCID mice even though the original clonogenic progenitors were transduced with an efficiency of 12% to 17%. These data have led us to speculate that the NOD/SCID-repopulating cells and multipotent progenitors are particularly susceptible to transduction by lentiviral vectors. Moreover, during 2 in vitro experiments, single progenitors able to generate a progeny of B, NK, and M cells appeared more efficiently transduced than monopotent and bipotent progenitors (Table 1), and CD34+CD38− cells exposed to the TRIP vector in the same conditions as CD34+ cells expressed EGFP in higher proportion (Figures 1 and 3). Alternatively, the transduction process by lentiviral vectors may alter the homing and/or survival/proliferation properties of transduced cells, thereby resulting in an increased engraftment in NOD/SCID mice.
We demonstrate here that the flap structure restores the nuclear import defect of HIV-1–based replacement vector genomes, thereby dramatically increasing the transduction efficiencies of human CD34+hematopoietic populations from different sources. TRIP, the flap-containing vector, is able to efficiently transduce human HSCs defined by their slow-dividing rate, multipotentiality, and ability to establish long-term active human hematopoiesis in the BM of NOD/SCID mice. Importantly, the transduction process of CD34+ cells did not alter their quantitative and qualitative differentiation potentials. The highly efficient transduction of human HSCs by the TRIP vector will undoubtedly open multiple gene therapy applications and help to resolve fundamental questions regarding the biology of human HSCs.
Acknowledgments
We would like to thank A. Rouchès and P. Ardouin for their expertise in the production of NOD/SCID mice, V. Schiavon for cell sorting, B. Izac for technical assistance, Dr Van Nifderick and colleagues for kindly providing CB samples, and Naomi Taylor for critical comments on the manuscript. We also thank Amgen for rhSCF, Kirin for rhPEG-MGDF, CILAG for rhEPO, and Immunex for rhFlt3-L.
A.S. and F.P. contributed equally to this work.
Supported by grant ACC-SV 96-335 from Ministère de la Recherche et de la Technologie, Paris; grants 1137 and 9225 from the Association de la Recherche contre le Cancer, Villejuif; and grants from the Institut National de la Santé et de la Recherche Médicale, Paris; from the Institut Gustave Roussy, Villejuif; from the Agence Nationale de Recherche contre le Sida, Paris; from the Association Française contre les Myopathies, Paris; and Electricité de France, Paris, France.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anne Dubart-Kupperschmitt, INSERM U474, Maternité de Port Royal, 123 Bd de Port Royal, 75014 Paris, France; e-mail: dubart@cochin.inserm.fr; or Pierre Charneau, Unité d'Oncologie Virale, Institut Pasteur, 25-28 rue du Dr Roux, 75724 Paris Cedex 15, France; e-mail:charneau@pasteur.fr.

![Fig. 5. The TRIP vector efficiently transduces NOD/SCID mice long-term reconstituting human cells. / (A) Phenotype of cells harvested from the BM of NOD/SCID mice grafted 15 weeks earlier with 7 × 104CD34+ CB cells transduced with the TRIP vector. At that time, human CD34+ cells were isolated by immunomagnetic selection. CD34+CD38− primitive cells were further sorted on a FACS Vantage. (B) Phenotype of cells differentiated from 104 CD34+CD38− cells, purified from reconstituted mice, following bulk culture in lymphomyeloid conditions. rhEPO was added to the bulk culture to allow terminal erythroid differentiation (glycophorin A [GPA+] cells). (C) The experiment was performed as described in panel B, except that the cells were cultured at 1 cell per well in the absence of rhEPO. Progeny of 39 of 120 cultures proliferated sufficiently to be phenotyped by FACS and were assessed 2 to 3 weeks later using flow cytometry, as illustrated in Figure 4B. Eight of 39 (20%) clones produced CD19+ B, CD56+ NK, and CD11b+ myeloid cells defining multipotential progenitors. Shown in panel C is a representative analysis of the progeny of an input multipotent progenitor.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/13/10.1182_blood.v96.13.4103/5/m_h82400496005.jpeg?Expires=1769150746&Signature=OlVbW~1XnNnsxEiQjzDrQzGW-aNn0cgMWxoittnCgDOX7IGGsB3M4ycDrQFYr1Vg8zURR2kGH6Hifm2jS4YHj2w-hwNBXjXOX4lEUymB7xUAzSB3Vhov5u9sJcD-9jFW7BtFd7MWe4fOoqBcfdzxe0drQ5EERsfDjdvRXaP~xuMki2QqdXmhl-A9dhsJ-VTME-wbPykjOn3EuZ4yYf4Nlo~~kQrHCFTI~CCx7GCu~l1piSqbWApaOhFCWCEs9d5zghQzmvptb9ISINwY4-j-u-pXqOOfdym219XT2cPP06iqhEVeBneEXfQux-Ruld4-gY3gmAbDJKT5XmM3bHsNHQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal