Abstract
Fanconi anemia (FA) is a clinically and genetically heterogeneous disorder. Clinical care is complicated by variable age at onset and severity of hematologic symptoms. Recent advances in the molecular biology of FA have allowed us to investigate the relationship between FA genotype and the nature and severity of the clinical phenotype. Two hundred forty-five patients from all 7 known complementation groups (FA-A to FA-G) were studied. Mutations were detected in one of the cloned FANC genes in 169 patients; in the remainder the complementation group was assigned by cell fusion or Western blotting. A range of qualitative and quantitative clinical parameters was compared for each complementation group and for different classes of mutation. Significant phenotypic differences were found. FA-G patients had more severe cytopenia and a higher incidence of leukemia. Somatic abnormalities were less prevalent in FA-C, but more common in the rare groups FA-D, FA-E, and FA-F. In FA-A, patients homozygous for null mutations had an earlier onset of anemia and a higher incidence of leukemia than those with mutations producing an altered protein. In FA-C, there was a later age of onset of aplastic anemia and fewer somatic abnormalities in patients with the 322delG mutation, but there were more somatic abnormalities in patients with IVS4 + 4A → T. This study indicates that FA patients with mutations in theFANCG gene and patients homozygous for null mutations inFANCA are high-risk groups with a poor hematologic outcome and should be considered as candidates both for frequent monitoring and early therapeutic intervention.
Introduction
Fanconi anemia (FA) is an autosomal recessive disorder characterized clinically by progressive pancytopenia, diverse congenital abnormalities, and predisposition to malignancy, particularly acute myelogenous leukemia (AML).1 FA patients exhibit extreme clinical heterogeneity and may have abnormalities in any major organ system.2 Common physical findings include abnormal skin pigmentation, growth retardation, radial ray or other skeletal malformations, microphthalmia, and renal or urinary tract malformations. Less common clinical features are genital, gastrointestinal or cardiac malformations, hearing loss, mental retardation, and central nervous system abnormalities.1,3,4 Aplastic anemia (AA) develops at an average age of 7 years, but a considerable range has been reported.5 Some FA patients have a relatively mild phenotype, with normal skeletal development, subclinical hematopoietic abnormalities, and survival into the third or fourth decade.6,7 Other patients have a more severe phenotype, with skeletal abnormalities, early onset AA, and cancer.8-10 Hematopoietic stem cell transplantation (SCT) offers the only current possibility of a cure for the hematologic complications.11
Cells cultured from FA patients exhibit increased spontaneous chromosomal aberrations and hypersensitivity to DNA cross-linking agents such as mitomycin C (MMC) and diepoxybutane (DEB).12 This is used as a diagnostic criterion for FA because of the difficulty of making the diagnosis on the basis of clinical manifestations alone. FA is also a genetically heterogeneous disorder, with 8 different complementation groups (A-H) having been described,13,14 which has subsequently been revised to 7.15 This heterogeneity has largely been verified by molecular cloning of FANCA on chromosome 16q24.3,16,17FANCC on 9q22.3,13FANCF on 11p13-p15,18 and FANCG on 9p13.19 The prevalence of the different complementation groups has been shown to vary considerably according to ethnic origin.20,21 FA-A is the most prevalent group, accounting for 65% of all cases.22
One possible explanation for the wide variety of clinical expression could be differences in complementation group, because the respective genes could function either at different points in the same pathway or in different pathways that lead to similar but not identical clinical conditions. Alternatively, variations in severity could result from different mutations within the same gene23 and other genetic or environmental factors.24-27 Early studies of FA-C patients showed that a clinical severity score was lower in patients with the 322delG mutation in FANCC than in patients with IVS4 + 4A → T.28,29 This was confirmed in a systematic study,23 which found that patients with the IVS4 + 4A → T mutation and mutations in exon 14 had an earlier onset of hematologic complications and poorer survival than those with mutations in exon 1.
The European Fanconi Anemia Research Group (EUFAR)30 has collected clinical, molecular, and cellular data on 245 FA patients from 24 countries, representing all 7 confirmed complementation groups. These data have been analyzed to determine whether specific complementation groups, mutations, or classes of mutation in FA genes are associated with differences in clinical outcome such as age of onset of aplastic anemia, survival, the incidence of leukemia, or the presence of somatic abnormalities. This paper presents the results of the study, which identified several highly significant associations among specific groups of patients and aspects of their clinical phenotype that may be relevant to the clinical management of the FA patient.
Patients, materials, and methods
Patients
A total of 245 FA patients from 179 families were recruited to this study during the period 1989-1999 via the framework of the European Fanconi Anemia Research Group, or were referred by hematology departments in their respective countries. The study included both incident and prevalent cases. Patients originated from 24 different countries, including South Africa (49), the United Kingdom (33), Germany (29), Turkey (27), Italy (20), France (18), The Netherlands (18), United Arab Emirates (7), Egypt (6), the United States of America (6), India (4), Pakistan (4), Spain (4), Bangladesh (3), China (3), Saudi Arabia (2), Greece (2), Lebanon (2), Poland (2), Russia (2), Africa (1), Austria (1), Brazil (1), and Hungary (1). Four patients were of Ashkenazi Jewish ancestry. The ages of patients ranged from infancy to 48 years. Inclusion criteria were (1) a clinical diagnosis of FA, supported by cellular hypersensitivity to MMC or DEB, or molecular data; (2) known complementation group, from cell fusion studies, mutation analysis, or Western blotting; and (3) availability of adequate clinical information. The clinical data were collected either from a questionnaire sent to the referring physician (238 patients), or from previous publications (7 patients).7-10 27 A blood sample was obtained from at least one affected individual from all families in the study with the consent of the patient or a guardian. The clinical information required included a range of qualitative and quantitative clinical parameters, including age of onset and severity of bone-marrow hypoplasia, length of survival, occurrence of leukemia or myelodysplasia, and the nature and extent of congenital abnormalities. All questionnaires were collated by one individual (L.F.) to avoid duplication of patients in the study.
Hematology and malignancies
Fifty-one siblings were excluded from the calculation of the mean age at diagnosis because they were diagnosed as a result of a family study. Onset of hematologic abnormalities was defined as the time at which one of the following laboratory parameters was observed: hemoglobin (Hb) level ≤ 10 g/dL, white blood count (WBC) ≤ 4 × 109/L or absolute neutrophil count (ANC) ≤ 1 × 109/L, and platelet count ≤ 100 × 109/L. Criteria used to define severe cytopenia were Hb ≤ 8 g/dL, ANC ≤ 0.5 × 109/L, and platelet count ≤ 20 × 109/L. A severity index was then defined by counting the number of variables with a severe reading, giving a value of 0, 1, 2, or 3 for each FA patient. No attempt was made to incorporate methods of treatment into the assessment of disease severity, because treatment varied over the years within and across different centers. The occurrence of an SCT was also excluded as a measure of severity of hematologic symptoms, as this depended on the existence of a suitable donor. Survival analyses were performed with and without censoring patients who received SCT. Because the results were similar in each case, the data are presented without censoring transplant recipients at the time of SCT. AML was defined as more than 30% myeloid blasts in the bone marrow, and myelodysplastic synrome (MDS) as patients with 5% to 30% myeloid blasts in the bone marrow. Specific dysgranulopoietic features, such as increase in size of myeloid cells, and characteristic chromosomal abnormalities were seen in the majority of MDS cases.
Somatic abnormalities
The prevalence of each specific somatic abnormality was compared in the different groups. The extent of the malformations was also assessed according to the number of anatomic sites involved, as previously described,31 excluding skin abnormalities and growth retardation for this purpose. The following were each considered as anatomic sites: (1) the head, including eyes (small eyes, strabismus, epicanthal folds, hypertelorism), ears (deafness, abnormal shape, atresia, dysplasia, low set, canal stenosis, abnormal middle ear), face (microcephaly, micrognathia, triangular face), and neck (short, sprengel); (2) the skeleton (including thumb and radial abnormalities); (3) the kidneys, (4) the gastrointestinal tract; (5) the urogenital tract, including small testes and cryptorchidism; (6) the cardiovascular system; and (7) the central nervous system (CNS). The CNS abnormalities included hydrocephaly, encephalocele, and spina bifida.
Molecular and cell biology
Complementation analysis.
Fusion of lymphoblastoid cell lines to genetically marked reference cell lines from known FA complementation groups was carried out as previously described.14
Western blotting.
This was carried out in selected cell lines using anti-FANCA, anti-FANCF, and anti-FANCGantibodies, as previously described.32
Mutation screening.
RNA was extracted from cell lines and genomic DNA from peripheral blood samples in 4 different laboratories and screened by SSCP analysis, heteroduplex analysis, chemical cleavage, or sequencing. Most of the mutations detected have been reported in previous publications16,17,19,33-37 or have been submitted to the International FA Mutation database [http://www.rockefeller.edu/fanconi/mutate]. Null mutations were classified as those that would be predicted to produce no FA protein (frameshifts, stop codons, and large deletions), and “altered protein” mutations as missense mutations and in-frame deletions. Also included in the latter group were frameshifts in the most distal 10% of the coding sequence of the genes, because C-terminal truncating mutations may be associated with functional protein.38 In 29 cases (11.8%), the predicted effect of the mutation was verified by Western blotting.
Statistical analysis
The proportion of patients with a specific abnormality was compared in the complementation groups with global and pairwise tests for differences, using the χ2 test or Fisher exact test for small numbers. The numbers of abnormalities and numbers of anatomic sites were compared across groups using the t test and analysis of variance. The predictive value of specific abnormalities for a patient's complementation group was assessed using logistic regression. Survival analysis was used to compare the age at onset of hematologic abnormalities, the age at diagnosis of AML or MDS, and survival after diagnosis by complementation group. Kaplan-Meier estimates were calculated and differences among the complementation groups were tested using the nonparametric log-rank test. Estimates of survival after diagnosis will contain a bias toward longer-surviving patients because patients diagnosed shortly before the start of the study who also died before the start of the study would not be included. Analyses of malformations and survival were performed by: (1) comparing the common mutations in complementation groups A and C with other patients within the complementation group and with all other FA patients; (2) comparing patients with 2 null mutations against patients with mutations causing an altered protein; and (3) comparing patients with 2 mutations at the 5′ end of the gene with those with 2 mutations at the 3′ end. Possible bias in the analysis of somatic abnormalities as a result of familial clustering was assessed by repeating the analysis with only the first diagnosed sibling in each family. Because of the complexity of the cross tabulation of the sites of somatic abnormalities and complementation group and to the relatively small sample size, no attempt was made to control study-wide error. Significant associations should therefore be confirmed in a future study, and nonsignificant associations that involved small numbers of observations should be viewed as inconclusive. All statistical testing used Splus5 (MathSoft Inc, Seattle, WA).
Results
Complementation group and mutations
Complementation group was assigned in 245 FA patients by cell fusion, by the detection of at least one clearly pathogenic mutant allele,39 by the absence of detectable FANCA, FANCF, or FANCG protein on a Western blot, or by the existence of a sibling classified by one or more of these methods. The number of patients assigned by each of these methods is listed in Table1. This also shows that 101 patients were assigned by 2 or more independent methods, and all of these results were concordant. This led to the classification of 172 patients as group A (70.2%), one as group B (0.4%), 34 as group C (13.9%), 3 as group D (1%), 5 as group E (2%), 6 as group F (2.5%), and 24 as group G (9.8%). The spectrum of mutations observed in the 169 patients with at least one mutation was highly heterogeneous, and the majority of patients from a nonconsanguineous mating were compound heterozygotes. Sixty-eight different pathogenic mutations were found inFANCA, 11 in FANCC, 11 in FANCG, and 5 in FANCF, 75.6% of which were predicted to generate null alleles and 24.3% to produce an altered protein. There were 159 patients in whom both mutations were identified, of which 105 were homozygous for null mutations (66.0%). The proportion of homozygous nulls in the 3 main complementation groups was not significantly different, with 75 of 114 in FA-A (65.8%), 22 of 34 in FA-C (64.7%), and 8 of 11 in FA-G (72.7%). Apparent founder mutations were noted in the Irish population (FANCA/deletion of exons 11-14 in 7 patients), in Ashkenazi Jews (FANCC/IVS4 + 4A → T in 5 patients), and in South African Afrikaners (FANCA/deletion of exons 12-31 in 44 patients, deletion of exons 11-17 in 10 patients, 3398delA in 7 patients). Two other common mutations were found without any obvious founder effect (FANCA/1263delF in 9 patients andFANCC/322delG in 17 patients).
Methods of assignment of Fanconi anemia complementation group
| Cell fusion | 43 |
| Mutation analysis | 73 |
| Western blot | 7 |
| Affected sibling* | 21 |
| Cell fusion + mutation analysis | 79 |
| Cell fusion + mutation + Western blot | 13 |
| Cell fusion + Western blot | 5 |
| Mutation + Western blot | 4 |
| Total | 245 |
| Cell fusion | 43 |
| Mutation analysis | 73 |
| Western blot | 7 |
| Affected sibling* | 21 |
| Cell fusion + mutation analysis | 79 |
| Cell fusion + mutation + Western blot | 13 |
| Cell fusion + Western blot | 5 |
| Mutation + Western blot | 4 |
| Total | 245 |
Affected sibling of patient assigned by one or more of the experimental methods.
Phenotypes in the complete patient cohort
Most patients had an abnormal blood count at presentation (93%), and the mean age at onset of hematologic abnormalities was 7.6 years. At the time of analysis, 72% of the patients were still living. The mean age at last observation was 14 years, including patients who died (14.4 years in FA-A, 16.2 years in FA-C, and 11.5 years in FA-G). The mean age at death for those patients who had died was 13.9 years. SCT had been carried out in 21% of patients, and 61% required transfusions. In the course of their disease, 9% of patients had AML develop, 8% MDS, and 6% nonhematologic malignancies. The pattern of somatic abnormalities in the full cohort of patients is listed in Table2.
Proportion of somatic abnormalities in Fanconi anemia complementation groups
| N = . | Complementation groups . | A/C/G . | Comparisons* . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All . | A . | B . | C . | D . | E . | F . | G . | A/C . | A/G . | C/G . | ||||||
| 246 . | 172 . | 1 . | 34 . | 3 . | 5 . | 6 . | 24 . | . | . | . | . | |||||
| . | N . | % . | N . | % . | N . | N . | % . | N . | N . | N . | N . | % . | . | Pvalue . | P value . | P value . |
| Skin | 166/235 | 71 | 124/167 | 74 | 1/1 | 15/30 | 50 | 3/3 | 3/5 | 3/6 | 16/24 | 66 | .0448 | .0250 | .4600 | .4000 |
| Growth retardation | 135/233 | 58 | 98/164 | 60 | 1/1 | 8/30 | 28 | 2/3 | 4/5 | 4/6 | 18/24 | 75 | .0009 | .0020 | .1800 | .0007 |
| Head | 132/237 | 56 | 97/170 | 53 | 1/1 | 7/31 | 23 | 3/3 | 5/5 | 2/6 | 17/22 | 77 | .0002 | .0007 | .1000 | .0002 |
| Radial ray | 115/243 | 47 | 83/170 | 49 | 1/1 | 6/34 | 18 | 3/3 | 5/5 | 4/6 | 13/24 | 54 | .0033 | .0010 | .6700 | .0090 |
| Kidneys | 53/243 | 22 | 33/170 | 19 | 0/1 | 8/34 | 24 | 1/3 | 4/5 | 2/6 | 4/24 | 17 | .7505 | — | — | — |
| Skeleton | 44/243 | 18 | 28/170 | 16 | 1/1 | 4/34 | 12 | 2/3 | 4/5 | 2/6 | 3/24 | 13 | .7497 | — | — | — |
| Urogenital tract | 29/243 | 12 | 22/170 | 13 | 1/1 | 0/34 | 0 | 2/3 | 3/5 | 1/6 | 1/24 | 4.2 | .0466 | .0300 | .3100 | .4200 |
| Gastrointestinal tract | 31/243 | 13 | 17/170 | 10 | 1/1 | 5/33 | 15 | 1/3 | 2/5 | 3/6 | 3/24 | 13 | .3957 | — | — | — |
| Mental retardation | 28/237 | 12 | 20/170 | 12 | 0/1 | 1/29 | 3.4 | 1/3 | 2/4 | 2/5 | 2/24 | 8.3 | .3741 | — | — | — |
| Heart | 25/243 | 10 | 16/170 | 9.4 | 0/1 | 2/34 | 6.1 | 3/3 | 0/5 | 2/6 | 1/24 | 4.2 | .5996 | — | — | — |
| Deafness | 22/237 | 9.3 | 19/159 | 11 | 0/1 | 0/29 | 0 | 1/3 | 0/4 | 1/5 | 1/24 | 4.2 | .1028 | — | — | — |
| Central nervous system | 11/243 | 4.5 | 4/170 | 2.4 | 0/1 | 4/34 | 12 | 0/3 | 3/5 | 0/6 | 0/24 | 0 | .0104 | .0080 | > .99 | .1300 |
| Mean number of abnormalities | 3.3 | 3.3 | 6 | 1.9 | 7.6 | 7.6 | 4.5 | 3.5 | < .0001 | < .0001 | .7800 | .0020 | ||||
| Mean number of sites | 1.8 | 1.8 | 3 | 1.1 | 4.3 | 4.6 | 3.2 | 1.9 | .0146 | .0060 | .6500 | .0400 | ||||
| N = . | Complementation groups . | A/C/G . | Comparisons* . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All . | A . | B . | C . | D . | E . | F . | G . | A/C . | A/G . | C/G . | ||||||
| 246 . | 172 . | 1 . | 34 . | 3 . | 5 . | 6 . | 24 . | . | . | . | . | |||||
| . | N . | % . | N . | % . | N . | N . | % . | N . | N . | N . | N . | % . | . | Pvalue . | P value . | P value . |
| Skin | 166/235 | 71 | 124/167 | 74 | 1/1 | 15/30 | 50 | 3/3 | 3/5 | 3/6 | 16/24 | 66 | .0448 | .0250 | .4600 | .4000 |
| Growth retardation | 135/233 | 58 | 98/164 | 60 | 1/1 | 8/30 | 28 | 2/3 | 4/5 | 4/6 | 18/24 | 75 | .0009 | .0020 | .1800 | .0007 |
| Head | 132/237 | 56 | 97/170 | 53 | 1/1 | 7/31 | 23 | 3/3 | 5/5 | 2/6 | 17/22 | 77 | .0002 | .0007 | .1000 | .0002 |
| Radial ray | 115/243 | 47 | 83/170 | 49 | 1/1 | 6/34 | 18 | 3/3 | 5/5 | 4/6 | 13/24 | 54 | .0033 | .0010 | .6700 | .0090 |
| Kidneys | 53/243 | 22 | 33/170 | 19 | 0/1 | 8/34 | 24 | 1/3 | 4/5 | 2/6 | 4/24 | 17 | .7505 | — | — | — |
| Skeleton | 44/243 | 18 | 28/170 | 16 | 1/1 | 4/34 | 12 | 2/3 | 4/5 | 2/6 | 3/24 | 13 | .7497 | — | — | — |
| Urogenital tract | 29/243 | 12 | 22/170 | 13 | 1/1 | 0/34 | 0 | 2/3 | 3/5 | 1/6 | 1/24 | 4.2 | .0466 | .0300 | .3100 | .4200 |
| Gastrointestinal tract | 31/243 | 13 | 17/170 | 10 | 1/1 | 5/33 | 15 | 1/3 | 2/5 | 3/6 | 3/24 | 13 | .3957 | — | — | — |
| Mental retardation | 28/237 | 12 | 20/170 | 12 | 0/1 | 1/29 | 3.4 | 1/3 | 2/4 | 2/5 | 2/24 | 8.3 | .3741 | — | — | — |
| Heart | 25/243 | 10 | 16/170 | 9.4 | 0/1 | 2/34 | 6.1 | 3/3 | 0/5 | 2/6 | 1/24 | 4.2 | .5996 | — | — | — |
| Deafness | 22/237 | 9.3 | 19/159 | 11 | 0/1 | 0/29 | 0 | 1/3 | 0/4 | 1/5 | 1/24 | 4.2 | .1028 | — | — | — |
| Central nervous system | 11/243 | 4.5 | 4/170 | 2.4 | 0/1 | 4/34 | 12 | 0/3 | 3/5 | 0/6 | 0/24 | 0 | .0104 | .0080 | > .99 | .1300 |
| Mean number of abnormalities | 3.3 | 3.3 | 6 | 1.9 | 7.6 | 7.6 | 4.5 | 3.5 | < .0001 | < .0001 | .7800 | .0020 | ||||
| Mean number of sites | 1.8 | 1.8 | 3 | 1.1 | 4.3 | 4.6 | 3.2 | 1.9 | .0146 | .0060 | .6500 | .0400 | ||||
FA indicates Fanconi anemia.
Significant P values are shown in bold. The prevalence of abnormalities was lower in FA-C than in FA-A or FA-G in all cases with the exception of the central nervous system.
Phenotypes in different complementation groups
Pairwise comparisons were carried out among groups A, C, and G because comparison against the total cohort would be dominated by FA-A, and these 3 groups were represented by a substantial number of patients.
Hematology and malignancies.
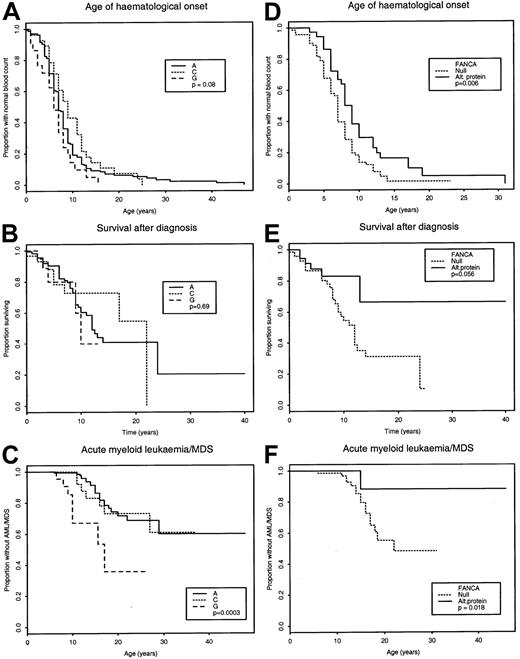
No significant difference among FA-A, FA-C, and FA-G was found for age at diagnosis of FA (P = .41), age at onset of hematologic abnormalities (P = .076, Figure1A), requirement for transfusions (P = .24), and solid tumors (P = .46). The mean number of parameters indicative of severe cytopenia (as defined in “Patients, materials, and methods”) were 1.56 in FA-A, 0.89 in FA-C, and 2.11 in FA-G. Analysis of variance showed a significant effect of complementation group (P = .035), and pairwise analysis showed that FA-G was more severe than FA-C (Wilcoxon rank-sum test, P = .016). Survival to age 10 years after diagnosis was 62% for FA-A, 73% for FA-C, and 50% for FA-G (Figure 1B), but postdiagnosis survival was not statistically significantly different either globally for groups A, C, and G (P = 0.69) or for pairwise analyses (P > .25). Postdiagnosis survival analysis for all complementation groups will be biased against patients with very short survival who died before the start of the study (described in “Patients, materials, and methods”). There were significant differences by complementation group in rates of AML (P = .001), MDS (P = .04), and AML or MDS (P = .0003), with FA-G having a higher rate of AML or MDS than FA-A and FA-C (Figure 1C). The proportion of cases with AML/MDS was 23 of 172 (13.4%) in FA-A, 7 of 34 (20.6%) in FA-C, and 8 of 24 (33.3%) in FA-G. Estimates from the survival analysis of the proportion of FA patients who had AML/MDS develop by age 10 years were less than 1% for FA-A (n = 158), 0% for FA-C (n = 25), and 33% for FA-G (n = 18). In the rare complementation groups, it is worth noting that the 3 FA-D patients presented with either very mild or no hematologic abnormalities at a mean age of 13.3 years, but the difference from all FA patients was not statistically significant.
Major complementation groups compared.
Comparison of age of onset of aplastic anemia, survival after diagnosis and incidence of AML or MDS in the major complementation groups (panels A through C), and in FA-A patients with mutations predicted to produce no FANCA protein (null) or an altered protein (panels D through F).
Major complementation groups compared.
Comparison of age of onset of aplastic anemia, survival after diagnosis and incidence of AML or MDS in the major complementation groups (panels A through C), and in FA-A patients with mutations predicted to produce no FANCA protein (null) or an altered protein (panels D through F).
Somatic abnormalities.
Three-way tests for differences in specific somatic abnormalities and in the total number of abnormalities across groups A, C, and G were carried out. If these were significant (P < .05), pairwise tests were then performed. The results are presented in Table2. Themean number of somatic abnormalities was significantly higher in FA-A than in FA-C, and in FA-G compared with FA-C, but FA-A and FA-G were similar. When considering specific somatic abnormalities, FA-C had significantly less growth retardation, fewer head abnormalities and radial ray defects than FA-A or FA-G, and fewer skin abnormalities and urogenital malformations than FA-A. More central nervous system defects were found in FA-C than FA-A, but these were all associated with a particular mutation (see below). Analysis of the number of anatomic sites, which excludes the mild and common abnormalities of skin and growth retardation,31 also showed a less severe phenotype in FA-C compared with FA-A or FA-G. These analyses were repeated by excluding any siblings of the index case to assess the possible effects of familial clustering. The results of the 3-way tests were very similar to those with all siblings, which suggests that differences across the groups are not related to familial clustering. Logistic regression analysis did not reveal differences in somatic abnormalities that were sufficiently marked to permit accurate prediction of the major complementation groups.
Although sample numbers were small for the rare complementation groups FA-D, FA-E, and FA-F, they showed a higher rate of involvement of anatomic sites. Significant results were obtained when each of these 3 groups was compared with the overall sample (P = .003 for FA-D, P < 10−7 for FA-E,P = .024 for FA-F). Furthermore, 3 of 5 FA-E patients presented with central nervous system malformations, and this number is significantly different when compared with the overall sample (P < .001).
Common mutations
Pairwise analyses were carried out for each of 7 common mutations in FANCA or FANCC against the remaining patients in the corresponding complementation group, and against all FA patients. Patients were scored as positive for a specific mutation if they had at least one copy of it.
Hematology and malignancies.
No significant differences were found for any of the common mutations in FANCA with regard to age of diagnosis, age of onset of hematologic symptoms, or the proportion of patients with severe cytopenia. A higher rate of AML or MDS was found in patients with at least one delE12-31 mutation (n = 44) compared with patients with other mutations in FANCA (P = .004). Survival analysis showed that by age 20 years an estimated 55.3% of patients with at least one delE12-31 mutation had AML or MDS develop, compared with 13.3% in other patients from FA-A. In patients who were homozygous for delG322 (n = 12), there was a later age of onset of hematologic symptoms compared with all FA patients (P = .036), with an estimated 33.3% of these patients developing symptoms by age 10 compared with 78.5% of all FA. It was not possible to analyze the hematologic course and survival in patients with the FANCC IVS4 + 4A → T mutation because 3 of the 5 cases were terminations of pregnancies.
Somatic abnormalities.
The prevalence of somatic abnormalities in patients with the common mutations was compared with rates in patients with other mutations in the same complementation group. Significant differences are shown in Table 3. Patients with at least one delE12-31 mutation in FANCA or with the FANCCmutation IVS4 + 4A → T had a higher prevalence for several different abnormalities, and for the number of abnormal anatomic sites. This was particularly significant for IVS4 + 4A →T, even though there were only 5 such patients in our cohort. In contrast, patients with the FANCC 322delG mutation had a lower number of abnormalities. Because half of the patients in group C had this particular mutation, we also compared the prevalence of somatic abnormalities in these patients with all other FA patients in our cohort. The prevalence of both the total number of abnormalities and of abnormal anatomic sites was lower in 322delG (P < .0001 for both). The results in Table 3 include all siblings, because removal of additional siblings to assess a possible effect of familial clustering reduces the number of observations to below the threshold for meaningful analysis.
Somatic abnormalities in Fanconi anemia patients with common mutations
| Mutation . | Abnormality . | Mutation present3-150 . | Mutation absent3-150 . | P value . |
|---|---|---|---|---|
| FANCA/1263delF | GIT | 3/9 | 10/121 | .047 |
| FANCA/delE12-31 | Skin | 38/44 | 56/83 | .032 |
| Head | 34/44 | 46/86 | .013 | |
| Radial ray | 32/44 | 39/86 | .005 | |
| Total number | 4.02 | 3.17 | .011 | |
| Anatomic sites | 2.25 | 1.66 | .013 | |
| FANCC/IVS4+4A>T | Radial ray | 4/5 | 2/28 | .002 |
| Kidney | 4/5 | 4/28 | .008 | |
| GIT | 4/5 | 1/28 | .0006 | |
| CNS | 4/5 | 0/28 | .0001 | |
| Total number | 5 | 1.32 | < .0001 | |
| Anatomic sites | 4 | 0.57 | < .0001 | |
| FANCC/322delG | Total number | 1.06 | 2.75 | .021 |
| Anatomic sites | 0.47 | 1.75 | .0096 |
| Mutation . | Abnormality . | Mutation present3-150 . | Mutation absent3-150 . | P value . |
|---|---|---|---|---|
| FANCA/1263delF | GIT | 3/9 | 10/121 | .047 |
| FANCA/delE12-31 | Skin | 38/44 | 56/83 | .032 |
| Head | 34/44 | 46/86 | .013 | |
| Radial ray | 32/44 | 39/86 | .005 | |
| Total number | 4.02 | 3.17 | .011 | |
| Anatomic sites | 2.25 | 1.66 | .013 | |
| FANCC/IVS4+4A>T | Radial ray | 4/5 | 2/28 | .002 |
| Kidney | 4/5 | 4/28 | .008 | |
| GIT | 4/5 | 1/28 | .0006 | |
| CNS | 4/5 | 0/28 | .0001 | |
| Total number | 5 | 1.32 | < .0001 | |
| Anatomic sites | 4 | 0.57 | < .0001 | |
| FANCC/322delG | Total number | 1.06 | 2.75 | .021 |
| Anatomic sites | 0.47 | 1.75 | .0096 |
Patients with at least one allele of the common mutation compared against all other Fanconi anemia patients from the same complementation group. The number of abnormalities is greater for these mutations, with the exception of 322delG.
GIT indicates gastrointestinal tract; CNS, central nervous system.
Proportion of patients with/without abnormality or mean number of abnormalities.
Class of mutation
Hematology and malignancies.
Patients with 2 FANCA mutations leading to a null allele had an earlier age of onset of hematologic abnormalities (7.2 years vs 10.3 years, P = .006, Figure 1D), and a shorter survival after diagnosis (P = .056, Figure 1E). Their survival 10 years postdiagnosis was 55.8% compared with 83% for patients with an altered FANCA protein. They also had a higher proportion of AML/MDS (P = .018, Figure 1F), when compared with patients with an altered protein. Significance levels were similar when patients with 2 null mutations were compared with patients with 2 mutations leading to an altered protein. There was no significant difference in the incidence of solid tumors in FANCA null homozygotes compared to those with at least one altered protein allele, but the number of malignancies observed was small (n = 6). No significant difference was found for any of the parameters for patients with mutations at the 5′ end of FANCA compared to those with 3′ mutations. The number of patients with different classes of mutations inFANCC and FANCG (n = 11) was too small for comparison.
Somatic abnormalities.
No significant difference was found in the pattern or the proportion of somatic abnormalities or anatomic sites when FA-A patients with 2 mutations leading to a null allele were compared with FA-A patients with at least one mutation leading to an altered protein, or for FA-A patients with 5′ mutations versus 3′ mutations. However, a lower rate of somatic abnormalities (P = .005) and anatomic sites (P = .006) was found in FA-C patients with 2 null mutations compared with those with at least one mutation leading to an altered protein.
Discussion
The diagnosis and treatment of Fanconi anemia is complicated by the great variability in the severity of the disease, particularly the wide range in the age of onset of hematologic abnormalities and survival. Monitoring the progression of the disease and knowing when to introduce major therapeutic interventions, such as SCT, presents a significant challenge. Previous work from our group showed that the extent of severe somatic abnormalities was a strong predictor of survival after matched, unrelated SCT.31 In the current study, we have investigated the possibility of defining risk groups in FA by examining the relationship between genotype and phenotype, first by separating patients by complementation group or type of mutation, and then comparing the clinical phenotypes of these subgroups.
Examination of the hematologic profiles of the most prevalent complementation groups (A, C, and G) did not reveal significant differences in age of onset of aplastic anemia, but cytopenia was more severe in FA-G, and AML or MDS occurred much earlier and in a higher proportion of patients in this group. There was no significant difference in the incidence of solid tumors in these groups, but numbers were small. Survival after diagnosis was lowest in FA-G, but this result was not statistically significant. This may reflect the lack of a genuine difference or insufficient power to detect a moderate effect. The overall finding was of a more severe hematologic course in FA-G, with the least severe course in FA-C. The extent of significant somatic abnormalities was similar in FA-A and FA-G, but very much lower in FA-C patients, particularly for abnormalities of the head and radial ray. There was also evidence of a higher rate of somatic abnormalities in the rare complementation groups, but this observation will need to be followed up in larger samples as more of these patients are identified.
Investigation of the clinical phenotypes associated with specific mutations in FA is complicated by great heterogeneity in the mutational spectrum of the known FA genes; thus numbers of patients with a specific mutation are generally small, and the majority are compound heterozygotes. However, within FA-A we found that the presence of at least one allele with a delE12-31 mutation was associated with a higher rate of AML or MDS and a high rate of severe somatic abnormalities. In FA-C, the 322delG mutation was associated with milder hematologic symptoms when compared with all FA patients, and there was a lower rate of somatic abnormalities, with 16 of 17 patients having normal thumbs and absence of deep organ malformations. This result is broadly similar to those from previous genotype-phenotype correlation studies in FA-C.23,28,29 In contrast, the IVS4 + 4A → T mutation in FANCC was associated with a high rate of somatic abnormalities. This is consistent with the previous analysis of Ashkenazi Jewish patients with this mutation.23 However, a recent study of Japanese FA patients detected IVS4 + 4A → T in 9 patients, 8 of whom were homozygous.40 Surprisingly, the clinical phenotype was not severe in these patients either in terms of hematologic onset or the extent of congenital abnormalities. Because our patients were of Ashkenazi origin, the Japanese data indicate that sequence changes in other regions of the FANCC gene, or indeed elsewhere in the genome, may have a protective or deleterious effect on the clinical expression of this mutation. The mutation was associated with multiple polymorphic haplotypes in the Japanese patients, but whether any of these correspond with the Ashkenazi haplotype is unknown. Additional variables are environmental effects such as exposure to pathogens, and differing standards of health care. This is exemplified by the lack of concordance of clinical phenotype in some affected sibling pairs.1,3 24-27
We also searched for a significant difference between mutations predicted to lead to the absence of an FA protein and those predicted to lead to an altered protein. In some cases, these predictions would not be fulfilled, as demonstrated by the detection of a FANCC protein with an amino terminal truncation in the 322delG mutation.29 However, when we compared FA-A patients with 2 null alleles with FA-A patients with at least one allele leading to an altered protein, we found striking differences in the hematologic phenotype. Complete loss of FANCA protein was associated with a severe phenotype, whereas patients with an altered protein had a milder phenotype with a later age at onset of aplastic anemia, longer survival after diagnosis, and a lower prevalence of AML or MDS. Because the spectrum of FANCA null mutations was heterogeneous, the effect is not driven by a single severe mutation. Thus, the class of mutation present in any individual patient presents an additional variable that may override the influence of the complementation group.
The phenotypic variations that we have observed among different complementation groups and different classes of mutation raises the question of whether they can be explained in terms of what is known about the biology of the FA proteins. The FANCG protein interacts with FANCA, and this complex is reduced or disrupted in several other complementation groups.32,41-43 It has been suggested that the FA proteins are involved in the fidelity of repair of double-stranded breaks in DNA.44 If the FANCG protein is a key molecule in a multiprotein complex, then the lack of this protein may result in the disruption of the complex and affect the function of multiple FA proteins. This might lead to a higher degree of chromosomal breakage than in the other groups, and thus a more severe hematologic profile and a higher incidence of leukemia. The milder clinical phenotype observed in FA-A patients with an altered FANCA protein would not be unexpected if these proteins had partial activity. The mutant protein FANCA-H1110P, for example, fails to correct MMC sensitivity in a cell survival assay,45 but does form a complex with FANCG.32 It may therefore enable some part of FANCG function to occur. The milder 322delG mutation produces a frameshift in exon 1 and would therefore not be expected to produce any FANCC protein. However, cell lines with this mutation are reported to produce a 50 kD protein by reinitiation at met55, which confers partial correction of MMC sensitivity when overexpressed in FA-C cells.29 A protective effect for this protein in vivo is not yet established, because patients with this mutation are reported to exhibit a high frequency of DEB-induced chromosomal breakage.23
We believe that the results of this study have important implications for the clinical management of the FA patient. FA-G patients and those with 2 null mutations in the FANCA gene appear to constitute high-risk groups, with a more severe hematologic course and a high incidence of AML or MDS. Conversely, most FA-C patients in our study, with the exception of those with the IVS4 + 4A → T mutation, had a milder clinical course, with fewer somatic abnormalities. Analysis of FA patients receiving stem cell transplants has shown that increased survival is associated with younger age at transplant and less severe hematologic disease before transplant.11 Early and frequent monitoring and early SCT should be recommended in the high-risk groups identified by this study. Early molecular classification of the FA patient would also provide additional useful criteria for patient selection in future trials of gene therapy.
Acknowledgments
We thank the families with Fanconi anemia and Ralf Dietrich (Deutsche Fanconi Anemia Hilfe e.V) for their cooperation.
European Fanconi Anemia Research Group (EUFAR) (in alphabetical order): Drs S. E. Ball, R. Calzone, J. Cavenagh, F. Desai, De Wolf, G. R. Evans, Giuang Ji, C. Gutteridge, A. V. Hoffbrand, Hyde, P. Jacobs, C. Jameson, C. Karabus, M. Karwacki, H. Kayserili, J. C. Llerena, S. P. Mohan, A. O'Marcaigh, J. C. Opperman, Phillips, I. Roberts, P. Roux, E. Samochatova, H. Schmitt, D. Schuler, C. E. M. De Die-Smulders, S. Temtamy, C. Tautz, A. Vora, Williams, and C. Zeilder.
A complete list of the members of the European Fanconi Anemia Research Group is given at the end of this article.
Submitted February 18, 2000; accepted August 14 2000.
Supported by the European Union BIOMED program and the Fanconi Anemia Research Fund (US). L.F. is the recipient of a grant from l'Association pour la Récherche contre le Cancer.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Christopher G. Mathew, Division of Medical and Molecular Genetics, 8th floor Guy's Tower, Guy's Hospital, SE1 9RT London, UK; email: christopher.mathew@kcl.ac.uk.