Abstract
Donor-derived hematopoiesis was assessed in 17 patients who received allogeneic marrow grafts from HLA-matched siblings between 1971 and 1980. Complete blood counts were normal or near normal in all patients except one. Chimerism analyses, using either dual-color XY-chromosome fluorescence in situ hybridization (FISH) or analysis of variable number tandem repeat loci, indicated that 15 out of 16 patients had greater than 97% donor-derived hematopoiesis, whereas 1 patient had indeterminate chimerism. All 12 recipients of grafts from female donors exhibited polyclonal hematopoiesis by X-linked clonal analysis with the use of molecular probes. Of the 17 recipients, 9 exhibited a less than 1.0-kilobase shortening of granulocyte telomere length compared with their respective donors, according to terminal restriction fragment analysis or flow-FISH with a fluorescein-labeled peptide nucleic acid probe. These data suggest that under standard transplantation conditions, the stem cell proliferative potential is not compromised during hematopoietic reconstitution.
Introduction
Hematopoietic reconstitution after allogeneic marrow transplantation relies on a relatively small number of hematopoietic stem cells (HSCs) compared with the estimated stem cell pool in the donor.1-4 However, given the suggested proliferative potential of HSCs, it has been assumed that even this significantly reduced number of HSCs could provide adequate hematopoiesis throughout the life of the patient.5Preclinical small animal studies have shown that small numbers of donor HSCs may give rise to polyclonal, oligoclonal, and even monoclonal hematopoiesis.1,2,6,7 In human transplantation patients, where both blood volume and life span are greater, polyclonal hematopoiesis is most commonly observed, but monoclonal and oligoclonal hematopoiesis has also been reported.3,4,8,9 According to the intrinsic-timetable model, HSCs have a finite number of divisions; therefore, repeated challenges to a reduced stem cell pool could exhaust the system, resulting in marrow failure.5 It is reasonable to speculate that extreme proliferative demand on a limited number of stem cells would result in significant telomere shortening. Significant differences might then exist between telomere length in granulocytes from the donor and those concurrently isolated from the transplant recipient. Only a few cases of patients more than 15 years after allogeneic hematopoietic stem cell transplantation (HSCT) have been described in the literature, and potentially conflicting observations have been reported.3,4 10-13 In the present study, we tested this hypothesis by evaluating blood samples from 17 long-term marrow transplantation survivors and their donors.
Study design
Patients
Seventeen patients who received transplants between 1971 and 198014-16 and their HLA-identical donors agreed to participate in the study (Table 1). Approval for the study was obtained from the Fred Hutchinson Cancer Research Center Institutional Review Board.
Patient and donor profiles
| Patient no. . | Diagnosis . | Years since HSCT . | Age at HSCT in years; patient/donor . | BM cells infused; dose × 108/kg . | Chimerism/assay . | Clonality/polymorphism . |
|---|---|---|---|---|---|---|
| 1 | AA | 28 | 13/14 | 2.9 | Donor/XY-FISH | Polyclonal/M27β |
| 2 | AA | 27 | 31/27 | 2.2 | Donor/XY-FISH | * |
| 3 | AA | 25 | 14/22 | 5.2 | Donor/XY-FISH | Polyclonal/PGK |
| 4 | AA | 25 | 15/19 | 6 | Donor/VNTR | Polyclonal/PGK |
| 5 | AA | 24 | 13/8 | 3.7 | Donor/XY-FISH | Polyclonal/M27β |
| 6 | AA | 24 | 9/14 | 10.4 | †/VNTR | Polyclonal/M27β |
| 7 | AA | 22 | 3/7 | 9.2 | Donor/XY-FISH | Polyclonal/M27β |
| 8 | ALL | 24 | 18/4 | 1.2 | Donor/VNTR | Polyclonal/M27β |
| 9 | AA | 23 | 23/22 | 8.9 | Donor/XY-FISH | * |
| 10 | CML | 22 | 13/13 | 5.0 | ‡ | * |
| 11 | ANL | 22 | 28/25 | 5.8 | Donor/VNTR | * |
| 12 | AA | 22 | 30/26 | 3.6 | Donor/VNTR | * |
| 13 | AA | 22 | 17/14 | 3.9 | Donor/VNTR | Polyclonal/M27β |
| 14 | AA | 22 | 30/30 | 2.5 | Donor/XY-FISH | Polyclonal/PGK |
| 15 | AA | 22 | 20/17 | 4.2 | Donor/VNTR | Polyclonal/PGK |
| 16 | AA | 22 | 24/20 | 3.7 | Donor/XY-FISH | Polyclonal/M27β |
| 17 | ALL | 19 | 25/17 | 2.2 | Donor/XY-FISH | Polyclonal/M27β |
| Patient no. . | Diagnosis . | Years since HSCT . | Age at HSCT in years; patient/donor . | BM cells infused; dose × 108/kg . | Chimerism/assay . | Clonality/polymorphism . |
|---|---|---|---|---|---|---|
| 1 | AA | 28 | 13/14 | 2.9 | Donor/XY-FISH | Polyclonal/M27β |
| 2 | AA | 27 | 31/27 | 2.2 | Donor/XY-FISH | * |
| 3 | AA | 25 | 14/22 | 5.2 | Donor/XY-FISH | Polyclonal/PGK |
| 4 | AA | 25 | 15/19 | 6 | Donor/VNTR | Polyclonal/PGK |
| 5 | AA | 24 | 13/8 | 3.7 | Donor/XY-FISH | Polyclonal/M27β |
| 6 | AA | 24 | 9/14 | 10.4 | †/VNTR | Polyclonal/M27β |
| 7 | AA | 22 | 3/7 | 9.2 | Donor/XY-FISH | Polyclonal/M27β |
| 8 | ALL | 24 | 18/4 | 1.2 | Donor/VNTR | Polyclonal/M27β |
| 9 | AA | 23 | 23/22 | 8.9 | Donor/XY-FISH | * |
| 10 | CML | 22 | 13/13 | 5.0 | ‡ | * |
| 11 | ANL | 22 | 28/25 | 5.8 | Donor/VNTR | * |
| 12 | AA | 22 | 30/26 | 3.6 | Donor/VNTR | * |
| 13 | AA | 22 | 17/14 | 3.9 | Donor/VNTR | Polyclonal/M27β |
| 14 | AA | 22 | 30/30 | 2.5 | Donor/XY-FISH | Polyclonal/PGK |
| 15 | AA | 22 | 20/17 | 4.2 | Donor/VNTR | Polyclonal/PGK |
| 16 | AA | 22 | 24/20 | 3.7 | Donor/XY-FISH | Polyclonal/M27β |
| 17 | ALL | 19 | 25/17 | 2.2 | Donor/XY-FISH | Polyclonal/M27β |
BM indicates bone marrow; HSCT, hematopoietic stem cell transplantation; AA, aplastic anemia; ALL, acute lymphocytic leukemia; ANL, acute nonlymphocytic leukemia; CML, chronic myelogenous leukemia.
Male donor.
Partially informative markers—indeterminate.
Syngeneic.
Cell preparation and DNA extraction
Granulocyte and mononuclear cell fractions were obtained from peripheral blood by Ficoll-Hypaque density gradient separation. Buccal samples were obtained from patients after mouthwash with 20 mL normal saline. High molecular weight (MW) DNA was extracted after lysis of cell pellets by means of DNazol (Molecular Research Center, Cincinnati, OH).
Chimerism studies
Chimerism was examined in granulocyte, mononuclear, and T-cell fractions of sex-mismatched recipients by dual-color XY-chromosome fluorescence in situ hybridization (FISH) analysis.17Variable number tandem repeat (VNTR) analyses were performed in sex-matched recipients with the use of polymorphisms for the ApoB, SE33, and 33.6 VNTR loci by a polymerase chain reaction (PCR) method as described.18 19 Pretransplant samples were unavailable; therefore, constitutional DNA was extracted from patient buccal mucosal samples.
Clonality analysis
Mean telomere length analysis
Terminal restriction fragment length (TRF) analysis on peripheral blood granulocytes and flow-FISH were performed on granulocytes and mononuclear cells as previously described.21-23
Statistical analyses
The Wilcoxon signed rank test was used to determine statistical significance of differences in recipient and donor complete blood cell count, mean corpuscular volume, and difference in TRF (dTRF; ie, the donor's TRF minus the patient's TRF).
Results and discussion
The absolute neutrophil counts (ANCs) of all the recipients except one were in the normal range. Patient 1 was pancytopenic with an ANC of 960/μL when contacted for the study. However, the average difference between recipients' and donors' ANCs was statistically significant, with recipients' ANCs lower than those of their donors (P = .03; Wilcoxon signed rank test). The significant difference in neutrophil counts may be related to posttransplant factors. All marrow recipients except one had normal hemoglobin (Hgb) levels (patient 1, Hgb = 12.1). The average difference between recipients' and donors' Hgb, was not statistically significant (P = .11; Wilcoxon signed rank test). In 15 evaluable recipients, the average difference between recipients' and donors' platelet counts was not statistically significant (P = .10; Wilcoxon signed rank test). However, 3 recipients had platelet counts lower than the normal range: 45 000/μL (patient 1), 116 000/μL (patient 9), and 131 000/μL (patient 12). Arnold et al24 and Li et al25found that the majority of recipients 2 months to 8.5 years after transplantation had normal leukocyte counts and normal hemoglobin levels. This further supports previous observations that long-term survivors of uncomplicated marrow transplants have normal hematopoiesis many years after bone marrow transpant (BMT).
Of the 17 patients, 15 had full (greater than 97%) donor-derived hematopoiesis. One patient had indeterminate chimerism owing to lack of informative markers (Table 1), and one patient/donor pair consists of monozygous twins.
Twelve evaluable recipients with female donors had polyclonal donor-derived hematopoiesis (Table 1). Furthermore, there was not a significant shift in the recipients' clonal ratios of the X-linked alleles compared with those from the donors. Our current findings further support our initial observations3 and demonstrate that hematopoietic reconstitution remains polyclonal many years after uncomplicated marrow transplantation.
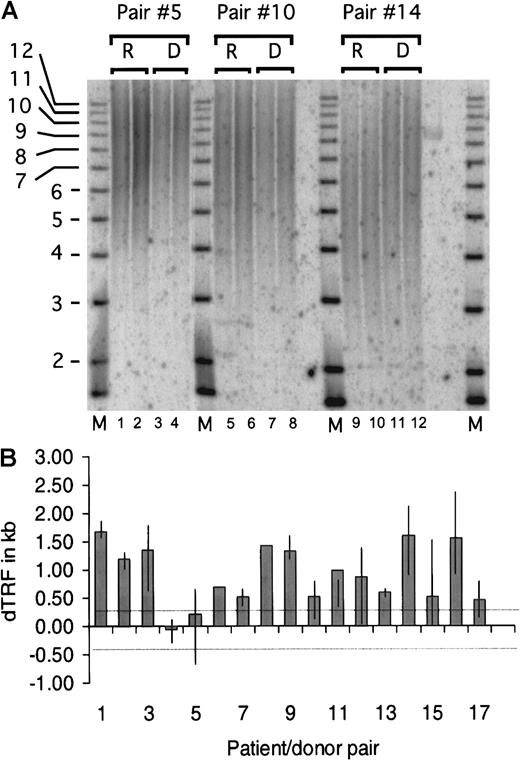
Results from a representative TRF experiment are shown in Figure1A, which depicts the analysis of 3 patient/donor pairs. We tested the null hypothesis that the average dTRF was equal to zero, where the variance of the estimated mean was adjusted to account for the fact that multiple experiments were done in individual patients. The average dTRF across all 50 experiments was estimated to be 0.94 kilobases (kb) (95% confidence interval, 0.69-1.20; P < .0001) (Figure 1B). When the null hypothesis that the average dTRF was equal to 0.369 kb was tested instead, the estimated average dTRF was still significantly different than 0.365 kb (P < .0001). Telomere length was also calculated by flow-FISH independently in recipient and donor pairs 1 and 9, and similar differences between donor and recipient telomere lengths were observed. Statistical analysis revealed no correlation between dTRF and total number of nucleated marrow cells infused, donor age, or time after transplant (data not shown).
Terminal restriction fragment length analysis on granulocyte DNA.
The donor's TRF minus the patient's TRF gives rise to the dTRF. TRF was calculated and compared between the duplicate samples. The average difference between each duplicate was calculated to be 104 base pairs (bp), and the SD was 88 bp. The lower limit of detection of the method was therefore selected as the average (104 bp) + 3 SD or 369 bp. Differences in TRF of fewer than 369 bp were considered not significant and ascribed to inherent experimental variability. One to 5 (median = 3) replicate experiments were performed with DNA from each of the 17 patient/donor pairs for a total of 50 TRF pairs (in 2 cases, only 1 experiment was done owing to limited DNA quantities). (A) Representative TRF analysis of patient/donor pairs 5, 10, and 14. High MW DNA was digested with HinfI and RsaI restriction enzymes and size-fractionated on a 0.8% agarose gel. Recipient and donor samples were loaded in duplicate lanes 1-4 (pair 5), 5-8 (pair 10), and 9-12 (pair 14), and a radiolabeled 1-kb DNA ladder MW marker was included (lanes M). In situ hybridization was performed with a P32 end-radiolabeled oligonucleotide probe comprising the telomere-specific sequence (TTAGGG)3. The signal was detected by scanning the gel by means of the Phosphorimager analysis system (Molecular Dynamics, Sunnyvale, CA). Mean TRF length was assigned to the MW corresponding to the distance of peak signal intensity from the origin of the electrophoresis. The MW was then calculated by means of the Imagequant (Molecular Dynamics) and Fragment (Molecular Dynamics) software. (B) dTRF data summary of each patient/donor pair. Each column represents the average dTRF of 1 to 5 (median 3) replicate experiments for each pair. The top and bottom ends of the vertical lines depict the maximum and minimum dTRF values obtained in each patient/donor pair. Single experiments were performed on pairs 6 and 8 because of limited amounts of DNA. The 2 thin dotted horizontal lines depict the experimental limit of detection, which was 369 bp as described in “Materials and methods.” When the average difference was computed across experiments for each individual patient, 15 of 17 recipients had dTRF more than 0.369 kb. Of the 15 recipients, 8 had dTRF of more than 1.0 kb and 7 had dTRF 0.3 to 1.0 kb. One of the remaining 2 recipients had dTRF of −0.1 kb, and the other, dTRF of 0.2 kb, both less than 0.369 kb.
Terminal restriction fragment length analysis on granulocyte DNA.
The donor's TRF minus the patient's TRF gives rise to the dTRF. TRF was calculated and compared between the duplicate samples. The average difference between each duplicate was calculated to be 104 base pairs (bp), and the SD was 88 bp. The lower limit of detection of the method was therefore selected as the average (104 bp) + 3 SD or 369 bp. Differences in TRF of fewer than 369 bp were considered not significant and ascribed to inherent experimental variability. One to 5 (median = 3) replicate experiments were performed with DNA from each of the 17 patient/donor pairs for a total of 50 TRF pairs (in 2 cases, only 1 experiment was done owing to limited DNA quantities). (A) Representative TRF analysis of patient/donor pairs 5, 10, and 14. High MW DNA was digested with HinfI and RsaI restriction enzymes and size-fractionated on a 0.8% agarose gel. Recipient and donor samples were loaded in duplicate lanes 1-4 (pair 5), 5-8 (pair 10), and 9-12 (pair 14), and a radiolabeled 1-kb DNA ladder MW marker was included (lanes M). In situ hybridization was performed with a P32 end-radiolabeled oligonucleotide probe comprising the telomere-specific sequence (TTAGGG)3. The signal was detected by scanning the gel by means of the Phosphorimager analysis system (Molecular Dynamics, Sunnyvale, CA). Mean TRF length was assigned to the MW corresponding to the distance of peak signal intensity from the origin of the electrophoresis. The MW was then calculated by means of the Imagequant (Molecular Dynamics) and Fragment (Molecular Dynamics) software. (B) dTRF data summary of each patient/donor pair. Each column represents the average dTRF of 1 to 5 (median 3) replicate experiments for each pair. The top and bottom ends of the vertical lines depict the maximum and minimum dTRF values obtained in each patient/donor pair. Single experiments were performed on pairs 6 and 8 because of limited amounts of DNA. The 2 thin dotted horizontal lines depict the experimental limit of detection, which was 369 bp as described in “Materials and methods.” When the average difference was computed across experiments for each individual patient, 15 of 17 recipients had dTRF more than 0.369 kb. Of the 15 recipients, 8 had dTRF of more than 1.0 kb and 7 had dTRF 0.3 to 1.0 kb. One of the remaining 2 recipients had dTRF of −0.1 kb, and the other, dTRF of 0.2 kb, both less than 0.369 kb.
Our findings indicate that, although telomere shortening was consistently seen in long-term marrow recipients, the degree of telomere shortening was variable among recipients and, on average, was not greater than what has been previously reported early after transplant.10-13,26,27 If we accept that telomere shortening reflects an increased number of stem cell divisions after transplantation, then the observed variability among patients suggests that either a variable number of HSCs contributed to engraftment and hematopoietic reconstitution or secondary demands in hematopoiesis vary among these recipients. Furthermore, assuming that telomeres lose 100 bp per cell division and given an average dTRF of 0.94 kb, transplanted HSCs would theoretically undergo an average of 9 to 10 extra divisions after transplant. In granulocytes, telomere length is estimated to decrease by 30 bp per year23; thus a decrease of 0.94 kb would correspond to 30 years of normal hematopoiesis, consistent with previous theoretical estimates.28 The similar degree of telomere shortening in both short- and long-term recipients is consistent with a model in which demand for HSC replication stabilizes following an initial accelerated period. Late after transplant, the demand for stem cell replication appears to be no greater than in the normal donor. A more rapid loss of telomere length over time relative to the donor may occur in a setting in which donor-derived hematopoietic reconstitution is monoclonal or oligoclonal.
In summary, our findings suggest that many years after BMT, despite increased demands early after transplantation, donor-derived hematopoiesis can sustain normal counts and remains polyclonal. These observations emphasize the extensive replicative reserve of HSCs.
Supported in part by National Institutes of Health grants HL36444, CA18221, CA15704, and CA09515, as well as by a Young Investigator Award presented to G.M. by the American Society of Clinical Oncology.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
George Mathioudakis, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, Mailstop D1-100, PO Box 19024, Seattle, WA 98109-1024; e-mail: gmathiou@fhcrc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal