Abstract
The t(11;19)(q23;p13.1) chromosomal translocation in acute myeloid leukemias fuses the gene encoding transcriptional elongation factor ELL to the MLL gene with consequent expression of an MLL-ELL chimeric protein. To identify potential mechanisms of leukemogenesis by MLL-ELL, its transcriptional and oncogenic properties were investigated. Fusion with MLL preserves the transcriptional elongation activity of ELL but relocalizes it from a diffuse nuclear distribution to the nuclear bodies characteristic of MLL. Using a serial replating assay, it was demonstrated that the MLL-ELL chimeric protein is capable of immortalizing clonogenic myeloid progenitors in vitro after its retroviral transduction into primary murine hematopoietic cells. However, a structure–function analysis indicates that the elongation domain is not essential for myeloid transformation because mutants lacking elongation activity retain a potent ability to immortalize myeloid progenitors. Rather, the highly conserved carboxyl terminal R4 domain is both a necessary and a sufficient contribution from ELL for the immortalizing activity associated with MLL-ELL. The R4 domain demonstrates potent transcriptional activation properties and is required for transactivation of a HoxA7 promoter by MLL-ELL in a transient transcriptional assay. These data indicate that neoplastic transformation by the MLL-ELL fusion protein is likely to result from aberrant transcriptional activation of MLLtarget genes. Thus, in spite of the extensive diversity of MLL fusion partners, these data, in conjunction with previous studies of MLL-ENL, suggest that conversion of MLL to a constitutive transcriptional activator may be a general model for its oncogenic conversion in myeloid leukemias.
Introduction
The development of more effective therapies for de novo and therapy-related acute myeloid leukemia (AML) represents a major challenge for oncology. Insights into the molecular pathogenesis of AML have come from the study of acquired chromosomal rearrangements specific to the leukemic clone. In particular, reciprocal translocations fusing the MLL (ALL-1, HRX, Htrx) gene on chromosome 11q23 with the ELL(MEN) gene at chromosome band 19p13.1 are found in a large proportion of AML arising in cancer patients previously treated with etoposide-based chemotherapy.1-3 The t(11;19)(q23;p13.1) translocation has also been identified in association with myeloid leukemias in infants and de novo AML and myelodysplastic syndromes in children and adults, suggesting that this rearrangement is not limited to chemotherapy-related mutagenesis.4,5 The consistent retention of a der(11) chromosome that encodes a translatableMLL fusion mRNA in the leukemic clone6 7 implies a transforming function for the chimeric protein encoded by the 5′-MLL-ELL-3′ transcript. However, the mechanism of transformation by MLL-ELL remains unclear.
ELL is one of more than 30 genes fused to the MLLgene in leukemia-associated translocations (reviewed in DiMartino and Cleary8). MLL normally functions as an upstream regulator of Hox gene expression9 and contains amino terminal DNA-binding domains that are retained by the chimeric proteins (Figure 1). Studies of leukemogenesis by MLL-AF9 in vivo and MLL-ENL in vitro provide compelling evidence that the amino terminal portion of MLL gains a transforming function from its partner proteins.10-12 No obvious structural features shared among the broader group of fusion partners, however, have emerged to explain their function in transformation. The lack of functional information about these proteins has made it difficult to generate testable hypotheses regarding their contributions to transformation as MLL fusion partners.
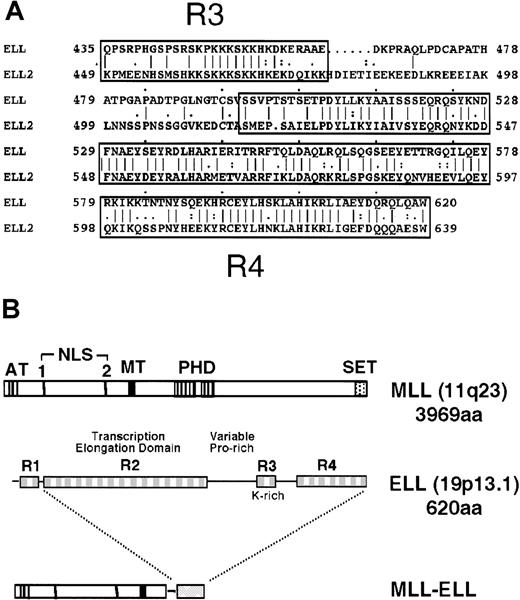
Diagrammatic representation of the ELL and MLL proteins.
(A) Sequence alignment of ELL and its closely related family member, ELL2, shows homology over their carboxyl-terminal sequences. (B) The MLL gene, located at chromosomal band 11q23, encodes a 3969-amino acid protein with A-T hooks (AT), 2 nuclear localization sequences (NLS), methyltransferase-like domain (MT), PHD-type zinc fingers, and a carboxy-terminal SET domain. ELL comprises 4 distinct domains (R1-R4) described in the text. Translocation t(11;19)(q23;p13.1) joins amino terminal regions of MLL to domains R2-R4 of ELL.
Diagrammatic representation of the ELL and MLL proteins.
(A) Sequence alignment of ELL and its closely related family member, ELL2, shows homology over their carboxyl-terminal sequences. (B) The MLL gene, located at chromosomal band 11q23, encodes a 3969-amino acid protein with A-T hooks (AT), 2 nuclear localization sequences (NLS), methyltransferase-like domain (MT), PHD-type zinc fingers, and a carboxy-terminal SET domain. ELL comprises 4 distinct domains (R1-R4) described in the text. Translocation t(11;19)(q23;p13.1) joins amino terminal regions of MLL to domains R2-R4 of ELL.
ELL is one of the few MLL fusion partners for which a substantial amount is known about its biochemical role. This 620-amino acid protein has been shown to facilitate transcriptional elongation by suppressing transient pausing of RNA polymerase II along the DNA template.13 In addition, the amino terminal 50 amino acids of ELL mediate negative regulation of promoter-specific transcription initiation by RNA polymerase II.14 ELL2, another member of the ELL family of proteins, was identified by its homology with ELL. These 2 proteins share extensive similarity in their transcriptional elongation domains (R2) and their lysine-rich (R3) and carboxyl-terminal (R4) domains (Figure 1A).15 The presence of an RNA polymerase II inhibition (R1) domain, however, is specific to ELL. Although highly conserved, the functions of the R3 and R4 domains are unknown.
The well-characterized biochemical functions and structural features of ELL provide an excellent opportunity to define its functional contributions to transformation by MLL-ELL. Leukemia-associated translocations at 11q23 join the R2-R4 domains of ELL to the putative DNA-binding domains of MLL (Figure 1B). It is unclear whether the loss of the RNA polymerase II inhibiting R1 motif contributes to the oncogenic properties of the chimeric protein. Moreover, it is unknown whether the elongation-promoting activity of ELL is retained after its fusion to MLL. In addition to the unknown effect(s) that fusion to MLL would have on the functional properties of ELL, it is unclear how its subcellular distribution would be affected. Fusion of several partner proteins to MLL results in their colocalization with MLL.16-18 Thus, 11q23 translocations involving ELL may create a state of haplo-insufficiency by sequestering ELL away from its normal location. To address these questions, we have assayed the transcriptional elongation activity of MLL-ELL fusion proteins, characterized their subnuclear localization, and defined the domains of ELL that are required for the immortalization of primary murine myeloid progenitors. We report that a novel transcriptional transactivation domain at the carboxyl terminus of ELL, rather than its elongation function, is essential for transformation by MLL-ELL.
Materials and methods
Plasmid constructs
The MLL5′ cloning vector was made by digesting the MSCVneo-HRX-ENL plasmid11 with PflmI andXhoI. ENL sequences flanked by these restriction sites were then replaced by a synthetic oligonucleotide containingNruI, PmlI, and HpaI sites and stop codons in all 3 reading frames. MLL-ELL, MLL-ELLΔ150-200, and MLL-ELLΔ374-620 were made by ligating the respective ELL clones14 into the PmlI site of MLL5′. For MLL-ELLC1 and MLL-ELLC2, 2 sets of 5′ end oligonucleotides with SalI restriction sites were used (C1 5′-GCACTGTCGA CAAGCCCAAGAAGAAGTCCAAG-3′and C2 5′-GCACTGTCGACGCGG CTGAGGACAAGCCCCGG-3′) with the same 3′ end oligo as that used to clone ELL in M13 vector.13 Polymerase chain reactions were set with the above appropriate oligos in 400 μL reactions with Tli polymerase (Promega, Madison, WI). Polymerase chain reaction products were digested with SalI, filled in with Klenow, phosphorylated with T4 polynucleotide kinase (Promega), and subcloned into murine stem cell virus (MSCV)–MLL vector digested withPmlI as described for other MLL-ELL chimeras. The FLAG-MLL expression construct was made by inserting a FLAG epitope tag into the MLL expression vector TK-88219 20 (a kind gift of Dr Masao Seto).
Transcriptional elongation assays
Briefly, pre-initiation complexes were assembled by pre-incubation of purified RNA polymerase II, recombinant TBP, recombinant TFIIB, recombinant TFIIE, and purified TFIIH with DNA template containing the adenoviral major late (AdML) promoter. Short, highly radioactive transcripts were synthesized during a brief pulse carried out in the presence of ATP, GTP, UTP, and a limiting concentration of γ-32P]-CTP. These short promoter-specific transcripts were then elongated to full-length runoff transcripts in the presence of MLL-ELL chimeras and an excess of nonradioactive CTP. Transcripts were analyzed by electrophoresis through a 6% polyacrylamide, 7.0 mol/L urea gel and visualized using a Molecular Dynamics PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Immunofluorescence staining
Mouse embryonic fibroblasts were transfected with 500 ng of either FLAG-MLL, ELL, MLL-ELL, MLL-ELLΔ150-200, or MLL-ELLΔ374-620 expression vectors in 4-well chamber slides using Lipofectamine (Life Technologies, Rockville, MD). Forty-eight hours later, cells were fixed with cold 4% paraformaldehyde in phosphate-buffered saline (PBS) and were permeabilized with 0.2% Triton X-100/PBS. Slides were blocked with 2% BSA/PBS and stained with the M2 FLAG monoclonal antibody (10 μg/mL) (Sigma, St Louis, MO) or a monoclonal antibody directed against ELL (1:400) in PBS/2% horse serum, followed by staining with goat antimouse immunoglobulin G conjugated to Alexa 488 in PBS/2% horse serum (1:400; Molecular Probes, Eugene, OR). Slides were washed 3 times with PBS/0.05% Triton X100 after each incubation period and were finally washed with 3 changes of PBS before they were cover-slipped. Slides were photographed with an Olympus (Tokyo, Japan) BX40 microscope equipped for immunofluorescence (original magnification, 1000 ×).
Myeloid immortalization assays
Transduction of murine bone marrow (BM) cells was performed essentially as described in Lavau et al.11 Briefly, 4-week-old C57B/6 mice were injected with 5-FU (150 mg/kg) by tail vein, and bone marrow was harvested from both femurs after 5 days. Transient retroviral supernatants were produced by transfecting φNX cells with MSCV vectors as described by Pear et al.21 Bone marrow cells were infected with recombinant virus by centrifugation at 2500×g for 2 hours at 32°C. Transduced cells were plated in 0.9% methylcellulose (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with 20 ng/mL stem cell factor (SCF) and 10 ng/mL each of interleukin-3 (IL-3), IL-6, and granulocyte macrophage–colony-stimulating factor (GM-CSF) (R&D Systems, Minneapolis, MN) in the presence or absence of G418 (1 mg/mL). After 7 to 10 days, colonies were counted and retroviral transduction efficiency was calculated for each construct as the ratio of G418-resistant (G418R) colonies to unselected colonies. Single-cell suspensions were made from pooled G418R colonies, and 104 cells were plated in secondary methylcellulose cultures without G418.
Cell surface phenotype analysis
Flow cytometry was performed by staining cells with phycoerythrin and fluorescein isothiocyanate isotype controls and with monoclonal antibody (mAb) for Gr-1, Mac-1, c-Kit, and other markers (Pharmingen, San Diego, CA). Stained cells were analyzed on a FACScan flow cytometer (Becton-Dickinson, Mountain View, CA).
Western blotting
A confluent 100-mm dish of φNX cells transiently transfected with each construct was lysed in 400 μL 1× sample buffer. Approximately 20μL lysate was loaded in each lane and electrophoresed through a 6% sodium dodecyl sulfate–polyacrylamide gel. Proteins were transferred to Immobilon-P membranes (Millipore, Bedford, MA) using CAPS transfer buffer as described in Slany et al.12 After blocking with 5% milk, membranes were probed with monoclonal antibody N6.3 directed against an MLL amino terminal epitope between residues 161 and 356.
Transcriptional activation assays
Four micrograms pGL3-HoxA7 (kind gift of Dr R. Slany) was cotransfected with 4 μg MSCVneo, MLL5′, MLL-ELL, MLL-ELLΔ374-620, or MLL-ELLC2 and 2 μg pCMVsportβgal (Gibco BRL, Grand Island, NY) into 293T cells by calcium-phosphate precipitation. After 48 hours, lysates were assayed for luciferase and β-galactosidase activity using commercially prepared reagents (Promega; Tropix, Bedford, MA) and a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA). Relative light units (RLU) from duplicate luciferase assays were corrected for transfection efficiency using RLU from their respective β-galactosidase controls.
Results
MLL-ELL stimulates transcriptional elongation by RNA polymerase II
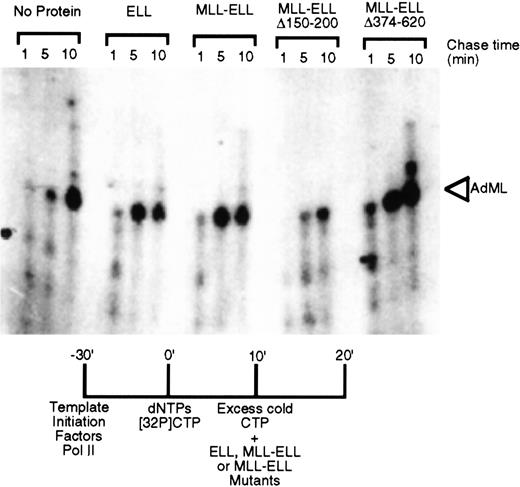
To determine whether MLL-ELL retained the transcriptional elongation activity of ELL, the chimeric protein was evaluated for its ability to increase the catalytic rate of transcription elongation from the AdML promoter in the presence of general initiation factors. Analysis of the kinetics of accumulation of full-length runoff transcripts revealed that MLL-ELL stimulated the rate of transcription elongation of promoter-specific transcription by RNA polymerase II as effectively as native ELL (Figure 2). We previously demonstrated that the elongation stimulation domain of both ELL and ELL2 resides in their respective R2 domains and that deletion of amino acids 150 to 200 resulted in the loss of transcriptional elongation stimulatory activity of these proteins.14 15Similarly, deletion of these amino acid residues from MLL-ELL significantly compromised its ability to stimulate the rate of transcriptional elongation (Figure 2). In contrast, the removal of C-terminal ELL amino acids 374 to 620 had no effect on elongation by either native ELL or its MLL chimera.
Transcriptional elongation activity of MLL-ELL chimeras.
ELL, MLL-ELL, or the indicated deletion mutants were tested for their ability to stimulate elongation of a labeled transcript by RNA polymerase II in vitro. Accumulation of the full-length transcript after 1, 5, and 10 minutes provides an indication of the stimulatory activity of the added proteins.
Transcriptional elongation activity of MLL-ELL chimeras.
ELL, MLL-ELL, or the indicated deletion mutants were tested for their ability to stimulate elongation of a labeled transcript by RNA polymerase II in vitro. Accumulation of the full-length transcript after 1, 5, and 10 minutes provides an indication of the stimulatory activity of the added proteins.
Subnuclear localization of ELL is altered by its fusion to MLL
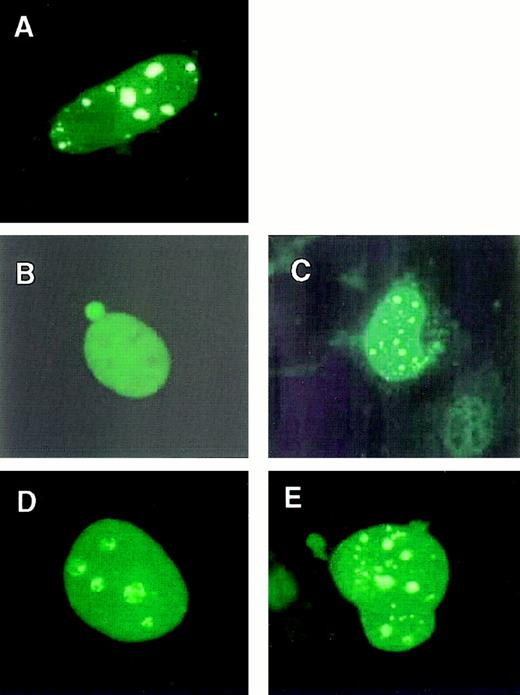
Because MLL-ELL retained an ability to stimulate transcriptional elongation, we determined whether this activity was present in the same nuclear compartment as that of native ELL. Mouse embryo fibroblasts were transfected with constructs expressing MLL-ELL fusion proteins or the respective wild-type proteins. Immunofluorescent staining with an ELL-specific mAb showed that ELL exhibited a diffuse nuclear distribution (Figure 3B). In contrast, MLL displayed a punctate and diffuse localization pattern as described16 (Figure 3A). Analysis of cells expressing either MLL-ELL or its mutants with an anti-ELL mAb showed a punctate and diffuse localization pattern identical to that of MLL (Figure3C-E). This indicated that fusion of ELL with MLL resulted in a relocalization of ELL from its normal diffuse nuclear distribution to the nuclear bodies characteristic of MLL localization.
Subnuclear localizations of MLL-ELL and wild-type ELL.
Mouse embryo fibroblasts were transfected with FLAG-tagged MLL (A), wild-type ELL (B), MLL-ELL (C), MLL-ELLΔ150-200 (D), or MLL-ELLΔ374-620 (E). Transfected mouse embryo fibroblasts were stained with a FLAG-specific mAb (A) to detect wild-type MLL or an ELL-specific mAb (B-E) to detect wild-type and chimeric ELL proteins. ELL exhibited a diffuse pattern, whereas ELL fusion proteins were distributed in a punctate and diffuse pattern indistinguishable from that of wild-type MLL.
Subnuclear localizations of MLL-ELL and wild-type ELL.
Mouse embryo fibroblasts were transfected with FLAG-tagged MLL (A), wild-type ELL (B), MLL-ELL (C), MLL-ELLΔ150-200 (D), or MLL-ELLΔ374-620 (E). Transfected mouse embryo fibroblasts were stained with a FLAG-specific mAb (A) to detect wild-type MLL or an ELL-specific mAb (B-E) to detect wild-type and chimeric ELL proteins. ELL exhibited a diffuse pattern, whereas ELL fusion proteins were distributed in a punctate and diffuse pattern indistinguishable from that of wild-type MLL.
MLL-ELL immortalizes primitive myeloid progenitors
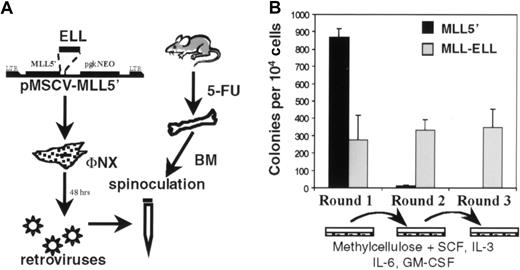
To study the oncogenic properties of MLL-ELL, the MSCV retroviral vector22 was used to transduce BM cells whose growth properties were then evaluated in a serial replating assay (Figure4A). Previously, we showed that retroviral transduction of primary BM cells with an MLL-ENLfusion cDNA under these conditions enhanced the self-renewal of a clonogenic myeloid progenitor that induced AML in transplanted mice.11 12 Using similar methodology,MLL-ELL–transduced BM cells were plated in methylcellulose cultures supplemented with SCF, IL-3, IL-6, and GM-CSF. Because the MSCV vector contained a pgkNeo cassette, G418 was also added to first-round cultures to select for transduced cells. In the first round of plating, numerous colonies were observed for cells transduced with empty MSCV vector (not shown) or MLL5′ cDNA (Figure 4B) reflecting the normal clonogenic potential of progenitors harvested from 5-FU–treated BM. Moreover, mock-transduced BM plated in the absence of G418 gave similar numbers of colonies as retrovirally transduced BM, suggesting that retroviral infection per se was not overtly toxic to these cells. Fewer Neo-resistant colonies were observed in first-round cultures of MLL-ELL–transduced BM (Figure 4B), consistent with the lower retroviral transduction efficiency obtained with this construct compared to MLL5′. As expected, the colonies were primarily of GM morphology, and their appearance did not vary appreciably betweenMLL-ELL– and MLL5′–transduced cultures (not shown). First-round colonies were then harvested, and 104cells were plated in secondary methylcellulose cultures. In contrast toMLL5′–transduced cells that had exhausted their self-renewal potential and produced only a few macrophage colonies,MLL-ELL–transduced cells produced hundreds of colonies with primitive GM morphology in second-round cultures. This effect was sustained through subsequent rounds of plating (Figure 4B and data not shown).
Immortalization of primary murine myeloid progenitors by MLL-ELL.
(A) Schematic representation of retroviral transduction protocol. (B) Colonies generated per 104 infected BM cells were determined in first-, second-, and third-round cultures for MLL5′ and MLL-ELL constructs. First-round bars represent G418Rcolonies. Bars represent the mean ± SD for 3 experiments.
Immortalization of primary murine myeloid progenitors by MLL-ELL.
(A) Schematic representation of retroviral transduction protocol. (B) Colonies generated per 104 infected BM cells were determined in first-, second-, and third-round cultures for MLL5′ and MLL-ELL constructs. First-round bars represent G418Rcolonies. Bars represent the mean ± SD for 3 experiments.
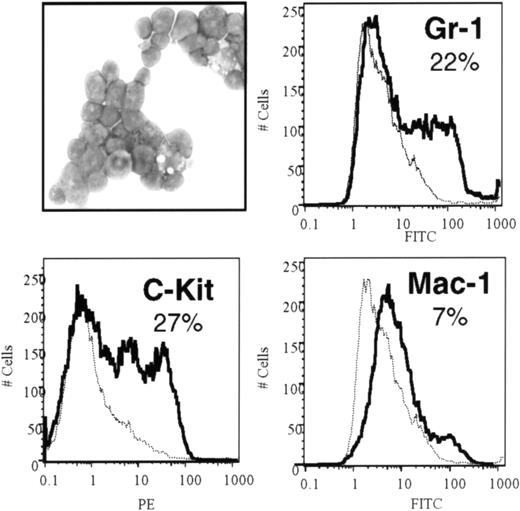
The ability to produce GM colonies in methylcellulose cultures is a property of primitive myeloid progenitors. May-Grünwald-Giemsa–stained cytospin preparations of these cells showed a range of morphologies. Myeloblastic cells with large nuclei and no obvious granules were seen along with cells showing morphologic evidence of monocytic and granulocytic differentiation (Figure5). Consistent with the degree of heterogeneity seen in stained cytospins, analysis of surface marker expression by flow cytometry revealed a range of myeloid differentiation (Figure 5). Less than 10% of cells expressed low levels of Mac-1, whereas 27% expressed substantial levels of c-Kit, a marker associated with primitive myeloid cells. Twenty-two percent of cells also expressed Gr-1, demonstrating a degree of granulocytic differentiation. Expression of erythroid (Ter119) or lymphoid (B220, Thy1.2) differentiation antigens was not observed (not shown). Thus,MLL-ELL–transduced cells were immortalized at a primitive stage of myeloid differentiation, consistent with their high clonogenic potential. Similar to MLL-ENL–transformed cells, however, MLL-ELL–transformed cells exhibited a range of myeloid differentiation characteristics.11 This suggested that primitive myeloid cells transduced with either of these MLL fusion proteins retained a limited capacity for myeloid differentiation in vitro.
Characterization of MLL-ELL–immortalized cells.
Mae-Grünwald-Giemsa–stained cytospin preparation (upper left) shows a range of primitive myeloid morphology. FACS analysis of MLL-ELL–transduced cells was performed after third-round cultures. Morphology and expression of c-Kit, Gr-1, and low levels of Mac-1 are consistent with primitive myeloid differentiation.
Characterization of MLL-ELL–immortalized cells.
Mae-Grünwald-Giemsa–stained cytospin preparation (upper left) shows a range of primitive myeloid morphology. FACS analysis of MLL-ELL–transduced cells was performed after third-round cultures. Morphology and expression of c-Kit, Gr-1, and low levels of Mac-1 are consistent with primitive myeloid differentiation.
ELL elongation activity is dispensable, but its C-terminal domain is necessary and sufficient for immortalization of primitive myeloid progenitors
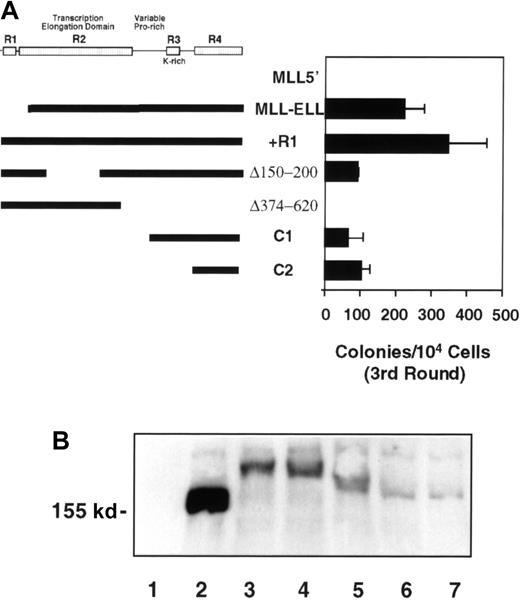
To identify domains in ELL that were required for the observed effects of MLL-ELL on the clonogenic potential of myeloid cells, a series of mutants was tested for immortalizing potential. Consistent production of third-round colonies, regardless of number, was considered to be indicative of immortalization activity. Construct MLL-ELLR1 contained all of ELL, including the RNA polymerase II inhibition domain (R1) that is not included in translocation-generated MLL-ELL chimeras. The presence of R1 in MLL-ELL did not inhibit its ability to enhance the clonogenic potential of primary 5-FU–treated BM cells (Figure6A). This observation correlated with our finding that the R1 domain no longer inhibited RNA polymerase II initiation as part of a chimeric MLL-ELLR1 protein (A. Shilatifard, unpublished observation). Deletion of ELL amino acids 150 to 200 (construct MLL-ELLΔ150-200), which removed most of the R2 domain and significantly diminished the transcriptional elongation activity of both native ELL and MLL-ELL (Figure 2), had only a modest effect on the number of third-round colonies produced (Figure6A). In contrast, deletion of amino acids 374 to 620 at the C-terminus of ELL (MLL- ELLΔ374-620), which removed both the lysine-rich R3 domain and the highly conserved R4 domain, completely abrogated immortalization. Conversely, a construct containing only the R3 and R4 domains of ELL fused to MLL (MLL-ELLC1) resulted in the production of substantial numbers of third-round colonies. A fusion construct in which the lysine-rich region (R3) of ELL was removed, leaving only the R4 domain fused to MLL (MLL-ELLC2), showed similarly efficient immortalizing properties. Taken together, these data showed that the R4 domain was both necessary and sufficient for the immortalization of primary murine myeloid progenitors.
Identification of ELL sequences required for immortalization of primary murine myeloid progenitors.
(A) Thick lines under the schematic of ELL indicate sequences fused to MLL5′; horizontal bars indicate the number of colonies generated per 104 cells plated in third-round cultures. Bars represent the mean ± SD of 3 independent experiments. Retroviral transduction efficiency for the various constructs was 60% (MLL-5′), 39% (MLL-ELL), 34% (+R1), 33% (Δ150-200), 29% (Δ374-620), 27% (C1), and 43% (C2). (B) Western blot analysis of fusion protein expression in transiently transfected φNX cells. Lane 1, MSCV vector; lane 2, MLL5′; lane 3, MLL-ELL; lane 4, MLL-ELLΔ150-200; lane 5, MLL-ELLΔ374-620; lane 6, MLL-ELLC1; lane 7, MLL-ELLC2.
Identification of ELL sequences required for immortalization of primary murine myeloid progenitors.
(A) Thick lines under the schematic of ELL indicate sequences fused to MLL5′; horizontal bars indicate the number of colonies generated per 104 cells plated in third-round cultures. Bars represent the mean ± SD of 3 independent experiments. Retroviral transduction efficiency for the various constructs was 60% (MLL-5′), 39% (MLL-ELL), 34% (+R1), 33% (Δ150-200), 29% (Δ374-620), 27% (C1), and 43% (C2). (B) Western blot analysis of fusion protein expression in transiently transfected φNX cells. Lane 1, MSCV vector; lane 2, MLL5′; lane 3, MLL-ELL; lane 4, MLL-ELLΔ150-200; lane 5, MLL-ELLΔ374-620; lane 6, MLL-ELLC1; lane 7, MLL-ELLC2.
The inability of MLL5′ and MLL-ELLΔ374-620 to immortalize myeloid progenitors was not the result of lack of expression. Western blotting of lysates from transiently transfected φNX cells confirmed that all the mutant MLL-ELL fusion proteins were expressed and that they displayed the expected molecular weights (Figure 6B). Interestingly, the nonimmortalizing MLL5′ protein consistently accumulated to very high levels relative to the MLL-ELL fusion proteins. It was unclear whether this was because of more efficient transcription/translation or enhanced stability of the transcript or protein. Nevertheless, this demonstrated that ELL did not contribute to transformation by simply conferring enhanced stability to the truncated MLL protein but rather, that ELL conferred a gain of function to MLL.
MLL-ELLC2 immortalizes cells in an IL-3–dependent manner
To determine whether MLL-ELLC2–transduced cells were capable of sustained growth in suspension cultures, colonies from third-round platings were placed in liquid media supplemented with various combinations of cytokines. Cells initially grew in RPMI 1640 media with 10% fetal calf serum (FCS) and recombinant SCF, IL-3, and IL-6 with a doubling time of 24 to 48 hours (not shown). Subsequently, MLL-ELLC2–transduced cells were passaged in RPMI 1640 with 20% FCS and 20% WEHI-conditioned media (WCM) as a source of IL-3. These cells have been maintained in continuous culture for more than 3 months without any change in their doubling time. Seven to 10 days after the removal of WCM, MLL-ELLC2–transduced cells cease proliferating. These data suggest that, similar to MLL-ENL–transformed myeloid progenitors, cells immortalized by MLL-ELLC2require exogenous IL-3 for sustained viability or growth in liquid culture. Unlike the former, however, the injection of 106MLL-ELLC2–immortalized cells into SCID mice has not yet produced myeloid leukemias 90 days after injection.
ELL R4 domain confers transcriptional activation activity to MLL-ELL
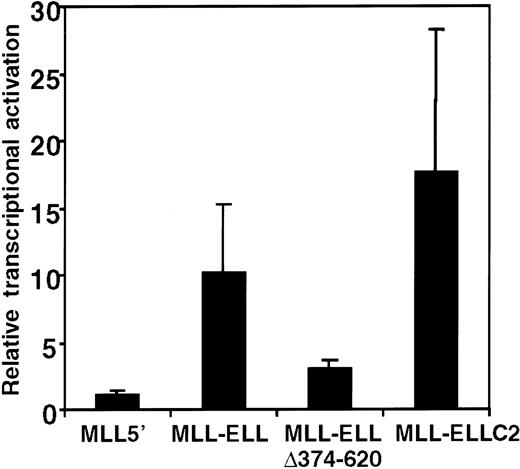
The foregoing structure–function analysis identified the portion of ELL that was required for the immortalizing activity of MLL-ELL; however, the biochemical function of the R4 domain remained unknown. Our previous studies of MLL-ENL showed that the portion of ENL required for MLL-ENL–mediated transformation exhibited transcriptional activation properties when fused to a GAL4 DNA-binding domain.12 Subsequently, MLL-ENL itself was shown to be capable of activating transcription of a luciferase reporter gene under the control of HoxA7 upstream sequences (pGL3-HoxA7) in a transient transcriptional assay.23 To determine whether the R4 domain of ELL may confer similar properties on MLL-ELL, the transcriptional properties of various MLL-ELL proteins were evaluated in 293T cells cotransfected with the pGL3-HoxA7 reporter gene. Under these conditions, MLL-ELL displayed significant transactivation potential, inducing luciferase expression 10- to 15-fold above the levels observed for MLL5′ or vector alone (Figure 7). This induction by MLL-ELL was not observed when a luciferase reporter gene under the control of the E1B promoter and GAL4 UAS was used (not shown), indicating that HoxA7 upstream sequences were required for MLL-ELL–mediated activation. The MLL-ELLΔ374-620 mutant protein lacking the 247 C-terminal residues of ELL was substantially compromised in its activation potential, which was reduced to levels near background. In contrast, MLL-ELLC2 containing the R4 domain of ELL alone fused to MLL retained transcriptional activation potential equal to or greater than the full-length fusion protein. The R4 domain of ELL was, therefore, both necessary and sufficient for transcriptional activation, and for immortalization of primary murine myeloid progenitors, when fused with 5′ MLL.
Transcriptional activation by MLL-ELL.
A luciferase reporter gene under control of HoxA7 upstream sequences (pGL3-HoxA7) was cotransfected into 293T cells with expression constructs encoding MLL5′ or various MLL-ELL fusion cDNAs. Luciferase activity was corrected for transfection efficiency based on the activity of a cotransfected β-galactosidase expression construct. The transcriptional activating potential for each MLL construct is expressed as the fold-induction relative to pMSCVneo. Bars represent the mean ± SD of 3 independent transfection experiments.
Transcriptional activation by MLL-ELL.
A luciferase reporter gene under control of HoxA7 upstream sequences (pGL3-HoxA7) was cotransfected into 293T cells with expression constructs encoding MLL5′ or various MLL-ELL fusion cDNAs. Luciferase activity was corrected for transfection efficiency based on the activity of a cotransfected β-galactosidase expression construct. The transcriptional activating potential for each MLL construct is expressed as the fold-induction relative to pMSCVneo. Bars represent the mean ± SD of 3 independent transfection experiments.
Discussion
The studies in this report evaluated the functional properties of MLL-ELL to establish the oncogenic contributions of ELL, a known transcriptional elongator. Transcriptional elongation assays using MLL-ELL showed that ELL retains its elongation function in the context of an MLL chimera. However, fusion of ELL to MLL alters its distribution within the nucleus, localizing it to the distinct nuclear bodies in which MLL is also localized. The latter is consistent with previous observations18 showing that MLL fusion partners, including those whose normal localization is cytoplasmic, are relocalized within the nucleus after fusion with MLL. Taken together, these findings appear to support a mechanism by which aberrant elongation activity at MLL target loci by MLL-ELL accounts for deregulated gene expression in myeloid leukemias. Based on our data, however, it is highly unlikely that this mechanism is responsible for the transforming function of MLL-ELL. Rather, our data provide evidence for a mechanism in which MLL-ELL must function as a chimeric transcriptional activator as opposed to an elongator to transform myeloid progenitors. As the second MLL fusion protein to demonstrate such activity, our data suggest that constitutive activation may be a general model for the oncogenic role of MLL chimeric proteins.
ELL has been shown in previous studies to be multifunctional, having both transcriptional elongation activity and inhibitory effects on transcriptional initiation by RNA polymerase II. The data in the current report demonstrate that the domains of ELL that confer these transcriptional activities are not necessary for the immortalization of primary murine myeloid progenitors by MLL-ELL. Rather, a new functional domain of ELL was defined by our studies. The carboxy-terminal R4 domain, which is highly conserved within the ELL family of proteins, was found to have potent transcriptional activation properties in transient transcriptional assays. The R4 domain does not display similarity with other known transcriptional activation domains, but its molecular role is likely to reflect an ability to interact with one or more components of the cellular machinery that regulates activated transcription. The fact that this activity did not read out in previous in vitro studies of ELL raises the possibility that it was masked by the inhibitory role of the amino-terminal R1 domain of ELL on RNA polymerase II initiation, which is not included in translocation-generated MLL-ELL fusion proteins. Alternatively, necessary cofactors may not have been present in the extracts used for previous in vitro studies, or the activation properties of ELL may be promoter-specific.
Identification of the ELL R4 domain as critical for immortalization is likely to be highly relevant to leukemogenesis induced by MLL-ELL. Generation of colonies in the third round of plating in our cultures reflects the ability of MLL-ELL to enhance the self-renewal of primitive myeloid progenitors. Such immortalizing activity may parallel the first steps of leukemogenesis in cells having undergone an 11q23 rearrangement. Indeed, the primitive morphology and surface phenotype of MLL-ELL–immortalized cells resemble those of leukemic blasts in human AML, further substantiating the link between immortalization in our in vitro assay and de novo leukemic transformation. Moreover, others have shown that primary murine BM cells transduced with MLL-ELL induce AML after approximately 150 days in transplanted mice.24 The failure of MLL-ELLC2 immortalized cells to produce AML in SCID mice after 90 days may be due to an insufficient latency period. These animals will continue to be monitored for the development of myeloid leukemia.
Our studies do not exclude the possibility that transcriptional elongation by MLL-ELL, though not essential, may contribute to myeloid transformation. We observed an approximately 50% decrease in the numbers of third-round colonies between full-length MLL-ELL and MLL-ELLΔ150-200, which lacks a function R2 elongation domain. However, in previous studies, we found no correlation between the number of third-round colonies and the ability of myeloid cells to induce tumors in animals either for MLL fusion genes or other oncogenes that immortalize myeloid progenitors in this assay. More significant was the failure of MLL- ELLΔ374-620 to induce immortalization of myeloid progenitors despite protein expression levels comparable to those of full-length MLL-ELL. This clearly indicates the presence of an essential domain for myeloid transformation within the carboxy-terminus of ELL. Of the 2 domains within this region, R4 alone was sufficient for immortalization. Thus, the R4 domain was necessary and sufficient for immortalization, indicating that the R2 elongation domain serves at best a subsidiary role and is nonessential.
We examined MLL-ELL for transcriptional activating function to address the general applicability of observations made with MLL-ENL, whose oncogenic contributions were shown previously to be intimately associated with its transcriptional activation function. The transactivation potential of ENL, when fused to a heterologous DNA-binding domain, and its contribution to the transformation activity of MLL-ENL co-localize to conserved motifs in the C-terminus of ENL.25,12 Recently, MLL-ENL has been shown to activate transcription in a more physiologic context with the HoxA7promoter used for our current studies.23 MLL-ELL now also proves to have significant transactivation activity on this promoter. Interestingly, despite the lack of sequence homology between ENL and ELL, both proteins are predicted to exhibit helical secondary structure at their carboxyl termini based on PHDsec computer modeling26 (not shown). These findings support the hypothesis that fusion of MLL to at least a subset of partner proteins results in the creation of hybrid transcription factors. MLL is thought to function as an epigenetic regulator of transcription rather than as a classical transcriptional activator. It does not activate expression of its target genes but ensures that they remain in an activated state through cell division. The enhancer-like activity displayed in our transient transfection assays, therefore, reflects a true gain of function by MLL. Perhaps more important, the replacement of PHD finger and SET domains of MLL by a constitutive transcriptional activation domain may preclude its ability to down-regulate target genes in response to differentiation signals. Thus, both gain and loss of MLL function may lead to aberrant expression of target genes, resulting in leukemogenic transformation.
Acknowledgments
We thank C. Lavau for assistance in establishing the myeloid immortalization assay in our laboratory and Y. Jacobs and B.-T. Rouse for expert assistance in producing MLL monoclonal antibodies. We also thank M. Seto and R. Slany for providing the TK-882 and pGL3-HoxA7 constructs, respectively.
Supported in part by grants from the National Cancer Institute (CA55029 and 5T32CA09151), the National Institutes of Health (RO1CA78815-01), and the American Cancer Society (RP69921801). A.S. is an Edward Mallinckrodt Young Investigator.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jorge F. DiMartino, Department of Pathology, Stanford University School of Medicine, 300 Pasteur Drive, L-216, Stanford, CA 94305; e-mail: jorged@leland.stanford.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal