Abstract
In B-cell lymphomas, loss of human leukocyte antigen (HLA) class I and II molecules might contribute to immune escape from CD8+ and CD4+ cytotoxic T cells, especially because B cells can present their own idiotype. Loss of HLA expression and the possible underlying genomic alterations were studied in 28 testicular, 11 central nervous system, and 21 nodal diffuse large B-cell lymphomas (DLCLs), the first two sites are considered as immune-privileged sites. The analysis included immunohistochemistry, loss of heterozygosity analysis, and fluorescent in situ hybridization (FISH) on interphase cells and isolated DNA fibers. Total loss of HLA-A expression was found in 60% of the extranodal cases and in 10% of the nodal cases (P < .01), whereas loss of HLA-DR expression was found in 56% and 5%, respectively (P < .01). This was accompanied by extensive loss of heterozygosity within the HLA region in the extranodal DLCLs. In 3 cases, retention of heterozygosity for D6S1666 in the class II region suggested a homozygous deletion. This finding was confirmed by interphase FISH that showed homozygous deletions in the class II genes in 11 of the 18 extranodal lymphomas but in none of the 7 nodal DLCLs (P < .001). Mapping by fiber FISH showed variable deletions that always included HLA-DQ and HLA-DR genes. Hemizygous deletions and mitotic recombinations often involving all HLA genes were found in 13 of 18 extranodal and 2 of 7 nodal lymphomas. In conclusion, a structural loss of HLA class I and II expression might help the B-cell lymphoma cells to escape from immune attack.
Introduction
Approximately 40% of all non-Hodgkin lymphomas (NHLs) are diffuse large B-cell lymphomas (DLCLs), and from these approximately 40% present at extranodal sites, most commonly the gastrointestinal tract. Both the origin and the homing of the tumor cells play an important role in the distribution at specific nodal and extranodal sites.1 Few DLCLs present in the testis or in the central nervous system (CNS) that, together with the eye and ovary, are considered as immune-privileged sites.2,3 These are defined as sites in which immune responses do not take place or that proceed in a manner different from other sites. Strikingly, a link between the testis, the CNS, and the eye is supported by the observation that testicular DLCL cells preferentially disseminate to the contralateral testis and the CNS4 and that at least 20% of the primary CNS lymphomas disseminate to the eye.5 6 This link suggests that, in addition to specific homing mechanisms, tumor cells may be locally selected by topographical differences in immune attack.
The existence of an in vivo immune response against human cancer cells has been demonstrated for a variety of human tumors, especially virally mediated tumors. In other neoplasias, tumor-specific antigens are supposed to be a target of cytotoxic cells. In B-cell lymphomas, the immunoglobulin idiotype has raised much interest as a unique target for immunotherapy.7-9
So far, most attention has been given to the role of CD8+ cytotoxic T cells and in consequence to loss of human leukocyte antigen (HLA) class I expression on tumor cells as a route for immune escape.10,11 However, recent observations indicate that HLA class II and CD4 play an important role as well. First, HLA class II molecules on the tumor cells may be a direct target for CD4+ cytotoxic T cells.12-14 Second, to mount a sufficient immune response, CD8+ cytotoxic T cells need assistance by antigen-presenting cells (APCs) and CD4+helper T cells.15 Because, like their normal counterparts, neoplastic B cells potentially express these molecules as well as CD40 and costimulatory molecules, they might have a dual role as APCs and target cells for cytotoxic T cells.16 In consequence, loss of HLA molecules on neoplastic B cells likely contributes to the natural poor in vivo antitumor immune response and the failure of T-cell-directed immunotherapy.10,11 Interestingly, in a subset of aggressive B-cell lymphomas (which mainly represent DLCL), loss of HLA class I and class II expression has been described, and this loss correlated with extranodal disease17 and poor survival.18-20 Similarly, in a preliminary analysis of 258 DLCL, we found a relatively frequent loss of class II expression in primary extranodal DLCL. Seven DLCLs of the testis, considered as an immune-privileged site, showed a coordinate loss of class I and class II expression (unpublished results). At immune-privileged sites, several factors mediate immune escape2,3; however, the privilege is not absolute,21 implying that loss of HLA expression on the tumor cells might additionally help them to escape from killing by cytotoxic T cells.
The molecular mechanisms leading to loss of HLA class I molecules in carcinomas and melanoma include defects in expression of β2-microglobulin (β2M)22 and TAP23 but also genetic aberrations in the HLA class I genes at 6p21.10 24 In contrast, little is known about the mechanisms that lead to loss of HLA class II expression in B-cell lymphomas. The coordinate loss of class I and II expression in the testicular lymphomas prompted us to search for a common mechanism in a large series of these tumors. Here, we report on the analysis of 60 DLCLs, including 28 primary testicular, 11 primary CNS, and 21 primary nodal cases. A very frequent loss of expression of HLA class I and II was found in both types of extranodal lymphomas but not in nodal DLCLs. Molecular studies revealed very frequent homozygous deletions of the HLA-DR and -DQ region as well as larger hemizygous deletions and mitotic recombinations in the HLA class I and III region.
Materials and methods
Tumor samples
Formalin-fixed paraffin-embedded tissue blocks of 60 lymphomas with the histopathologic diagnosis of DLCL (Revised European-American Lymphoma classification)25 were collected. Assessment criteria for DLCL of the testis were age older than 15 years, original complaint of a unilateral or bilateral testicular mass with a histological diagnosis of a large B-cell lymphoma, and a complete staging procedure. Primary DLCLs of the CNS were diagnosed in patients without a history of immune deficiency, who presented with one or more cerebral or cerebellar masses with a histological diagnosis of a large B-cell lymphoma and who underwent complete staging procedures. In all cases, a B-cell origin was confirmed by immunohistological staining for CD19, CD20, CD22, or CD79a. From these DLCLs, 21 were of primary nodal origin, 28 of primary testicular origin, and 11 of primary CNS origin. Tissue blocks from 12 of the 60 cases were obtained from the tissue bank of the Pathology Department at the Leiden University Medical Center (LUMC; Leiden, The Netherlands), and 12 testicular DLCLs were collected by Dr L. Looijenga from the Josephine Nefkens Institute (Rotterdam, The Netherlands). The other 35 cases were obtained from the NHL Registry of the Comprehensive Cancer Center West in The Netherlands between 1981 and 1989.26
Immunohistochemistry
Immunohistochemical staining was performed on freshly cut 3-μm thick buffered formalin-fixed paraffin-embedded tissue sections according to standard procedures.27 Slides were incubated overnight with mouse monoclonal antibodies (MoAbs): HCA2 (anti-HLA-A; Dr J. Neefjes, NKI, Amsterdam, The Netherlands), anti-HLA-DR (cloneTAL.1B5; DAKO, Copenhagen, Denmark), the primary rabbit polyclonal anti-β2M (A 072; DAKO), and the rabbit MoAb anti-TAP1.28
Frozen sections were fixed in acetone, washed in phosphate-buffered saline, and subsequently incubated with the mouse MoAb W6/32, a pan-class I MoAb, SPV-L3 directed against DQ,29 and B8.11.2 directed against DR30 for 1 hour. For immunodetection, the same protocol was used for the paraffin sections.
In each tumor, T cells, endothelial cells, macrophages, and dendritic cells served as a positive control for class I expression, whereas macrophages and dendritic cells served as a positive control for class II expression. Tumor cells were only scored negative if absolutely no staining was present as compared to a strong staining of internal control cells. If some staining was present but reactive cells stained much stronger, tumor cells were scored weakly positive.
Microdissection and DNA extraction
DNA was extracted according to the protocol described by Isola et al32 with some adjustments. Paraffin-embedded tissue was cut in 10-μm sections and stained with hematoxylin and eosin. Before the normal dehydration steps, the staining procedure was interrupted for microdissection. To enrich for tumor cells, selected areas that contained more than 70% tumor cells were microdissected by using a needle under direct light microscopic visualization. Normal control tissue was obtained by using the same procedure. DNA was extracted by incubation for 72 hours at 56°C in 1 mL of isolation buffer (100 mmol/L NaCl, 10 mmol/L Tris-HCl, 25 mmol/L EDTA, pH 8, 0.5% sodium dodecyl sulfate). An aliquot of proteinase K (30 μL) was added, and this step was repeated 24 and 48 hours later. DNA was isolated by using phenol-chloroform-isoamyl alcohol, 20 μg/mL glycogen, and 250 μL 7.5 mol/L ammonium acetate and precipitated with 1 mL of 100% ethanol. DNA was dissolved in Tris-EDTA (10 mmol/L Tris, 0.1 mmol/L EDTA, pH 7.6), and 1 μL was used as a template for polymerase chain reaction (PCR).
Loss of heterozygosity (LOH) analysis
DNA from normal and tumor-microdissected material was analyzed for LOH by PCR amplification. For most microsatellite markers, the primers have been described before,32 except for C125, MICA, TY2A, BAT2, C47, and X87 344 that were retrieved from submitted sequences (GenBank). The sequences of the latter markers are as follows: C125.R, 5′AAGTCAAGCATATCTGCCATTTGG; C125.F, 5′CCCCAAACCCTGAAACTTG; MICA.R, 5′GGTGCTTCAGAGTCATTGG; MICA.F, 5′CTTTTTTTCAGGGAAAGTGC; TY2A.R, 5′TCAAACCAATCAGGGTGGC; TY2A.F, 5′AGAAGCAGTATACAGGGGC; BAT2.R, 5′AAGGGCTTTAGGAGGTCTG; BAT.F, 5′CCAGCCTGGATAACAGAAC; C47.R, 5′TCCTCCAGGTTCATCCATG; C47.F, 5′GTCTGTCCTGCATCAAATGG; X87 344.R, 5′CTCTAACTCCTTTCATGCTGC; and X87 344.F, 5′CAAGCAGAGGAACAAAGTCA.
Standard PCR amplifications33 were carried out in a 12-μL reaction volume that contained 1 μL purified template DNA, 6 pmol of each primer, 2 mmol/L dNTP-C, 0.1 mg/mL bovine serum albumin, Taq polymerase buffer (10 mmol/L Tris-HCl, 1.5 mmol/L MgCl, 50 mmol/L KCl, 0.01% (w/v) gelatin, 0.1% Triton), 0.06 units SuperTaq polymerase (Sphaero Q, HT Biotechnology, Cambridge, UK), and 1 μCi [α-32P]-CTP (Amersham, Buckinghamshire, UK). Samples were denatured for 5 minutes and amplified for 33 cycles, consisting of 1 minute denaturation at 94°C, 2 minutes primer annealing at 55°C, 1 minute elongation at 72°C, and followed by a final extension of 6 minutes at 72°C. The amplification reactions were carried out in 96-wells microtiter plates (DYNAX DPC, Breda, The Netherlands) with the use of a thermal cycler (MJ Research, Watertown, MA). Radiolabeled products were denatured in formamide-loading dye and analyzed on 6% polyacrylamide gels. Dried gels were autoradiographed at room temperature for 18 to 24 hours. The Molecular Dynamics Phosporimager 445SI (Molecular Dynamics, Sunnyvale, CA) was used for quantification of the PCR products. An allelic imbalance factor was calculated by the quotient of the ratios of the peak heights from normal and tumor DNA. A calculated factor more than 1.7 was considered as allelic imbalance, whereas a factor less than 1.7 was regarded as retention. Loci showing microsatellite instability were not scored as allelic imbalance. Each PCR reaction was performed at least twice.
Interphase FISH
From the 60 cases used in immunohistochemical and LOH studies, 14 testicular, 4 CNS, and 7 nodal DLCLs with available frozen tissue material were analyzed by interphase FISH as previously described.34 The α-satellite centromeric 6-probe (D6Z1, Oncor, Gaithersburg, MD) was biotin-16dUTP-labeled. PAC 223H1 was isolated from the RCPI-1 Human PAC Library of the Roswell Park Cancer Institute (Dr J. den Dunnen, Genome Technology Center, LUMC, Leiden) with the use of a TAP1 complementary DNA (cDNA) probe.23According to known DNA sequences, PACs 93N13 and 172K2 were directly obtained from the RCPI-1 library (GenBank accession No: PAC 93N13, Z84 489; PAC 172K2, Z84 814). Cosmids 619pWE1, DV19, and U16 were kindly provided by Dr H. Inoko (Tokai University School of Medicine, Kanagawa, Japan), Dr J. Trowsdale (University of Cambridge, Cambridge, UK), and Dr G. Blanck (University of South Florida, Tampa, FL), respectively. Cosmid c109K2118 derived from the ICRF flow-sorted chromosome 6 library was obtained from the Resource Center/Primary Database of the German Human Genome Project (RZPD, Berlin, Germany) and cosmid M31A was obtained from the American Tissue Culture Center. All cosmid and PAC probes were labeled with digoxenin-12-dUTP (ROCHE, Basel, Switzerland) by standard nick translation. Hybridization was performed as previously described.34 Hybridization mixture (5 μL) that contained 3 ng/μL of the centromere 6 probe combined with 3 ng/μL of each cosmid or PAC probe, 1.5 μg human Cot-1 DNA, and 3.5 μL hybridization mix (50% formamide, 10% dextran sulphate, 50 mmol/L sodium phosphate, pH 7.0, 2 × sodium chloride/sodium citrate [SSC]) was denatured for 8 minutes at 80°C and then pre-annealed for 30 minutes at 37°C. After denaturation for 3 minutes at 80°C, nuclei were hybridized overnight at 37°C in a moist chamber that contained 60% formamide in 2 × SSC. Immunodetection was performed as previously described.34
In 10 tonsils of healthy individuals, for each probe combination the signal ratio in 200 nuclei was determined; the cut-off level for homozygous or hemizygous loss was set at the average of the controls plus 3 times the SD. Hemizygous deletions were defined by the presence of a lower number of locus-specific PAC or cosmid signals relative to the number of centromere 6 signals (usually 1 locus-specific signal and 2 centromeric signals). Homozygous deletions were defined as the complete absence of PAC or cosmid signals in cells with one or more preserved centromere 6 signals. Thus, in all cases, technical artifacts were as much as possible excluded by inclusion of the centromere 6 probe in each hybridization experiment, by simultaneous hybridization with other probes for the HLA region, and by the analysis of 10 normal controls. Additionally, in all cases, cell counts were performed by two independent investigators (SAR and ESJ).
Fiber FISH
DNA fibers were prepared according to the halo technique.35 The probe set used to study the genomic abnormalities in extranodal lymphomas consisted of PAC clones 223H1, 93N13, and 172K2 and cosmid clones DV19 and U16 (see section “Interphase FISH”). Additional PAC clones 214F11, 122L3, 60C22, and 71I17 were isolated from the RCPI-1 Human PAC Library with the use, respectively, of a TAP1 cDNA probe (see section “Interphase FISH”) or PCR-generated probes for unique sequences (GenBank) in the vicinity of microsatellite marker C47 and the genes TNXB and Hsp70. A normal “bar code” was generated on DNA fiber preparations obtained from normal peripheral blood leukocytes. The hybridization solution consisted of 30% formamide, 10% dextran sulphate, 50 mmol/L sodium phosphate, pH 7.0, 2 × SCC, 3 ng/μL of each probe, and a 50-fold excess of human Cot-1 DNA. Hybridization and immunodetection were performed as previously described.35
Fluorescence microscopy
Slides were analyzed with a Leica DM-RXA fluorescence microscope (Leica, Wetzlar, Germany). Images were captured with the use of a COHU 4910 series monochrome CCD camera (COHU, San Diego, CA) attached to the fluorescence microscope equipped with a PL Fluotar 100×, NA 1.30 to 0.60 objective and I3 and N2.1 filters (Leica) and Leica QFISH software (Leica Imaging Systems, Cambridge, UK). Images were processed with Paintshop Pro and Corel Draw 8.0.
Statistical analysis
The Fisher exact test was used for determining the significance of differences in immunohistochemistry and LOH data between extranodal and nodal DLCL cases. Statistical analysis for interphase FISH was performed with the use of the Mann-Whitney test for independent samples. Two-sided tests were used in all calculations. A Pvalue <.05 was considered statistically significant for both tests.
Results
Loss of HLA class I and II expression in DLCL of the testis and CNS
Tissue sections of 21 nodal, 28 testicular, and 11 CNS DLCLs were stained for HLA-A and HLA-DR expression (Figure1). Loss of HLA-A expression was observed in 61% and 55% of the primary testicular and CNS lymphomas, respectively, as compared with 10% of the primary nodal cases (difference between extranodal and nodal DLCLs significant,P < .01; Table 1). A similar trend was observed for W6/32, a well-established pan-class I antibody, which could only be applied on 25 cases of which frozen material was available. The TAP1 and β2M proteins are essential for respectively transport and stabilization of HLA class I molecules, and loss of expression of either results in very low or undetectable levels of class I expression.22 23 In 7 of 12 cases with total loss of HLA class I expression as assessed for W6/32, absence of β2M or TAP1 expression provided an explanation for the loss of class I expression.
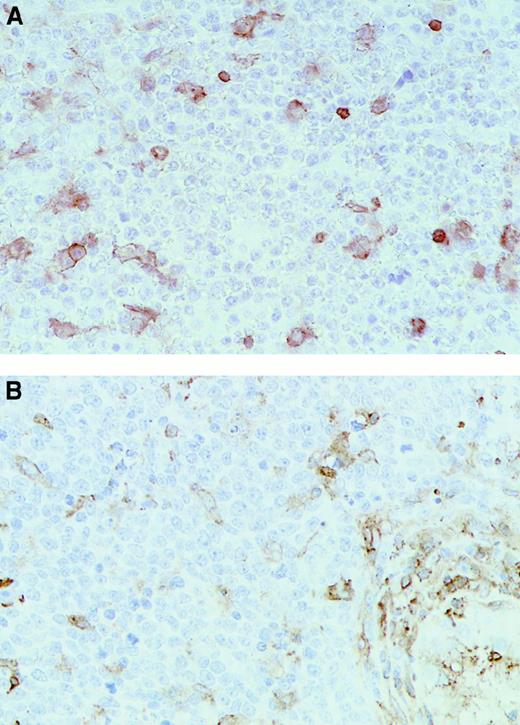
Immunohistochemical analysis of representative testicular DLCL cases.
Immunohistochemical staining was performed with the use of (A) the MoAb HCA2 (anti-HLA-A) (T20) and (B) the MoAb clone B8.11.2 (HLA-DR) (T18) (original magnification × 40). Note the positive (brown) staining in the internal control cells scattered through the mass of negative lymphoma cells.
Immunohistochemical analysis of representative testicular DLCL cases.
Immunohistochemical staining was performed with the use of (A) the MoAb HCA2 (anti-HLA-A) (T20) and (B) the MoAb clone B8.11.2 (HLA-DR) (T18) (original magnification × 40). Note the positive (brown) staining in the internal control cells scattered through the mass of negative lymphoma cells.
HLA class I and class II expression
| Antibody . | Extranodal DLCL* . | Nodal DLCL* % of cases (n = 21) . | Total extranodal vs nodal DLCL† . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Testicular % of cases (n = 28) . | CNS % of cases (n = 11) . | |||||||||
| − . | +/− . | + . | − . | +/− . | + . | − . | +/− . | + . | P . | |
| HLA-A | 61 | 29 | 11 | 55 | 18 | 27 | 10 | 33 | 57 | 0.008 |
| β2M‡ | 23 | 46 | 31 | 18 | 27 | 55 | 11 | 5 | 84 | 0.7 |
| TAP‡ | 26 | 33 | 41 | 30 | 40 | 30 | 0 | 9 | 91 | 0.02 |
| HLA-DR | 61 | 29 | 11 | 46 | 18 | 36 | 5 | 10 | 85 | 0.004 |
| W6/32‡ | 62 | 0 | 38 | 75 | 0 | 25 | 25 | 13 | 62 | 0.4 |
| Antibody . | Extranodal DLCL* . | Nodal DLCL* % of cases (n = 21) . | Total extranodal vs nodal DLCL† . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Testicular % of cases (n = 28) . | CNS % of cases (n = 11) . | |||||||||
| − . | +/− . | + . | − . | +/− . | + . | − . | +/− . | + . | P . | |
| HLA-A | 61 | 29 | 11 | 55 | 18 | 27 | 10 | 33 | 57 | 0.008 |
| β2M‡ | 23 | 46 | 31 | 18 | 27 | 55 | 11 | 5 | 84 | 0.7 |
| TAP‡ | 26 | 33 | 41 | 30 | 40 | 30 | 0 | 9 | 91 | 0.02 |
| HLA-DR | 61 | 29 | 11 | 46 | 18 | 36 | 5 | 10 | 85 | 0.004 |
| W6/32‡ | 62 | 0 | 38 | 75 | 0 | 25 | 25 | 13 | 62 | 0.4 |
HLA indicates human leukocyte antigen; DLCL, diffuse large B-cell lymphoma; CNS, central nervous system; β2M, β2-microglobulin.
The − indicates no detectable staining of tumor cells compared with staining of internal control cells (respectively, T lymphocytes, macrophages, and dendritic cells for HLA class I, β2M, and TAP as well as macrophages and dendritic cells for class II) within each tumor. The +/− indicates weaker or heterogeneous staining of tumor cells compared with internal control cells.
The P value was determined by using a two-sided Fisher exact test that compared the total loss of expression in extranodal (testis and CNS) and nodal DLCL.
Frozen tissue sections of 13 testicular, 4 CNS, and 8 nodal DLCLs were stained for W6/32. One testicular and one CNS lymphoma were not interpretable for TAP. Two testicular and 3 nodal lymphomas were not interpretable for β2M.
Significant differences between extranodal and nodal lymphomas were also found for expression of HLA class II molecules, with 61% and 46% of the testicular and CNS lymphomas completely lacking HLA-DR expression compared with only 5% of the nodal DLCLs (difference between extranodal and nodal DLCLs significant; P < .05; Table 1). Similar results were obtained with antibodies that recognized HLA-DR and HLA-DQ on frozen tissue sections (Table2). Interestingly, 15 of the 23 cases analyzed on paraffin sections showed a coordinate loss of both class I and class II expression.
Interphase FISH results in DLCL and comparison with HLA protein expression
| . | Homozygous deletion . | Hemizygous deletion . | Expression* . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 619pWE1 DPB . | 223H1 TAP . | 93N13 DQA1 . | 172K2 DRA . | M31A TNF-α . | C109K2118 HLA-A . | 619pWE1 DPB . | 223H1 TAP . | 93N13 DQA1 . | 172K2 DRA . | M31A TNF-α . | C109K2118 HLA-A . | HLA-DQ . | HLA-DR . | HLA-A . | |
| T2 | 3.0 | 1.5 | 48.5 | 49.0 | 1.0 | 3.0 | 27.0 | 12.5 | 16.0 | 14.0 | 19.0 | 14.0 | H | H | − |
| T3* | 0.0 | 1.0 | 5.0 | 1.5 | 3.0 | 1.0 | 5.0 | 15.0 | 30.0 | 10.0 | 10.0 | 2.0 | + | + | + |
| T11 | 1.5 | 0.0 | 9.0 | 15.5 | 3.5 | 1.0 | 13.0 | 5.5 | 58.0 | 49.5 | 35.0 | 20.0 | − | + | − |
| T13 | 1.5 | 2.5 | 0.5 | 16.0 | 1.0 | 0.0 | 24.0 | 25.5 | 26.5 | 40.0 | 42.0 | 35.5 | − | − | − |
| T14 | 1.5 | 2.5 | 69.0 | 58.5 | 2.5 | 0.0 | 15.0 | 48.0 | 13.5‡ | 5.0‡ | 57.5 | 58.5 | − | − | − |
| T16 | 0.0 | 0.0 | 22.5 | 8.0 | 2.5 | 3.0 | 23.5 | 18.5 | 15.0 | 25.5 | 27.0 | 27.5 | − | − | − |
| T18† | 0.0 | 7.0 | 64.5 | 71.5 | 1.0 | 2.5 | 18.5 | 17.0 | 2.0 | 2.0 | 7.5 | 23.5 | − | − | H |
| T19 | 2.5 | 3.0 | 16.5 | 3.0 | 0.5 | 1.5 | 19.0 | 15.0 | 11.0 | 11.0 | 13.5 | 14.0 | − | − | − |
| T20† | 0.0 | 0.0 | 59.5 | 3.0 | 2.0 | 6.5 | 70.0 | 70.0 | 25.5 | 79.5 | 75.5 | 53.0 | − | − | − |
| T22 | 1.0 | 0.5 | 1.5 | 6.0 | 1.0 | 0.0 | 8.5 | 1.0 | 11.5 | 10.0 | 6.0 | 7.0 | − | W | + |
| T23† | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.5 | 4.0 | 14.5 | 14.5 | 21.0 | 4.0 | − | W | + |
| T25† | 0.0 | 6.0 | 60.5 | 11.5 | 2.5 | 12.0 | 58.0 | 33.5 | 9.0 | 45.0 | 52.0 | 26.0 | − | − | − |
| T26 | 0.0 | 0.0 | 2.0 | 5.5 | 1.0 | 0.0 | 11.0 | 10.0 | 2.5 | 6.5 | 9.5 | 12.5 | ND | H | H |
| T30 | 2.0 | 1.5 | 0.5 | 0.5 | 0.5 | 0.0 | 16.0 | 12.0 | 10.0 | 15.5 | 9.0 | 30.5 | − | + | − |
| C1 | 0.0 | 0.5 | 27.5 | 26.0 | 1.0 | 2.0 | 3.0 | 16.0 | 32.5 | 15.0 | 22.0 | 17.0 | − | − | − |
| C2 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 1.0 | 8.5 | 9.5 | 72.0 | 72.5 | 68.5 | 68.5 | − | − | − |
| C9 | 0.0 | 30.0 | 44.0 | 52.0 | 1.0 | 0.0 | 10.0 | 13.5 | 27.0 | 22.0 | 56.0 | 8.5 | ND | − | − |
| C11† | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 14.0 | 15.0 | 3.5 | 5.0 | 6.0 | + | + | − |
| N3 | 2.0 | 0.0 | 0.0 | 0.5 | 2.0 | 0.0 | 5.0 | 12.5 | 12.0 | 24.0 | 16.5 | 9.0 | − | − | − |
| N4 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 1.5 | 9.5 | 5.0 | 32.0 | 16.5 | 9.5 | 9.0 | − | + | − |
| N8† | 0.0 | 1.0 | 0.0 | 0.0 | 0.5 | 1.0 | 15.0 | 9.5 | 7.0 | 7.0 | 17.0 | 16.0 | + | + | + |
| N9 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 4.5 | 12.0 | 8.5 | 8.5 | 21.0 | 20.0 | 13.0 | + | + | + |
| N13 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.0 | 9.0 | 10.5 | 9.0 | 13.0 | 8.0 | + | + | H |
| N15 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 11.0 | 12.0 | 6.0 | 5.0 | 4.5 | 7.0 | + | + | + |
| N18 | 0.0 | 0.0 | 0.5 | 0.0 | 2.0 | 0.0 | 9.0 | 7.0 | 5.0 | 4.0 | 16.5 | 9.0 | ? | H | + |
| Co1 | 0.0 | 0.5 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 | 1.5 | 8.0 | 5.0 | 9.0 | 2.0 | ND | ND | ND |
| Co2 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 8.0 | 1.5 | 3.5 | 8.5 | 7.5 | ND | ND | ND |
| Co3 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 4.0 | 9.0 | 8.0 | 9.0 | 4.0 | ND | ND | ND |
| Co4 | 0.0 | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 4.0 | 5.0 | 9.0 | 5.0 | 7.0 | 5.0 | ND | ND | ND |
| Co5 | 0.0 | 0.0 | 1.0 | 7.0 | 0.0 | 0.0 | 5.5 | 5.0 | 2.0 | 1.0 | 4.5 | 5.5 | ND | ND | ND |
| Co6 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 2.5 | 3.5 | 6.0 | 7.0 | 4.0 | 4.0 | ND | ND | ND |
| Co7 | 0.0 | 3.0 | 1.0 | 1.0 | 1.0 | 0.0 | 6.0 | 7.0 | 5.0 | 4.0 | 3.5 | 6.0 | ND | ND | ND |
| Co8 | 2.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 5.0 | 5.5 | 1.0 | 6.5 | 10.5 | ND | ND | ND |
| Co9 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 3.0 | 5.0 | 8.0 | 8.0 | 9.5 | ND | ND | ND |
| Co10 | 0.0 | 1.0 | 0.0 | 0.0 | 0.5 | 0.0 | 1.5 | 3.0 | 3.0 | 10.5 | 19.0 | 9.5 | ND | ND | ND |
| . | Homozygous deletion . | Hemizygous deletion . | Expression* . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 619pWE1 DPB . | 223H1 TAP . | 93N13 DQA1 . | 172K2 DRA . | M31A TNF-α . | C109K2118 HLA-A . | 619pWE1 DPB . | 223H1 TAP . | 93N13 DQA1 . | 172K2 DRA . | M31A TNF-α . | C109K2118 HLA-A . | HLA-DQ . | HLA-DR . | HLA-A . | |
| T2 | 3.0 | 1.5 | 48.5 | 49.0 | 1.0 | 3.0 | 27.0 | 12.5 | 16.0 | 14.0 | 19.0 | 14.0 | H | H | − |
| T3* | 0.0 | 1.0 | 5.0 | 1.5 | 3.0 | 1.0 | 5.0 | 15.0 | 30.0 | 10.0 | 10.0 | 2.0 | + | + | + |
| T11 | 1.5 | 0.0 | 9.0 | 15.5 | 3.5 | 1.0 | 13.0 | 5.5 | 58.0 | 49.5 | 35.0 | 20.0 | − | + | − |
| T13 | 1.5 | 2.5 | 0.5 | 16.0 | 1.0 | 0.0 | 24.0 | 25.5 | 26.5 | 40.0 | 42.0 | 35.5 | − | − | − |
| T14 | 1.5 | 2.5 | 69.0 | 58.5 | 2.5 | 0.0 | 15.0 | 48.0 | 13.5‡ | 5.0‡ | 57.5 | 58.5 | − | − | − |
| T16 | 0.0 | 0.0 | 22.5 | 8.0 | 2.5 | 3.0 | 23.5 | 18.5 | 15.0 | 25.5 | 27.0 | 27.5 | − | − | − |
| T18† | 0.0 | 7.0 | 64.5 | 71.5 | 1.0 | 2.5 | 18.5 | 17.0 | 2.0 | 2.0 | 7.5 | 23.5 | − | − | H |
| T19 | 2.5 | 3.0 | 16.5 | 3.0 | 0.5 | 1.5 | 19.0 | 15.0 | 11.0 | 11.0 | 13.5 | 14.0 | − | − | − |
| T20† | 0.0 | 0.0 | 59.5 | 3.0 | 2.0 | 6.5 | 70.0 | 70.0 | 25.5 | 79.5 | 75.5 | 53.0 | − | − | − |
| T22 | 1.0 | 0.5 | 1.5 | 6.0 | 1.0 | 0.0 | 8.5 | 1.0 | 11.5 | 10.0 | 6.0 | 7.0 | − | W | + |
| T23† | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.5 | 4.0 | 14.5 | 14.5 | 21.0 | 4.0 | − | W | + |
| T25† | 0.0 | 6.0 | 60.5 | 11.5 | 2.5 | 12.0 | 58.0 | 33.5 | 9.0 | 45.0 | 52.0 | 26.0 | − | − | − |
| T26 | 0.0 | 0.0 | 2.0 | 5.5 | 1.0 | 0.0 | 11.0 | 10.0 | 2.5 | 6.5 | 9.5 | 12.5 | ND | H | H |
| T30 | 2.0 | 1.5 | 0.5 | 0.5 | 0.5 | 0.0 | 16.0 | 12.0 | 10.0 | 15.5 | 9.0 | 30.5 | − | + | − |
| C1 | 0.0 | 0.5 | 27.5 | 26.0 | 1.0 | 2.0 | 3.0 | 16.0 | 32.5 | 15.0 | 22.0 | 17.0 | − | − | − |
| C2 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 1.0 | 8.5 | 9.5 | 72.0 | 72.5 | 68.5 | 68.5 | − | − | − |
| C9 | 0.0 | 30.0 | 44.0 | 52.0 | 1.0 | 0.0 | 10.0 | 13.5 | 27.0 | 22.0 | 56.0 | 8.5 | ND | − | − |
| C11† | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 14.0 | 15.0 | 3.5 | 5.0 | 6.0 | + | + | − |
| N3 | 2.0 | 0.0 | 0.0 | 0.5 | 2.0 | 0.0 | 5.0 | 12.5 | 12.0 | 24.0 | 16.5 | 9.0 | − | − | − |
| N4 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 1.5 | 9.5 | 5.0 | 32.0 | 16.5 | 9.5 | 9.0 | − | + | − |
| N8† | 0.0 | 1.0 | 0.0 | 0.0 | 0.5 | 1.0 | 15.0 | 9.5 | 7.0 | 7.0 | 17.0 | 16.0 | + | + | + |
| N9 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 4.5 | 12.0 | 8.5 | 8.5 | 21.0 | 20.0 | 13.0 | + | + | + |
| N13 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.0 | 9.0 | 10.5 | 9.0 | 13.0 | 8.0 | + | + | H |
| N15 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 11.0 | 12.0 | 6.0 | 5.0 | 4.5 | 7.0 | + | + | + |
| N18 | 0.0 | 0.0 | 0.5 | 0.0 | 2.0 | 0.0 | 9.0 | 7.0 | 5.0 | 4.0 | 16.5 | 9.0 | ? | H | + |
| Co1 | 0.0 | 0.5 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 | 1.5 | 8.0 | 5.0 | 9.0 | 2.0 | ND | ND | ND |
| Co2 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 8.0 | 1.5 | 3.5 | 8.5 | 7.5 | ND | ND | ND |
| Co3 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 4.0 | 9.0 | 8.0 | 9.0 | 4.0 | ND | ND | ND |
| Co4 | 0.0 | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 4.0 | 5.0 | 9.0 | 5.0 | 7.0 | 5.0 | ND | ND | ND |
| Co5 | 0.0 | 0.0 | 1.0 | 7.0 | 0.0 | 0.0 | 5.5 | 5.0 | 2.0 | 1.0 | 4.5 | 5.5 | ND | ND | ND |
| Co6 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 2.5 | 3.5 | 6.0 | 7.0 | 4.0 | 4.0 | ND | ND | ND |
| Co7 | 0.0 | 3.0 | 1.0 | 1.0 | 1.0 | 0.0 | 6.0 | 7.0 | 5.0 | 4.0 | 3.5 | 6.0 | ND | ND | ND |
| Co8 | 2.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 5.0 | 5.5 | 1.0 | 6.5 | 10.5 | ND | ND | ND |
| Co9 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 3.0 | 5.0 | 8.0 | 8.0 | 9.5 | ND | ND | ND |
| Co10 | 0.0 | 1.0 | 0.0 | 0.0 | 0.5 | 0.0 | 1.5 | 3.0 | 3.0 | 10.5 | 19.0 | 9.5 | ND | ND | ND |
FISH indicates fluorescent in situ hybridization; DLCL, diffuse large B-cell lymphomas; HLA, human leukocyte antigen; TNFα, tumor necrosis factor α; T, testicular DLCL; C, central nervous system DLCL; N, nodal DLCL; Co, normal control (tonsil) tissue.
The table shows the percentages of nuclei with loss of one or two HLA region-specific signals in combination with retention of the centromeric signals for chromosome 6. In each experiment, 200 nuclei were counted. Shaded boxes indicate loci with an abnormal percentage of cells with homozygous or hemizygous deletions. These percentages were significantly higher than those obtained in the 10 normal controls. For homozygous deletions, the cut-off level was > 6% cells, for hemizygous deletions > 21% (see text).
Immunohistochemistry (see also Table 1) was performed on frozen sections (HLA-DR and HLA-DQ) or paraffin-embedded tissue sections (HLA-A), whereas FISH analysis was done on nuclei obtained from frozen tissue sections. The − indicates negative; +, positive; W: weakly positive; H: tumor heterogeneity with distinct tumor parts being completely negative; ?, undeterminable; ND, not done.
In case of aneuploidy for chromosome 6, this was corrected per cell for the number of centromere 6 specific signals.
These and many other percentages are relatively low because the tumor was also affected by a homozygous deletion.
Increased LOH at 6p21 in DLCLs of the testis and CNS
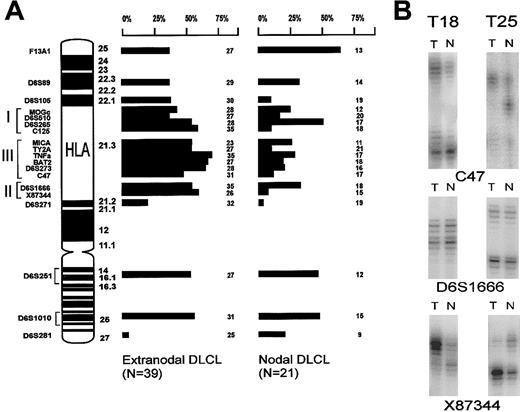
To investigate whether genetic alterations of the HLA region at 6p21.3 contributed to loss of class I or class II expression, the same 60 cases were studied by LOH analysis. Nineteen microsatellite markers on chromosome 6, including 12 markers in the HLA region, were used (Figure 2A). In contrast to allelic imbalances at 6q,36 which were equally frequent in nodal and extranodal DLCLs of our series, the testicular and the CNS lymphomas frequently showed allelic imbalance in the HLA region. The differences between these extranodal and nodal DLCLs were statistically significant for C125, TY2A, BAT2, and X8344 (Figure 2A). However, the individual markers are closely linked; therefore, allelic imbalances for adjacent markers are not independent from each other.37 Furthermore, homozygous deletions will counterbalance allelic imbalance and thus will give rise to discontinuous patterns of allelic imbalance if a set of adjacent markers are used.38
LOH analysis.
(A) Ideogram depicting the chromosome 6p and q arms. All microsatellite markers used for LOH analysis and their location are indicated. On the right-hand side, the LOH data of the extranodal and nodal series of DLCLs are summarized. Each bar represents the percentage of informative cases with LOH (their number depicted at the right). (B) Example of 2 testicular lymphomas (T18 and T25) that show retention of heterozygosity at marker D6S1666 near HLA-DQB1, and allelic imbalance is shown at both flanking markers C47 and X87 344. This suggests a homozygous deletion. Allelic imbalance was determined by comparing the ratio between alleles of tumor DNA (T) and normal DNA (N).
LOH analysis.
(A) Ideogram depicting the chromosome 6p and q arms. All microsatellite markers used for LOH analysis and their location are indicated. On the right-hand side, the LOH data of the extranodal and nodal series of DLCLs are summarized. Each bar represents the percentage of informative cases with LOH (their number depicted at the right). (B) Example of 2 testicular lymphomas (T18 and T25) that show retention of heterozygosity at marker D6S1666 near HLA-DQB1, and allelic imbalance is shown at both flanking markers C47 and X87 344. This suggests a homozygous deletion. Allelic imbalance was determined by comparing the ratio between alleles of tumor DNA (T) and normal DNA (N).
Three testicular lymphomas (T18, T20, and T25) showed retention of heterozygosity at marker D6S1666 but allelic imbalance at the flanking markers C47 and X87 344 (Figure 2B). This finding suggested the presence of a homozygous deletion at D6S1666 in the class II region, with the remaining signal resulting from contaminating normal cells in the tumor sample.
Interphase FISH shows frequent homozygous deletions in the class II region and larger hemizygous deletions in DLCLs of the testis and CNS
To evaluate homozygous deletion of HLA class II genes, we applied two-color FISH on isolated nuclei of all cases with available frozen tissue material (ie, 14 testicular, 4 CNS, and 7 nodal lymphomas). Six digoxigenin-labeled PAC and cosmid clones specific for the HLA region, each in combination with a biotin-labeled centromere 6 probe, were used (Table 2; Figure 3A). The panel consisted of 3 PAC clones specific for the class II region, including PAC 93N13 that contains marker D6S1666 and covers the DQA1 and DRB1 genes. For each probe combination, the cut-off level was determined as the average plus 3 times the SD as assessed on 10 normal tonsils (Table 2, homozygous and hemizygous deletion columns). For the detection of homozygous deletions (loss of all HLA allele-specific signals in combination with retention of chromosome 6 centromeric signals), this cut-off level varied between 0% and 5.8%. To exclude the possibility that hybridization artifacts accounted for loss of signals, we fixed the cut-off level for homozygous deletion at more than 6% cells. With the use of this threshold, the homozygous deletion was confirmed in all 3 cases, 64.5% (T18), 59.5% (T20), and 60.5% (T25) of the cells showing loss of both allelic signals for PAC 93N13 (Table 2; Figures3B-D). In fact, PAC 93N13 was homozygously lost in 8 of 14 testicular (T2, T11, T14, T16, T18, T19, T20, and T25) and 2 of 4 CNS lymphomas (C1 and C9) but in none of the nodal cases (difference between extranodal and nodal DLCLs significant; P < .001; Table2). Similarly, PAC 172K2 that covers the HLA-DRA gene was homozygously lost in respectively 7 of 14 testicular DLCLs (T2, T11, T13, T14, T16, T18, and T25) and in the same 2 CNS lymphomas (C1 and C9) but in none of 7 nodal lymphomas (difference between extranodal and nodal DLCLs significant, P < .001; Table 2). Two lymphomas (T18 and C9) showed extension of the homozygous deletion to 223H1, a PAC that covers TAP1 (Table 2). In case C9, a similar fraction (ie, 30%, 44%, and 52%) of the cells showed a homozygous deletion for probes 223H1, 93N13, and 172K2, whereas, in T18, 223H1 was deleted only in a small fraction (7%) of the cells.
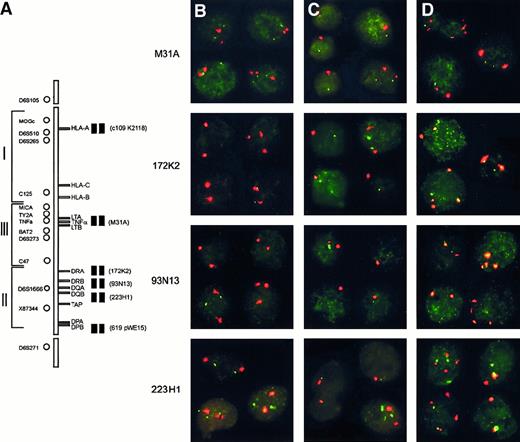
Homozygous and hemizygous deletions within the HLA region determined by interphase FISH.
(A) Schematic representation of the HLA region on chromosome 6p21.3. On the left-hand side, the localization of the microsatellite markers used for LOH and on the right-hand side the 6 PAC/cosmid probes used for interphase FISH. (B-D) Examples of interphase FISH. Each panel represents a composite of 3 to 4 individually captured nuclei, including at least 1 control nucleus without a deletion. In all cases, the red signal is derived from centromere 6, and the green signal is derived from 1 of the 6p21.3-specific probes as indicated on the left. (B) Testicular lymphoma T18 showing homozygous deletions of PACs 93N13 and 172K2 indicated by the presence of 2 centromere 6 signals and no detectable PAC signal. The homozygous deletion was also present in the 20% of nuclei with trisomy 6. (C) Testicular lymphoma T20 showing monosomy 6 and a deletion of PAC 93N13 in 75% of the nuclei with monosomy 6. (D) Testicular lymphoma T25 showing a homozygous deletion of PAC 93N13, including all cells with tetrasomy (25% of cells), and a large hemizygous deletion, including PACs 223H1, 172K2, and cosmid M31A.
Homozygous and hemizygous deletions within the HLA region determined by interphase FISH.
(A) Schematic representation of the HLA region on chromosome 6p21.3. On the left-hand side, the localization of the microsatellite markers used for LOH and on the right-hand side the 6 PAC/cosmid probes used for interphase FISH. (B-D) Examples of interphase FISH. Each panel represents a composite of 3 to 4 individually captured nuclei, including at least 1 control nucleus without a deletion. In all cases, the red signal is derived from centromere 6, and the green signal is derived from 1 of the 6p21.3-specific probes as indicated on the left. (B) Testicular lymphoma T18 showing homozygous deletions of PACs 93N13 and 172K2 indicated by the presence of 2 centromere 6 signals and no detectable PAC signal. The homozygous deletion was also present in the 20% of nuclei with trisomy 6. (C) Testicular lymphoma T20 showing monosomy 6 and a deletion of PAC 93N13 in 75% of the nuclei with monosomy 6. (D) Testicular lymphoma T25 showing a homozygous deletion of PAC 93N13, including all cells with tetrasomy (25% of cells), and a large hemizygous deletion, including PACs 223H1, 172K2, and cosmid M31A.
The percentage of cells with homozygous deletion varied between 7.0% and 71.5%. The occasionally low percentages of cells with homozygous deletion might have been caused by either the presence of many reactive cells in the frozen tissue sample or by the presence of tumor heterogeneity. The first could be the case in T16, T19, and C1 in which maximally 32.5% of all cells showed any abnormality (loss of 1 or 2 spots; see Table 2). Tumor heterogeneity was likely present in cases T11, T13, T16, T18, and T25, showing variable percentages of cells with loss of one or both HLA allele specific signals (Table 2). For instance, in case T11, a small fraction (9% to 15.5%) of all cells contained a homozygous deletion for probes 93N13 and 172K2, whereas a larger fraction (35% to 58%) contained a hemizygous deletion covered by the probes 93N13, 172K2, and M31A (Table 2).
For the hemizygous deletions (loss of one HLA-specific signal while both centromere signals were retained), the individual cut-off levels on the normal tonsils varied between 8.5% and 20.4%. With the use of a general cut-off level of more than 21%, 7 of 18 extranodal DLCLs and 1 of 7 nodal cases (N4) contained a hemizygous deletion covered by PAC 93N13. Similarly, 9 of 18 extranodal and 1 of 7 nodal DLCLs contained a hemizygous deletion covered by PAC 172K2. In all but one case (T3), these deletions were part of much larger hemizygous deletions also involving the region that contains the HLA-DP genes and/or the HLA class III and even the HLA class I region (see below). The percentage of cells with hemizygous deletions varied between 24% and 79.5% (Table 2).
We combined all data for the HLA class II region covered by PAC 93N13 and/or 172K2 and found 4 of 18 extranodal cases contained exclusively homozygous deletions, 2 cases showed only hemizygous deletions, and 7 cases showed combined homo- and hemizygous deletions. In contrast, only 2 of 7 nodal DLCLs contained a small hemizygous deletion in this region.
HLA class I and class III genes were mainly affected by large hemizygous deletions (Table 2). Only case T25 contained 12% of cells with loss of both signals for C109K2118 that covers HLA-A, suggesting a small tumor subclone with a homozygous deletion in the class I region. With respect to the class III region covered by M31A, 9 of 18 extranodal versus none of 7 nodal DLCLs contained hemizygous deletions. For the class I region covered by C109K2118, these numbers were 8 of 18 extranodal versus none of 7 nodal DLCLs.
Aneuploidy of chromosome 6 was observed in 7 cases, including one nodal case. Five centromeric signals were seen in T3 and C11 (data not shown). T20 showed monosomy for chromosome 6. T23 showed trisomy 6 and loss of one allele on LOH analysis, indicating that 1 of the 2 chromosomes was lost and the other was present in triplicate. In addition, T18, T25, and N8 contained subclones with aneuploidy for chromosome 6 (Figure 3).
DNA fiber FISH mapping of the homozygous deletions in the HLA-DR and -DQ region
To determine the size of the deletions of each allele, 10 cases were also analyzed by high-resolution DNA fiber FISH with the use of a probe set covering approximately 900 kilobase (kb) of the class II region (Figure 4). Within the normal HLA class II region, the genomic organization of the different DR haplotypes differs in size between HLA-DRB1 and HLA-DRA. The size of DR 4, 7, and 9 haplotypes is approximately 110 kb, and DR 1, 2, and 10 haplotypes are 30 kb larger compared with the size of DR 3, 5, and 6 haplotypes.39 To determine these polymorphisms, HLA-DR and -DQ typing of the patients was performed with the use of PCR and oligo-hybridization according to standard protocols (courtesy Prof F.H.J. Claas, Dept. of Immunohematology, LUMC).40 41
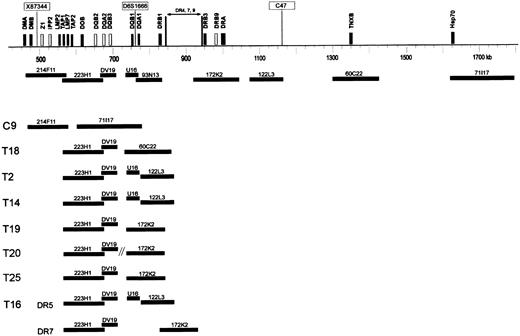
DNA fiber FISH detection of HLA-DQ and -DR deletions.
Schematic representation of part of the HLA class II and class III region of a DR 4, 7, or 9 haplotype, showing the approximate localization of clones used for fiber FISH. Black bars represent expressed genes, and white bars represent pseudogenes in the class II region. Microsatellite markers X87 344, D6S1666, and C47 are boxed. DNA-fiber FISH of 8 extranodal lymphoma cases with various deletions in the HLA class II region is depicted. In all cases except case T16, 1 allele contained a deletion involving all used probes. In these cases, the allele with a small deletion is shown. In case T16, both alleles DR5 and DR7 contained a deletion as illustrated.
DNA fiber FISH detection of HLA-DQ and -DR deletions.
Schematic representation of part of the HLA class II and class III region of a DR 4, 7, or 9 haplotype, showing the approximate localization of clones used for fiber FISH. Black bars represent expressed genes, and white bars represent pseudogenes in the class II region. Microsatellite markers X87 344, D6S1666, and C47 are boxed. DNA-fiber FISH of 8 extranodal lymphoma cases with various deletions in the HLA class II region is depicted. In all cases except case T16, 1 allele contained a deletion involving all used probes. In these cases, the allele with a small deletion is shown. In case T16, both alleles DR5 and DR7 contained a deletion as illustrated.
In cases T11 and T13, no abnormal fibers were observed, likely because of the too low percentage of aberrant cells (15.5% and 16% as determined by interphase FISH for 172K2; Table 2). In all other 8 cases (T2, T14, T16, T18, T19, T20, T25, and C9), the deletions were confirmed (Figure 4). In all cases except T16, one allele was affected by a deletion involving the entire probe set. In 2 cases (C9 and T18), the deletion of the other allele involved all functional HLA-DQ and HLA-DR genes, and the other HLA-DQ genes outside the deletion are pseudogenes. Furthermore, in 2 cases (T2 and T14), the deletion involved HLA-DQA1 and all HLA-DR genes; in 3 cases (T19, T20, and T25), it involved both HLA-DQ genes and a part of the HLA-DR genes. In one case (T16), the allele with the smallest deletion involved only the DQB1, DQA1, and DRB1 genes covered by cosmid DV19 and PAC 93N13, whereas the other allele had a deletion involving DQA1 and all DR genes. In cases T16 and T25, interphase FISH showed homozygous deletion of the area covered by PAC 93N13 and in a small fraction (8% and 11.5%) also of the area covered by PAC 172K2. Probably, because of the relative insensitivity, this deletion of PAC 172K2 was not detected by using fiber FISH.
As shown in Figure 4, the deletions were relatively variable at the telomeric side but never extended to the class III genes covered by PAC71 117. In T20 (HLA-DR typed DR6) with monosomy 6 (as detected by interphase FISH for the centromere of chromosome 6), a deletion of PAC 93N13 and cosmid U16 was detected by fiber FISH. However, PAC 172K2 was not on the same fiber as PAC 223H1, suggesting a chromosomal breakpoint in addition to the deletion. This finding was in agreement with the absence of co-localization of these PACs as determined by interphase FISH (not shown).
Correlation between homozygous deletions and loss of HLA class II expression
Table 3 shows the association between the homo- and hemizygous deletions in the class II region and the loss of HLA-DR and HLA-DQ expression as assessed on frozen tissue sections. Most importantly, in 8 of 15 cases without HLA-DQ expression, homozygous deletions of PAC 93N13 were present, and in 3 additional cases a hemizygous deletion was found. In 4 cases, we did not find any gross genetic alterations. Similarly, 10 of 14 DLCL cases with loss of HLA-DR expression showed homozygous deletions of PAC 172K2, whereas in 2 cases extensive hemizygous deletions were present. In only 2 DLCL cases, we did not find any explanation for loss of expression.
Correlation between homozygous deletions and loss of HLA class II expression
| . | HLA-DQ expression (n = 22) . | HLA-DR expression (n = 25) . | ||
|---|---|---|---|---|
| + . | − . | + . | − . | |
| No deletion | 6 | 7 | 8 | 6 |
| Homozygous deletion | 0 | 9 | 1 | 10 |
| . | HLA-DQ expression (n = 22) . | HLA-DR expression (n = 25) . | ||
|---|---|---|---|---|
| + . | − . | + . | − . | |
| No deletion | 6 | 7 | 8 | 6 |
| Homozygous deletion | 0 | 9 | 1 | 10 |
For correlation analysis of loss of HLA-DQ expression, homozygous deletions of PAC 93N13 covering the only functional HLA-DQA gene DQA1 were taken into account (differences significant as determined by the chi-square test; P < .01); similarly, for HLA-DR expression PAC 172K2, covering the only HLA-DRA gene was used (P < .01). HLA indicates human leukocyte antigen.
Discussion
We show that homozygous deletions within the HLA class II region of primary testicular and CNS lymphomas account for an important mechanism of loss of HLA-DR and HLA-DQ expression. In addition, in most cases large hemizygous deletions and/or mitotic recombination involved the entire HLA region.
In solid tumors and corresponding cell lines, investigators have mainly focused on loss of HLA class I expression as a route for tumor cells to escape killing by class I-restricted CD8+ cytotoxic T cells.10,11 According to these studies using pan-class I, locus- or allele-specific antibodies, between 39% and 88% of the epithelial tumors are completely or partially deficient in HLA class I expression. Mainly on the basis of studies of cell lines, distinct phenotypes can be related to separate molecular mechanisms: (a) absence or strongly reduced class I expression is often attributed to the absence of β2M molecules or the defects in TAP expression, respectively22,23; (b) loss of an haplotype can result from loss of one copy of chromosome 6 or from large deletions or mitotic recombination42-44; (c) decreased or completely abolished expression of HLA-A or -B may result from transcriptional down-regulation45-47; (d) finally, absence of expression of a single allele can result from point mutations, partial deletion, or somatic recombination within the HLA locus.24,44 48
In our series of primary DLCLs of the testis and CNS, aberrant class I expression was very frequent with more than half of these lymphomas showing complete loss of HLA-A expression. Our data are in line with previous studies49 50 that show loss of class I expression in 6% to 30% of aggressive B-cell lymphomas. In those studies, loss of expression was associated with extranodal presentation, and also with a relatively poor prognosis. In the present study, this loss could be explained by absence of β2M or TAP1 expression in more than half of the cases assessed for class I expression (W6/32). In 5 class I negative extranodal lymphomas without loss of β2M or TAP1, large hemizygous deletions involving the entire class I region were present. Loss of expression of the remaining haplotype was probably due to mutations, small homozygous deletions, or alterations in the methylation status of the genes, but it could also be explained by defects in TAP2 or LMP-2 and LMP-7 expression, especially because these genes are often involved in hemizygous deletions or mitotic recombination.
So far, little attention has been given to loss of HLA class II expression in cancer, probably because class II molecules are not constitutively expressed on normal epithelial cells. We showed loss of HLA class II expression in approximately half of the testicular and CNS lymphomas but in only 5% of the nodal cases. Previously, loss of HLA class II expression has been reported in DLCLs,18-20especially those presenting at extranodal sites.17 In primary testicular and CNS lymphomas, homozygous deletions are a very important mechanism for loss of class II expression because more than half of these DLCLs contained such homozygous deletions (Table 2). In addition to these homozygous deletions, several class II negative DLCLs showed large hemizygous deletions. Similar to the situation in the class I region, other structural abnormalities, such as point mutations or small homozygous deletions undetectable by FISH, or methylation of the promoter region might have caused loss of expression of the other allele. Of note, several cases showed tumor heterogeneity with different subpopulations that contain homo- and hemizygous deletions. In few cases, this was also represented by heterogeneous staining pattern for HLA-DR and -DQ.
The homozygous deletions within the HLA class II region covered a minimal region of approximately 100 kb and always included the HLA-DQA1 and HLA-DRB1 genes (Figure 4). Because HLA-DQB2, -DQA2, and -DQB3 are pseudogenes, loss of HLA-DQA1 and in consequence the inability to form a heterodimer with HLA-DQB1 is sufficient to explain the total loss of expression of HLA-DQ. The situation is almost similar but more complex for HLA-DR, because the smallest homozygously deleted areas involved PAC 93N13 that contains HLA-DRB1 and a variable area downstream of this PAC that contains DRB3-5, the other functional alleles.39The presence of a length of polymorphism in this area and the lack of appropriate probes for this polymorphic region hampered the exact deletion mapping within this region. However, it should be noted that at least 4 cases showed homozygous deletion of all HLA-DR genes.
Interestingly, in Epstein-Barr virus–transformed irradiated human B-lymphoblastoid cell lines selected for absence of HLA class II expression, similar homozygous deletions within the HLA class II region have been observed.51 In one cell line, the homozygously deleted region only contained the HLA-DRB1 and HLA-DQA1 genes, whereas in two other cell lines the homozygous deletion also included the HLA-DQB1 gene. No other genes have been identified in the recently published complete sequence of this region.52 This suggests that the HLA-DR and HLA-DQ genes are the real target of the deletion and that loss of these genes results in a growth advantage through immune selection. Furthermore, in none of the cases the class III region was affected, suggesting that genes essential for B-cell lymphomagenesis reside in this region.
What could be the implication of loss of HLA class II expression in lymphoma cells? HLA class I expression on tumor cells is essential for killing by CD8+ cytotoxic T cells. However, recent studies also suggest an important role for cytotoxic CD4+ effector T cells and thus for HLA class II expression on tumor cells.12-14 Additionally, cytotoxic CD8+ cells are only effective after activation by CD4+ helper T cells (ie when both CD8+ and CD4+ cells recognize tumor-specific antigenic determinants on the same APCs).16,53,54 Like normal B cells, neoplastic B cells may function as APCs as they can express both class I and II molecules, CD40, and costimulatory molecules.55 This is supported by the observation that murine lymphoma cells can present their own idiotype to CD4+ T cells.56 In consequence, loss of both HLA class I and II expression may result in an impaired activation of cytotoxic CD8+ T cells and may facilitate the growth of the tumor cells.
Tumor cells growing in immune-privileged sites, such as the CNS, testis, and the eye, experience a relative protection from immune attack. Various mechanisms may account for this effect, including the presence of an organ-blood barrier and the specific microenvironment with local production of immunosuppressive neuropeptides and cytokines2,3 as well as expression of Fas ligand on preexisting cells.2 However, immune protection at immune-privileged sites is not absolute as inoculated tumors are eventually killed by adoptively transferred cytotoxic T cells.21 This suggests that not only host factors but also tumor cell characteristics, such as loss of HLA class I and II expression, contribute to the phenomenon of immune privilege. Interestingly, primary DLCLs of the CNS preferentially express the immunoglobulin heavy chain gene V4-34.57-60 The V4-34 gene product, which is implied in several autoimmune disorders, might, therefore, function as a specific-tumor antigen in these lymphomas, and one could speculate that the lack of presentation in HLA molecules might give the tumor cells a selective immune privilege.
Strategies for the treatment of B-cell lymphomas are currently focusing on T-cell-mediated antitumor responses based on recognition of tumor-specific antigens.9 Our study shows that assessment of the HLA expression status and irreversible HLA gene defects is important to choose the optimal therapeutic regimen as the extranodal lymphomas we describe here likely will not be susceptible to T-cell-mediated immunotherapy.
Supported by grant RUL99-1997 from the Dutch Cancer Society.
S.A.R. and E.S.J. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sietske A Riemersma, Department of Pathology, (L1-Q), Leiden University Medical Center, Albinusdreef 2, 2333 ZA Leiden, The Netherlands; e-mail:s.a.riemersma@lumc.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal