Abstract
CCR5 and CXCR4 are the major coreceptors that mediate human immunodeficiency virus 1 (HIV-1) infection, while most simian immunodeficiency virus (SIV) isolates use CCR5. A number of alternative coreceptors can also mediate infection of some virus strains in vitro, although little is known about their in vivo relevance. Therefore, we characterized the expression pattern and coreceptor activity of one of these alternative coreceptors, STRL33/Bonzo, using a newly developed monoclonal antibody. In addition to being highly expressed (approximately 1000-7000 STRL33 ABS [antibody binding sites]) on specific subsets of natural killer cells (CD3−/CD16−/low/CD56+ and CD3−/CD16low/CD56−) and CD19+ B lymphocytes (approximately 300-5000 STRL33 ABS), STRL33 was expressed at levels sufficient to support virus infection on freshly isolated, truly naive CD4+/CD45RA+/CD62L+cells (6000-11 000 ABS). STRL33 expression on peripheral blood mononuclear cells (PBMCs) was increased by mitogenic stimulation (OKT3/IL-2 [interleukin-2] had a greater effect than phytohemaglutinin (PHA)/IL-2), but it was dramatically decreased upon Ficoll purification. Infection of CCR5− human peripheral blood lymphocytes (PBLs) showed that 2 different SIV envelope (Env) proteins mediated entry into STRL33+cells. More importantly, the preferential infection of STRL33+ cells in CCR5− PBLs by an R5/X4/STRL33 HIV-1 maternal isolate in the presence of a potent CXCR4 antagonist (AMD3100) suggests that STRL33 can be used as a coreceptor by HIV-1 on primary cells. Rhesus macaque (rh) STRL33 was used less efficiently than human STRL33 by the majority of SIV Env proteins tested despite similar levels of expression, thereby making it less likely that STRL33 is a relevant coreceptor in the rhesus macaque system. In summary, the expression pattern and coreceptor activity of STRL33 suggest its involvement in trafficking of tumor-infiltrating lymphocytes and indicate that STRL33 may be a relevant coreceptor in vivo.
Productive viral entry by primate lentiviruses requires the cooperative interaction of the viral envelope (Env) protein with CD4 and a coreceptor.1-4 CCR5 and CXCR4 are the major human immunodeficiency virus 1 (HIV-1) coreceptors, while most simian virus (SIV) isolates use CCR5 but not CXCR4 for viral entry.5Most SIV isolates can be propagated in T cell lines (eg, CEMx174) that express CXCR4 but not CCR5. As a result, coreceptors other than CCR5 and CXCR4 must be responsible for effecting viral entry into these cells.6-8 Receptors such as APJ, ChemR23, GPR1, GPR15/BOB, and STRL33/Bonzo, which support infection for some HIV and SIV strains in vitro,9-14 have been identified, although their in vivo relevance is not clear.
STRL33/Bonzo was originally cloned from a tumor infiltrating cell line by degenerate polymerase chain reaction (PCR) during a search for novel chemokine receptors.15 It was independently isolated in a functional expression screening assay by its ability to support viral infection by some SIV strains.9 Subsequent testing revealed that STRL33 supports efficient cell-cell fusion by a wide variety of SIV Env proteins,11 although it is generally inefficient at mediating infection by HIV-1 isolates.11,16 However, most HIV-1 isolates are derived from peripheral blood, and studying the coreceptor usage patterns of these isolates may not be optimal for correlation with the myriad aspects of HIV pathogenesis. Of all the HIV-1 isolates screened thus far, studies have only described a maternal-infant pair from a clear case of vertical transmission that uses STRL33 as efficiently as the major HIV-1 coreceptors CCR5 and CXCR4.17 18
Given the use of STRL33 by a wide variety of SIV Env proteins and the prominence of the SIV infection model in the study of HIV pathogenesis, we sought to finely characterize the expression pattern and coreceptor activity of STRL33. Expression studies using a monoclonal antibody (mAb) developed against STRL33 found that STRL33 was expressed on CD4+ truly naive (CD45RA+/CD62L+) T cells, B lymphocytes, and specific subsets of natural killer (NK) cells that are found predominantly in the human decidua during pregnancy.19 These NK cells have also been implicated in solid tissue surveillance.20 Infection ofccr5Δ32 homozygous peripheral blood lymphocytes (PBLs) was detected in STRL33+ cells using either SIV Env protein–pseudotyped Green Fluorescent Protein (GFP) reporter viruses or the maternal STRL33-using primary HIV-1 isolates described above. The preferential infection of STRL33+, CCR5− PBLs by the R5/X4/STRL33–tropic maternal isolate in the presence of a CXCR4 antagonist (AMD3100) strongly suggests that STRL33 can mediate viral entry into primary cells. In summary, the restricted but specific expression of STRL33 on cellular subsets provides potential insights into the physiologic role of STRL33, and the characterization of its coreceptor activity on primary PBLs provides insights into its role in SIV and HIV-1 pathogenesis.
Materials and methods
Cell culture conditions and cell lines
All cell lines (GHOST-STRL33 CELLS) (American Type Culture Collection and the National Institutes of Health AIDS Research and Reference Reagent Program, Bethesda, MD) were maintained according to the suppliers' recommendations. Primary PBLs from either wild-type orccr5Δ32 homozygous individuals, determined by PCR genotyping, were purified by Ficoll-Hypaque gradient (Pharmacia; Uppsala, Sweden) and resuspended in Roswell Park Memorial Institute medium (RPMI 1640) supplemented with 1% penicillin-streptomycin and 10% fetal calf serum (FCS). For 7-10 days, the cells were left in either unstimulated medium or medium stimulated with interleukin-2 (IL-2) alone, 10 μg/mL phytohemagglutinin (PHA) plus 20 U/mL IL-2 (PHA/IL-2), or 2 μg/mL goat-antimouse cross-linked anti-CD3 plus (20 U/ml) IL-2 (OKT3/IL-2).
Infection studies
GFP was cloned in place of the luciferase gene in pNL-R-E-luc21,22 to create GFP reporter viruses when the pNL-R-E-GFP backbone was cotransfected with plasmids expressing the appropriate Env protein. GFP reporter viruses were used in infection assays in a similar manner to luc reporter viruses.21 22All infections were performed in the presence of 8 μg/mL diethylaminoethyl-dextran (DEAE-dextran). Approximately 48 hours after infection, infected target cells were harvested with 5 mmol/L ethylenediamine tetraacetic acid (EDTA) or Versene, washed once with phosphate-buffered saline (PBS) and either fixed directly with 2% paraformaldehyde or stained with the appropriate antibodies and processed for fluorescence activated cell sorter (FACS) analysis. pcDNA3 transfected cells infected with the same amount of GFP reporter virus were used as background controls for GFP expression. Inhibition experiments were carried out by preincubating the target cells with indicated amounts of appropriate antibodies for 30 minutes prior to virus exposure. Infection using the maternal-fetal HIV-1 viral isolates (M6v3 and P6v3) was performed as above, but the analysis was done 1 week after infection using intracellular p24-antigen staining and FACS analysis.
Generation of rhesus and human STRL33 expression vectors
Rhesus macaque PBMCs (rhPBMCs) were isolated using lymphocyte separation medium (Biochrom, Berlin, Germany) and stimulated for 3 days in RPMI 1640 medium containing 4 μg PHA, 100 U/mL IL-2, and 20% FCS. RNA was extracted from 107 cells using Trizol (Gibco Life Technologies, Karlsruhe, Germany) and reverse transcribed using an Avian Myeloblastosis Virus (AMV)-RT–based (AMV–reverse transcription–based) cDNA synthesis kit (Boehringer Mannheim, Mannheim, Germany). The primer used for amplification of STRL33/p5Bonzo was 5′-CCGGATCCGGTGTT CATCAGAACAGACACCATG-3′ and for STRL33/p3Bonzo, the primer was 5′-CCGAATTCTTTCGAAACCCT GGCAAGGCCT-3′. The BamHI (Bacillus amyloliquefaciens H) and EcoRI (Escherichia coli RY13) restriction sites in the multiple cloning site (MCS) were used for cloning into pcDNA3 (Invitrogen, San Diego, CA). STRL33/Bonzo variants with the AU1-tag (DTYRYI) at the N-terminal were constructed by PCR mutagenesis using primers p5BonzoAU1 5′-CCGGATCCACCATG GACA CCTACAGATACATAGCAGAGTATGATCACTATGA and p3- Bonzo (for N-terminal tag). The PCR-amplicons were inserted into the BamHI-EcoRI site in the multiple cloning site of pcDNA3. All cloned PCR products were completely sequenced to confirm that only the introduced changes were present. Our sequence was identical to the GenBank entry AF124380 except that the codon for the last amino acid was TTA instead of CTA, both of which encode for leucine. Human STRL33 was PCR-amplified from genomic DNA using previously described primers11 and cloned into the EcoR1/XbaI MCS of pcDNA3. The ORF of the cloned human STRL33 was sequenced and was found to be identical to the deposited GenBank sequence (U73529).
Antibodies
The antibodies used in the study included phycoerythrin-conjugated (PE-conjugated) CD4 (Q4120, Sigma); allophycocyanin-conjugated (APC-conjugated) anti-CD3, Tricolor-conjugated anti-CD4 (S3.5), anti-CD8(3B5), anti-CD45RA(MEM56), anti-CD56 (NKI-nbl-1), fluorescence isothiocyanate–conjugated (FITC-conjugated) anti-CD19 (SJ35-C1), anti-CD16(3G8), and anti-CD62L (DREG-56) (clone names noted in parentheses; Caltag, Burlingame, CA); FITC- and PE-conjugated anti-p24 (KC57; Coulter, Miami, FL); and PE-conjugated anti-CCR52D7(PharMingen, San Diego, CA). Biotinylated anti-AU1 mAb (Babco, Berkeley, CA) was detected by secondary staining with PE-conjugated streptavidin (PharMingen). The mAb 699 (mouse anti-STRL33 clone 56811, R&D Systems, Minneapolis, MN) was made by immunizing Balb/c mice using syngeneic mouse myeloma (NSO) cells stably expressing the full-length human STRL33 sequence as described.23 Purified mAb 699 was directly conjugated to PE (by Molecular Probes, Eugene, OR), and only chromatographic fractions with 565 λ/280 λ absorbance ratios of 3.4 ± 0.2 (indicating a fluorochrome-to-conjugate ratio of 1) were used.
FACS analysis
Multicolor FACS analysis was performed essentially as described.24 Whole blood lysis was carried out using CAL-Lyse (Caltag, Burlingame, CA) according to manufacturer's instructions. Results are presented as indicated in the appropriate figure legends. Quantitative FACS (QFACS) analysis was performed by converting the geometric mean channel fluorescence to the mean number of equivalent soluble fluorochrome units (MESFUs) using a standardized microbead kit (Quantibrite, Becton Dickinson, San Jose, CA). For STRL33 quantification, only antibodies with a fluorochrome-to-conjugate ratio of 1 were used. Therefore, MESFUs are equivalent to the number of ABS on a cell. The actual number of antigens (STRL33) on a cell may vary from the number of ABS by 2-fold or more depending on whether the antibody exhibits monomeric or bivalent binding or whether all STRL33 molecules are conformationally accessible to mAb 699.
Results
Specificity of the anti-STRL33 mAb
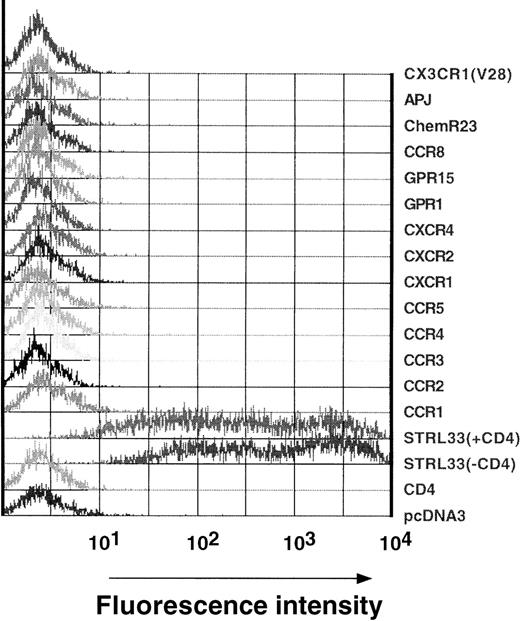
An anti-STRL33 mAb, mAb 699, was derived by immunization of Balb/c mice with STRL33-stable transfectants. To confirm its specificity, human 293T cells transiently expressing high levels of CD4 or various chemokine or orphan 7 transmembrane domain receptors (CCR1, CCR2, CCR3, CCR4, CCR5, CXCR1, CXCR2, CXCR4, GPR1, GPR15, CCR8, ChemR23, APJ, and CX3CR1), many of which have been shown to have in vitro HIV-1 or SIV coreceptor activity, were stained with mAb 699 (Figure1). The mAb only reacted against STRL33 transfectants, and its reactivity was not affected by coexpression of CD4 (Figure 1). Therefore, mAb 699 was specific for STRL33 among the known coreceptors for HIV and SIV. Like many other antibodies to chemokine receptors, mAb 699 recognizes a conformation-dependent epitope since it did not recognize STRL33 by Western blot analysis (data not shown).
Specificity of anti-STRL33 mAb 699.
293T cells were transfected with CD4 and/or the chemokine/orphan receptor constructs listed in the figure. Transfected cells were lifted off the plate with 5 mmol/L EDTA 18 hours after transfection and were stained with PE-conjugated anti-STRL33 antibody as indicated in “Materials and methods.” Data are representative of 2 independent experiments.
Specificity of anti-STRL33 mAb 699.
293T cells were transfected with CD4 and/or the chemokine/orphan receptor constructs listed in the figure. Transfected cells were lifted off the plate with 5 mmol/L EDTA 18 hours after transfection and were stained with PE-conjugated anti-STRL33 antibody as indicated in “Materials and methods.” Data are representative of 2 independent experiments.
STRL33 expression on fresh whole blood
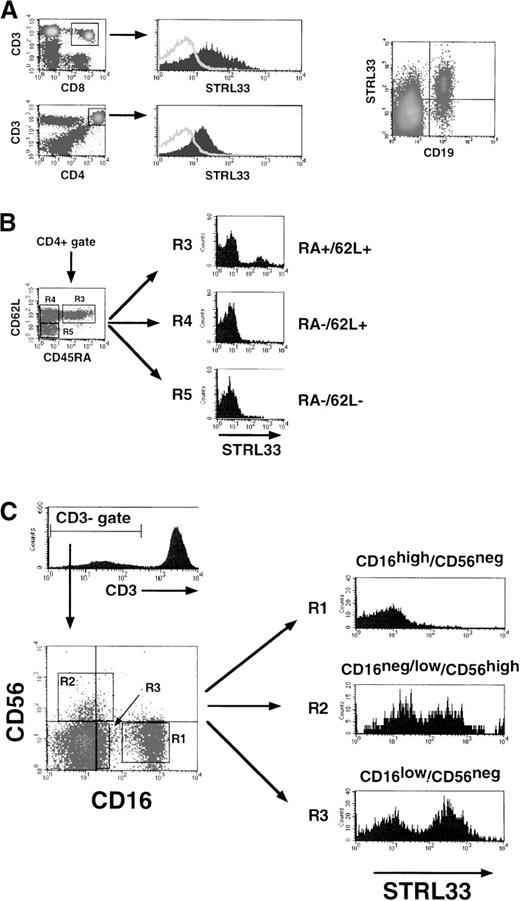
For multicolor and QFACS analysis, the STRL33 mAb was directly conjugated to PE, and only high-pressure liquid chromatography (HPLC) fractions with 565 λ/280 λ absorbance ratios of 3.4 ± 0.2 (indicating a fluorochrome-to-conjugate ratio of 1) were used. Fresh whole blood from 8 healthy HIV−donors was obtained in acid citrate dextrose tubes and stained within the hour for the cell surface markers indicated. There was minimal and variable expression of STRL33 on bulk CD4+ or CD8+ T lymphocytes (CD3+). A high expressing donor is shown in Figure 2A. The majority (more than 80%) of STRL33+ cells were in the B-lymphocyte (CD19+) gate. Among CD4+ cells, a distinct subpopulation of truly naive CD45RA+/CD62L+cells highly expressed STRL33. True memory cells (CD45RA−/CD62L−) or memory cells targeting to the lymph node (CD45RA−/CD62L+) were essentially negative for STRL33 (Figure 2B).
STRL33 expression on PBMCs.
We used 4-color FACS analysis to identify the various subpopulations of PBMCs indicated. Fresh whole blood was stained with fluorochrome-conjugated mAb within an hour of venupuncture, and FACS analysis was performed after ammonium chloride–mediated selective red blood cell lysis. STRL33 expression on (A) bulk CD4+, CD8+ T cells (CD3+), and CD19+ B cells; (B) CD4+ naive (CD45RA+/CD62L+) or memory (CD45RA−) T cells; and (C) NK cell subsets (R1, R2, and R3).
STRL33 expression on PBMCs.
We used 4-color FACS analysis to identify the various subpopulations of PBMCs indicated. Fresh whole blood was stained with fluorochrome-conjugated mAb within an hour of venupuncture, and FACS analysis was performed after ammonium chloride–mediated selective red blood cell lysis. STRL33 expression on (A) bulk CD4+, CD8+ T cells (CD3+), and CD19+ B cells; (B) CD4+ naive (CD45RA+/CD62L+) or memory (CD45RA−) T cells; and (C) NK cell subsets (R1, R2, and R3).
Because STRL33 was originally cloned from a tumor-infiltrating lymphocyte cell line,15 we investigated the expression of STRL33 on tumor-infiltrating lymphocyte subsets. NK cells, although comprising only 5%-15% of circulating lymphocytes, can account for up to 45% of tissue- or tumor-infiltrating lymphocytes.25 At least 3 subsets of NK cells have been recognized based on the cell surface markers CD16 and CD56 (Figure 2C: R1, R2, and R3).26 STRL33 expression was confined to the CD3−/ CD16−/low/CD56+(R2) or CD3−/CD16low/CD56− (R3) NK subsets, which have been implicated in solid tissue surveillance20 and are present in high numbers in the human decidua during pregnancy.19 STRL33 was not expressed on CD16high/CD56± NK cells, which are thought to mediate the classical non–major histocompatibility complex–restricted (non-MHC–restricted) natural immunity to virally infected cells.25 27
STRL33 expression levels required for efficient infection
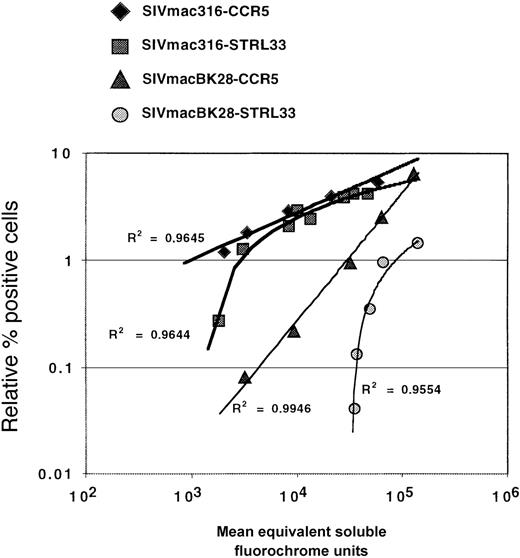
Coreceptor expression levels are important for efficient viral entry.28,29 This has been shown for CCR528 and CCR3,30 and it has also been suggested for STRL33 because infection of GHOST cells stably expressing STRL33 is typically much less efficient than transient STRL33 transfectants.11,16 Using QFACS analysis,24 we determined that transient STRL33 transfectants expressed at least 1 to 2 logs more surface STRL33 than the stable GHOST cells (more than 30 000 vs less than 3000 STRL33 ABS), perhaps accounting for the inability of GHOST-STRL33 cells to support efficient infection in some cases. To directly compare the efficiency of infection mediated by CCR5 versus STRL33, we titrated the amount of STRL33 and CCR5 transfected onto 293T cells and infected them in parallel with SIV Env protein–pseudotyped (SIVmacBK28 and SIVmac316) GFP reporter viruses. Two days after infection, the CCR5 and STRL33 levels were quantified only on transfected cells to directly correlate the efficiency of infection with the levels of coreceptor expression. In separate experiments, we found that infection of cells with ourenv- and nef-deleted reporter viruses did not down-modulate receptor expression (data not shown). For both SIVmacBK28 and SIVmac316, we found that below a threshold level of coreceptor molecules, STRL33 was much less efficient than CCR5 in supporting infection (Figure 3). This was in stark contrast to CCR5, where the efficiency of infection was linearly reduced as far as the threshold of our QFACS assay (approximately 200-400 ABS) (Figure 3).31 Therefore, efficient infection via STRL33 requires a higher threshold level than CCR5, at least for the virus strains tested here.
Effects of receptor expression levels on virus infection.
Serial dilutions (1 μg to 0.009 μg per 12 wells) of STRL33 and CCR5 expression plasmids were transiently transfected into 293T cells along with a constant amount of 1 μg CD4. Two days later, cells were harvested with Versene and stained for either CCR5 (mAb 2D7) or STRL33 (mAb 699). CCR5 and STRL33 levels on gated GFP+ cells (ie, infected cells) were quantified using QFACs as described. The relative percentage (%) of GFP+ cells was linearly regressed against CCR5 or STRL33 levels (presented as the number of ABS). Data from 1 representative experiment out of 3 experiments with SIVmac316 and SIVmacBK28 env protein–pseudotyped reporter viruses are presented. The linear trend for the CCR5 infections can be extrapolated back to approximately 400 ABS as previously shown.31
Effects of receptor expression levels on virus infection.
Serial dilutions (1 μg to 0.009 μg per 12 wells) of STRL33 and CCR5 expression plasmids were transiently transfected into 293T cells along with a constant amount of 1 μg CD4. Two days later, cells were harvested with Versene and stained for either CCR5 (mAb 2D7) or STRL33 (mAb 699). CCR5 and STRL33 levels on gated GFP+ cells (ie, infected cells) were quantified using QFACs as described. The relative percentage (%) of GFP+ cells was linearly regressed against CCR5 or STRL33 levels (presented as the number of ABS). Data from 1 representative experiment out of 3 experiments with SIVmac316 and SIVmacBK28 env protein–pseudotyped reporter viruses are presented. The linear trend for the CCR5 infections can be extrapolated back to approximately 400 ABS as previously shown.31
Quantification of STRL33 levels on primary PBLs
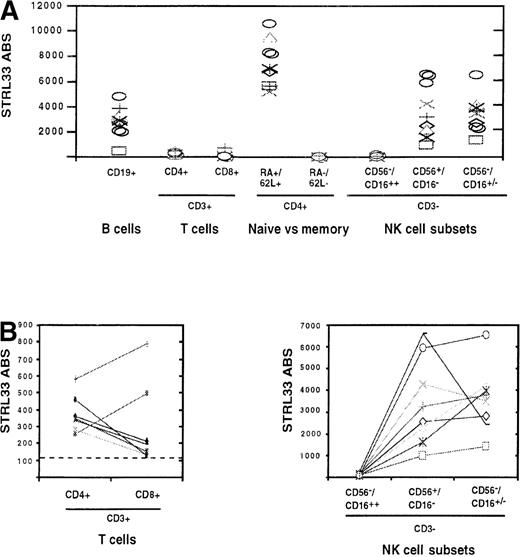
To determine if the minimal levels of STRL33 required for efficient infection were present on primary PBLs, the levels of STRL33 on the various PBL subsets described in Figure 2 were quantified using QFACS analysis.24 We found that expression levels could vary by more than 10-fold between donors, depending on the lymphocyte subset analyzed (Figure 4A). STRL33 levels were highest on a subset of naive CD45RA+/CD62L+/CD4+ T cells, ranging from approximately 6000-11 000 ABS per cell. The relative differences between subsets were not always the same between donors. For example, on most donors, STRL33 expression was higher on bulk CD4+ T cells compared with CD8+ T cells, but the opposite was true for 2 of 8 donors (Figure 4B). Variable expression was also seen on NK subsets. While STRL33 expression was uniformly absent on the CD3−/CD16highsubset of NK cells, there was a differential expression of STRL33 among donors between the remaining 2 NK subsets (Figures 2C and 4B). Whether this differential expression of STRL33 on specific NK subsets can be correlated to the activation status or functional activity of these NK cells is a matter for future studies. Nevertheless, at least on some subsets and in some volunteers, there were sufficient cell surface levels of STRL33 to mediate efficient virus infection.
Quantification of STRL33 levels on PBL subsets.
Levels of STRL33 expression on various PBL subsets delineated in Figure2 were quantified using QFACS as described in “Materials and methods.” (A) STRL33 ABS from 8 different donors on the various PBL subsets are shown. (B) Variation in STRL33 expression levels within each donor among bulk CD4+/CD8+ T cells (CD3+) and NK cell subsets are shown.
Quantification of STRL33 levels on PBL subsets.
Levels of STRL33 expression on various PBL subsets delineated in Figure2 were quantified using QFACS as described in “Materials and methods.” (A) STRL33 ABS from 8 different donors on the various PBL subsets are shown. (B) Variation in STRL33 expression levels within each donor among bulk CD4+/CD8+ T cells (CD3+) and NK cell subsets are shown.
STRL33 expression can be modulated by isolation and stimulation protocols
Ficoll-Hypaque purification of PBMCs can acutely affect chemokine receptor expression.24,32 Indeed, we found that STRL33 expression on CD19+ B cells (the most abundant STRL33+ cell type) was almost completely down-regulated after Ficoll-Hypaque purification and overnight incubation in media supplemented with 10% FCS (Figure 5A). Ficoll-Hypaque–induced transient down-regulation also occurred in bulk T cells (data not shown) and appears to be true for STRL33, CCR5, and CXCR4.24 32 Therefore, until this phenomenon is better characterized, we believe it is more accurate to use whole blood for native chemokine receptor expression studies.
Modulation of STRL33 expression levels by isolation and stimulation protocols.
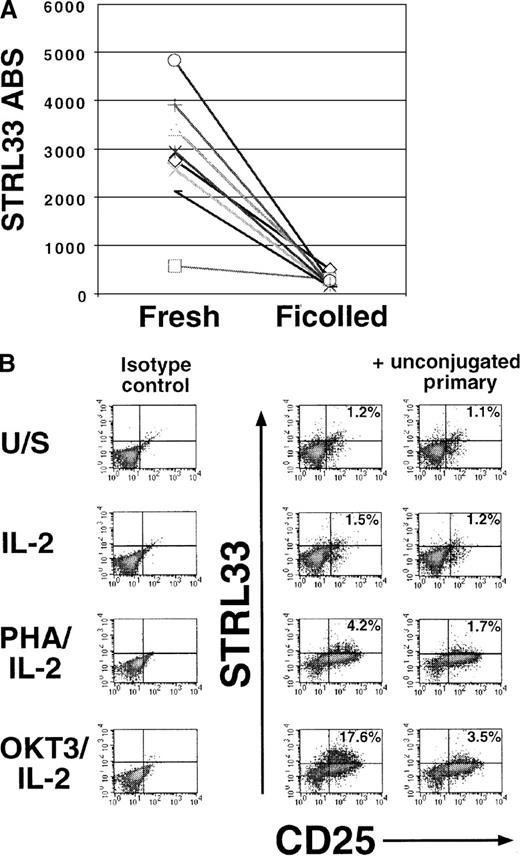
(A) STRL33 levels were quantified on fresh whole blood (CD19+ B-cell subset) and after Ficoll-Hypaque gradient purification and overnight incubation in RPMI 1640 supplemented with 10% FCS. Each symbol represents a different donor out of 8 total donors. (B) PBLs from a homozygous ccr5Δ32 donor were Ficoll-Hypaque purified and either left unstimulated (U/S) in media (RPMI 1640 plus 10% FCS) or stimulated with IL-2, PHA/IL-2, or cross-linked anti-CD3/IL-2 (OKT3/IL-2) for 7 days. Cells were then costained for STRL33 and CD25 (as an activation marker). Note the increase in STRL33 levels only on the CD25+ subset. Similar data were obtained from 8 donors who had a wild-type for CCR5.
Modulation of STRL33 expression levels by isolation and stimulation protocols.
(A) STRL33 levels were quantified on fresh whole blood (CD19+ B-cell subset) and after Ficoll-Hypaque gradient purification and overnight incubation in RPMI 1640 supplemented with 10% FCS. Each symbol represents a different donor out of 8 total donors. (B) PBLs from a homozygous ccr5Δ32 donor were Ficoll-Hypaque purified and either left unstimulated (U/S) in media (RPMI 1640 plus 10% FCS) or stimulated with IL-2, PHA/IL-2, or cross-linked anti-CD3/IL-2 (OKT3/IL-2) for 7 days. Cells were then costained for STRL33 and CD25 (as an activation marker). Note the increase in STRL33 levels only on the CD25+ subset. Similar data were obtained from 8 donors who had a wild-type for CCR5.
To determine if STRL33 expression can be up-regulated upon in vitro stimulation, we purified PBMCs with Ficoll-Hypaque and stimulated them for 7-10 days as shown in Figure 5B. Mitogenic stimulation was required for the induction of STRL33 expression in the presence of IL-2, and OKT3 was a more potent inducer than PHA (Figure 5B, middle column). By contrast, IL-2 treatment alone resulted in minimal STRL33 expression. STRL33 up-regulation was confined to acutely activated lymphocytes (CD25++; Figure 5B, upper right quadrant). As an additional specificity control, a 50-fold excess of unconjugated anti-STRL33 mAb 699 was used as a competitor and was able to compete off the positive STRL33 staining in the CD25++ gate (Figure 5B). Our results show that STRL33 expression in cultured PBLs is highly dependent on the stimulation protocol used, and studies into the use of STRL33 as a coreceptor on primary cells will need to take this important variable into account.
Infection of STRL33+ CCR5− PBLs
A number of SIV strains that use CCR5 but not CXCR4 have been shown to infect CCR5− human PBMCs or T cell lines, which indicates that an alternative coreceptor, such as STRL33, may be used.6,8 To determine if SIV strains can infect CCR5−, STRL33+ human cells, SIV Env protein–pseudotyped GFP reporter viruses were used to infect stimulated PBLs from ccr5Δ32 homozygous donors. Infection was obtained with SIV and Murine Leukemia Virus (MLV) (amphotropic) Env protein–pseudotyped viruses but not with an HIV-1 R5 Env protein–pseudotyped virus (Figure 6A, first column). The addition of 20 μg/mL neutralizing anti-CD4 antibody (Leu3A) completely inhibited infection by SIVmacBK28, but the anti-CD4 antibody only inhibited infection by SIVsmΔB670clone3 by about 50% (Figure 6A, second column). We have previously shown that SIVsmΔB670clone3 exhibits a considerable degree of CD4 independence with regard to CCR5-mediated infection.31,33 The addition of 20 μg/mL STRL33 mAb had no significant effect on SIV infection. However, infection inhibition experiments on transfected cells indicated that our STRL33 antibody was nonneutralizing (Figure 6B), a not uncommon property of antichemokine receptor antibodies.23,34,35 In addition, preliminary evidence indicates that the mAb 699 epitope is distinct from the regions in STRL33 that are required for coreceptor activity.54 Therefore, the lack of inhibition by mAb 699 does not imply that STRL33 is not used as a coreceptor on primary cells. Nevertheless, we costained the infected ccr5Δ32homozygous PBLs for STRL33 and found GFP+ cells within the STRL33+ populations (Figure 6C). This provides at least circumstantial evidence that STRL33 can be used as a coreceptor on stimulated ccr5Δ32 homozygous PBLs.
SIV infection of homozygous ccr5▵32 PBLs.
We stimulated ccr5Δ32 PBLs with OKT3/IL-2. (A) One week after stimulation, the indicated pseudotyped GFP reporter viruses were used to infect the stimulated cells in the presence or absence of the indicated antibodies (10 μg/mL anti-CD4 [leu3A], 20 μg/mL anti-CCR52D7, and 20 μg/mL anti-STRL33 [mAb 699]). The cells were fixed and analyzed by FACS for GFP expression 3 days later. (B) Pseudotyped GFP reporter viruses were also used to infect CD4- and CCR5- or STRL33-transfected 293T cells in the presence or absence of 20 μg/mL of the indicated antibodies (anti-CD4 [leu3A], 2D7 [R5 mAb], and anti-STRL33 [mAb 699]). The cells were harvested, fixed, and analyzed for GFP expression 2 days later. The results are presented as the percent of infected cells (GFP+) normalized against the negative control (percent of infected cells in the presence of 20 μg/mL mouse IgG). (C) Infected ccr5Δ32 PBLs from panel A were costained with anti-STRL33. GFP+ cells can clearly be seen in the STRL33+ gate (compare with uninfected/isotype control, at far right dot-plot). Data shown is representative of results from 2 donors.
SIV infection of homozygous ccr5▵32 PBLs.
We stimulated ccr5Δ32 PBLs with OKT3/IL-2. (A) One week after stimulation, the indicated pseudotyped GFP reporter viruses were used to infect the stimulated cells in the presence or absence of the indicated antibodies (10 μg/mL anti-CD4 [leu3A], 20 μg/mL anti-CCR52D7, and 20 μg/mL anti-STRL33 [mAb 699]). The cells were fixed and analyzed by FACS for GFP expression 3 days later. (B) Pseudotyped GFP reporter viruses were also used to infect CD4- and CCR5- or STRL33-transfected 293T cells in the presence or absence of 20 μg/mL of the indicated antibodies (anti-CD4 [leu3A], 2D7 [R5 mAb], and anti-STRL33 [mAb 699]). The cells were harvested, fixed, and analyzed for GFP expression 2 days later. The results are presented as the percent of infected cells (GFP+) normalized against the negative control (percent of infected cells in the presence of 20 μg/mL mouse IgG). (C) Infected ccr5Δ32 PBLs from panel A were costained with anti-STRL33. GFP+ cells can clearly be seen in the STRL33+ gate (compare with uninfected/isotype control, at far right dot-plot). Data shown is representative of results from 2 donors.
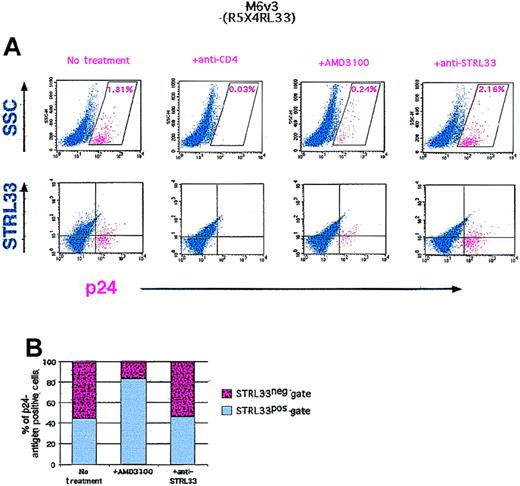
Zhang and colleagues17,18 have shown that a maternal-infant (R5/X4/STRL33–R5/STRL33) HIV-1 isolate pair derived from a case of vertical transmission uses STRL33 as efficiently as CCR5 and/or CXCR4. We used the R5/X4/STRL33 maternal isolate to infect PHA/IL-2 stimulated ccr5Δ32 homozygous PBLs. Intracellular p24-antigen staining 1 week after infection clearly indicated p24-antigen–positive cells (Figure 7A). Infection was inhibited completely by neutralizing anti-CD4 antibodies, but it was only inhibited by about 85% in the presence of excess AMD3100 (a potent CXCR4 inhibitor that abrogates X4-mediated infection by this and other virus strains)36,37 (Figure 7A, upper panel). Although CD4 is largely down-regulated upon productive viral infection,38 if one assumes that p24-antigen–positive cells were all initially CD4+, costaining with CD4 indicated that close to 10% of the initial CD4+ population was infected with the M6 virus (data not shown). Of these cells, up to 15% may be infectable via STRL33; AMD3100 only inhibited infection by about 85%, and the remaining p24-antigen–positive cells were largely in the STRL33 gate (Figure 7A, lower panel, and 7B). It should be noted that while this strain can also use the APJ coreceptor, the expression of APJ on PBLs has not been confirmed.10 14
HIV-1 infection of homozygous ccr5▵32 PBLs.
We stimulated ccr5Δ32 PBLs with PHA/IL-2 and infected with approximately 1200 TICD50 units of M6-v3 in the presence or absence of the indicated amounts of antibodies or the CXCR4 antagonist (AMD3100). The cells were stained for cell surface STRL33 and intracellular p24 antigen 1 week after infection. The p24-antigen–positive gate was defined as the region giving less than 0.01% of p24-antigen–positive cells when the p24-antigen antibody was used on uninfected donor cells (negative control). The p24-antigen–positive cells are indicated in red. (A) Upper panel: The percent of p24-antigen–positive cells in the presence or absence of the indicated antibodies or AMD3100 are shown in their respective dot-plots. Lower panel: The distribution of p24-antigen–positive cells in the STRL33+ or STRL33− gates under the same conditions. (B) Lower panel: Graphic representation of part A. The total number of p24-antigen–positive cells in the presence or absence of AMD3100 or the nonneutralizing anti-STRL33 antibody was normalized to 100%, and the proportion of p24-antigen–positive cells in the STRL33+ or STRL33− gates are shown.
HIV-1 infection of homozygous ccr5▵32 PBLs.
We stimulated ccr5Δ32 PBLs with PHA/IL-2 and infected with approximately 1200 TICD50 units of M6-v3 in the presence or absence of the indicated amounts of antibodies or the CXCR4 antagonist (AMD3100). The cells were stained for cell surface STRL33 and intracellular p24 antigen 1 week after infection. The p24-antigen–positive gate was defined as the region giving less than 0.01% of p24-antigen–positive cells when the p24-antigen antibody was used on uninfected donor cells (negative control). The p24-antigen–positive cells are indicated in red. (A) Upper panel: The percent of p24-antigen–positive cells in the presence or absence of the indicated antibodies or AMD3100 are shown in their respective dot-plots. Lower panel: The distribution of p24-antigen–positive cells in the STRL33+ or STRL33− gates under the same conditions. (B) Lower panel: Graphic representation of part A. The total number of p24-antigen–positive cells in the presence or absence of AMD3100 or the nonneutralizing anti-STRL33 antibody was normalized to 100%, and the proportion of p24-antigen–positive cells in the STRL33+ or STRL33− gates are shown.
Human STRL33 functions more efficiently as a coreceptor than rhesus STRL33
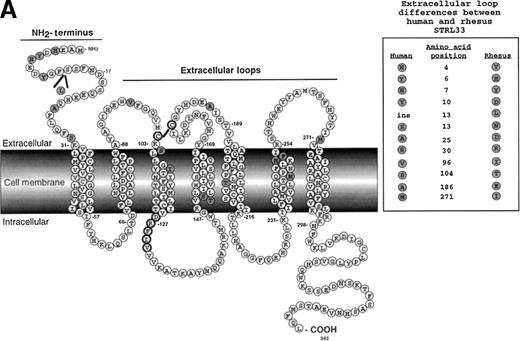
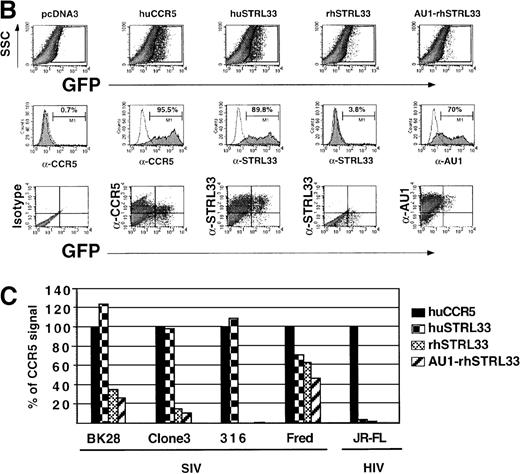
Rhesus CCR5 functions more effectively as a coreceptor than human CCR5 for many SIV isolates when CD4 levels are limiting or nonexistent.31 We sought to determine if this was also true for rhesus STRL33. An alignment profile shows 94% amino acid identity between human and rhesus STRL33, with most of the differences being in the N-terminal region (Figure 8A). Using SIVmacBK28 Env protein–pseudotyped GFP reporter viruses in infection assays on transfected cells, we found that rhesus STRL33 was used far less efficiently than huSTRL33 (Figure 8B, top row, and 8C). However, because rhesus STRL33 was not recognized by mAb 699, we generated an epitope-tagged rhesus STRL33 to ensure adequate cell surface expression (Figure 8B, middle row). Both tagged and untagged rhesus STRL33-supported SIV Env protein–mediated infection to a similar degree, albeit far less efficiently than human STRL33. Additional SIV Env proteins were also tested for their abilities to use human and rhesus STRL33 to infect cells. We found that SIVmac316 was unable to use rhesus STRL33, while SIVmac/17-E-Fr (Fred) used rhesus STRL33 at about 40%-60% efficiency compared with human STRL33 (Figure 8C). SIVsmΔB670clone3 minimally used rhesus STRL33. Preliminary studies have identified key residues in the divergent STRL33 N-terminal that are critical for coreceptor activity54.
Rhesus STRL33 is a less efficient coreceptor than human STRL33.
(A) Schematic diagram of human STRL33. Human and rhesus STRL33 are 94% identical at the amino acid level, with most of the extracellular loop differences (inset) clustered at the amino terminus. Differences between human and rhesus STRL33 are indicated by the shaded residues. The conserved “DRY” motif in chemokine receptors implicated for coupling to G-proteins is modified in human and rhesus STRL33 (filled circle residues). (B) SIVmac251(BK28 clone) env protein–pseudotyped GFP reporter virus was used to infect cells expressing CD4 and the indicated coreceptor (top row). Receptor expression was confirmed by staining with the indicated antibodies (middle row). Note that mAb 699 does not recognize rhesus STRL33, but AU1-tagged rhesus STRL33 indicated that rhesus STRL33 was expressed at similar levels to human STRL33. Infection was seen only on the transfected cell population (bottom row). Results were similar for SIVΔB670 clone 3 (Clone3), SIVmac/17E-Fr (Fred), SIVmac316 (316) and HIV-1 JR-FL and are quantified in panel C, where infection efficiency (% of GFP+ cells) was normalized against that obtained for human CCR5.
Rhesus STRL33 is a less efficient coreceptor than human STRL33.
(A) Schematic diagram of human STRL33. Human and rhesus STRL33 are 94% identical at the amino acid level, with most of the extracellular loop differences (inset) clustered at the amino terminus. Differences between human and rhesus STRL33 are indicated by the shaded residues. The conserved “DRY” motif in chemokine receptors implicated for coupling to G-proteins is modified in human and rhesus STRL33 (filled circle residues). (B) SIVmac251(BK28 clone) env protein–pseudotyped GFP reporter virus was used to infect cells expressing CD4 and the indicated coreceptor (top row). Receptor expression was confirmed by staining with the indicated antibodies (middle row). Note that mAb 699 does not recognize rhesus STRL33, but AU1-tagged rhesus STRL33 indicated that rhesus STRL33 was expressed at similar levels to human STRL33. Infection was seen only on the transfected cell population (bottom row). Results were similar for SIVΔB670 clone 3 (Clone3), SIVmac/17E-Fr (Fred), SIVmac316 (316) and HIV-1 JR-FL and are quantified in panel C, where infection efficiency (% of GFP+ cells) was normalized against that obtained for human CCR5.
Discussion
All primary strains of HIV-1 studied to date use CCR5, CXCR4, or both of these major coreceptors in conjunction with CD4 to enter cells.5 Alternative coreceptors, such as CCR2, CCR3, CCR8, CX3CR1, STRL33, APJ, ChemR23, GPR1, GPR15, and STRL33,9-13,30,39,40 can also support virus infection, but typically do so less efficiently than the major coreceptors and are used by smaller numbers of virus strains.5,11,16,17,30,41 The fact that SIV strains (which do not use CXCR4) can infect CCR5− human PBMCs6,8 indicates that at least some alternative coreceptors are expressed at levels sufficient to support virus infection on primary cell types. Our evidence indicates that STRL33 is a likely candidate for supporting infection of primary cells. In addition to being expressed on B cells and some NK cell subsets, STRL33 was expressed on freshly isolated truly naive CD45RA+/CD62L+ T lymphocytes at levels (approximately 6000-11 000 ABS) sufficient to support virus infection. In fact, STRL33 was more highly expressed on this CD4+T-cell subset than CXCR4 (approximately 2000-5000 ABS24). Furthermore, mitogenic (PHA or OKT3) stimulation of primary PBLs in the presence of IL-2 increased STRL33 expression on bulk CD4+/CD25+ T cells (Figure 5B and data not shown), potentially rendering additional cell types susceptible to infection via this coreceptor.
Coreceptor expression levels can impact the efficiency of viral entry, and indeed we found that STRL33 expression levels strongly impacted its coreceptor function. While there was a linear relationship between CCR5 expression and infection efficiency down to very low levels of coreceptor molecules (less than 103 CCR5 ABS), STRL33-dependent infection fell sharply below a threshold level of ABS. This threshold differed depending on the virus strain used (approximately 3000 ABS for SIVmac316 and approximately 30 000 for SIVmacBK28), and the differing threshold most likely accounts for the reported differences in the ability of HIV-1 strains to use STRL33 as a coreceptor since transient transfectants express far more STRL33 (at least 30 000 STRL ABS) than GHOST-STRL33 stable cells (less than 3000 STRL33 ABS).11,16 Our results suggest that expression levels of alternative coreceptors, such as STRL33 and CCR3,30 may more strongly impact coreceptor activity compared with the major coreceptors CCR5 and CXCR4.
Our STRL33 mAb did not block infection, which is not an uncommon feature of chemokine receptor antibodies. Therefore we used GFP reporter viruses bearing different Env proteins to infect stimulated CCR5− PBLs and asked whether STRL33+ cells became infected. We found that infection mediated by the SIVmacBK28 and SIVsmΔB670clone3 Env proteins, which do not use CXCR4, was clearly but not exclusively detected in STRL33+ cells (Figure 6C). More importantly, infection of STRL33+, CCR5− cells was also seen when using a maternal HIV-1 isolate (M6-v3), which was previously determined to be R5/X4/STRL33-tropic.18 M6-v3 infection of STRL33+, CCR5− cells in the presence of a potent CXCR4 antagonist (AMD3100)37,42,43resulted in the majority of p24-antigen–positive cells appearing in the STRL33+, CXCR4− gate, which suggests that STRL33 can mediate viral entry on primary PBLs. This observed difference from the published report18 is likely due to donor variability and the conditions under which the cells were grown. (We observed a 10-fold donor variability with regard to STRL33 expression.) To our knowledge, this is the first report of an HIV-1 infection of human PBMCs that occurs independently of CCR5 and CXCR4.
It is interesting to note that the only HIV-1 isolates known to use STRL33 as efficiently as the major HIV-1 coreceptors (CCR5 and CXCR4) were isolated from a case of maternal-infant transmission.17,18 There is evidence that bidirectional traffic of leukocytes may occur across the placental barrier during pregnancy,44 and contact with infectious maternal blood may be a route for HIV transmission. Naive CD45RA+/CD62L+ cells that express STRL33 are much more abundant in umbilical cord blood than in adult peripheral blood.43 It is interesting to note that CD3−/CD56+/CD16−/low NK cells (which express STRL33) are also present in high numbers in the human decidua during pregnancy19,45 and are known to be intimately associated with the fetal trophoblast cells during placental invasion.45 Although CD4 is not known to be expressed on NK cells, soluble33,46 or membrane-bound47 CD4 can in some circumstances mediate viral infection in trans. Whether transactivation by membrane-bound CD4 from neighboring CD4+lymphocytes in the crowded cellular environment of the maternal-fetal placental interface allows infection of STRL33-expressing NK cells remains to be determined.
Given the broad use of STRL33 by SIV strains,5,11 we were surprised to find that rhesus STRL33 was used much less efficiently than human STRL33, a deficiency that cannot be explained simply by expression levels. Rhesus and human STRL33 share a 94% identity at the amino acid level, with most of the differences clustered at the N-terminal (Figure 8A). Preliminary evidence indicates that single amino acid changes in the N-terminal from rhesus to human residues can make rhesus STRL33 as efficient as human STRL33 in terms of coreceptor activity and that the mAb 699 epitope is distinct from residues required for coreceptor activity.54 These results call into question the in vivo importance of STRL33 in the rhesus macaque system and underscores the importance of using coreceptors derived from appropriate nonhuman primates when studying primary SIV strains.
The expression pattern of STRL33 suggests possible in vivo functions for this orphan receptor. While the expression of STRL33 tended to track more with CXCR4 than CCR5, its pattern was certainly unique. In addition to naive CD45RA+/CD62L+ cells, STRL33 was expressed on B cells and on CD56+/CD16−/low or CD56−/CD16low NK cells, but it was not expressed not on CD16high/CD56± NK cells that were thought to target virally infected cells. STRL33 was originally cloned from a tumor-infiltrating cell line.15 NK cells possess non-MHC–restricted cytolytic activity and are thought to be the first line of defense against both virus infected cells and nascent tumorgenesis.25,27 Tumors known to elicit high numbers of infiltrating NK cells include bronchiogenic carcinomas and melanomas.48-50 We speculate that the immune system may have evolved a receptor (such as STRL33) to home in on specific tumor-secreted or membrane-associated products. This hypothesis can be tested by transplanting STRL33 knock-out mice with TIL-attracting tumors.
The proliferation of alternative coreceptors for primate lentiviruses, as determined by in vitro assays, underscores the need for understanding the roles these coreceptors may play in the immunopathogenesis of acquired immunodeficiency syndrome (AIDS). Whether these receptors support infection of primary cells is not yet clear. But reports that infection of CCR5− primary human cells can be invariably inhibited by CXCR4 antagonists18,51,52 argue that if alternative coreceptors do contribute to viral pathogenesis in vivo, it is apt to be via infection of very specific cell subsets. However, given the recent development of CCR5 and CXCR4 antagonists, it is important to ask if HIV-1 could evolve to use receptors other than CCR5 or CXCR4 in the face of strong selective pressure. A naturally occurring example of adaptive coreceptor use is provided by studies of Red-capped mangabeys, many of which are CCR5− due to the presence of a common CCR5 polymorphism (ccr5Δ24). SIV isolates taken from CCR5− Red-capped mangabeys (SIVrcm) exhibit highly efficient use of CCR2.53 We are not aware of other examples in which primary SIV strains can use CCR2 to infect cells in conjunction with CD4, which indicates that unexpected patterns of coreceptor use may arise if use of major coreceptors is prevented.
Interestingly, the only other coreceptor that can be detectably used by SIVrcm is STRL33. In dissecting the expression profile of STRL33 and delineating its responses to various mitogenic stimuli, we provide evidence that STRL33 may be a relevant coreceptor for HIV-1 in vivo. It is expressed on some CD4+ cells at levels sufficient to support virus infection. In addition, mitogenic stimulation could well result in additional cell types becoming positive for STRL33. Importantly, we have found a primary HIV-1 strain that can infect human PBLs independently of CCR5 and CXCR4, most likely via STRL33. Does this mean that inhibitors to STRL33 will have to be developed? At this point it is not clear. Only a single HIV-1 isolate studied thus far uses STRL33 efficiently, and this is the first reported case of HIV infection of human PBLs in a CCR5- and CXCR4-independent manner. In addition, STRL33 is expressed on a much smaller fraction of CD4+ T cells than are CCR5 or CXCR4. However, the use of CCR2 by SIV strains isolated from Red-capped mangabeys clearly demonstrates that unexpected coreceptor use patterns might occur in the face of strong selective pressure. Thus, studies into the coreceptor activity, expression patterns, and biologic function of STRL33, as well as other alternative coreceptors, will clearly be of value in complementing the development of effective coreceptor inhibitors.
Acknowledgments
We thank Drew Weissman, Luis Montaner, and Jim Hoxie for helpful discussions. We would also like to thank Y. J. Zhang and John Moore for providing the maternal-fetal viral isolates used in this study and for sharing unpublished results. The technical help provided by George Leslie is also appreciated. We also thank Mary Dietz, Dorothy Shroeder, John Humphrey, James David, and Frank Mortari at R&D Systems for invaluable help in generating the hybridoma.
Supported in part by K08 grant HL03923-01 (B.L.) from the National Heart, Lung, and Blood Institute, the National Institutes of Health (NIH), Bethesda, MD; by grant R01 40880 (R.W.D.) from the NIH; by a Burroughs Wellcome Fund Award for translation research, Burroughs Wellcome Fund, Research Triangle Park, NC; and by Small Business Innovation Grant (SBIR) R44 AI41299 (collaboration between R&D Systems and R.W.D.). R.W.D. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
Submitted January 17, 2000; accepted February 22, 2000.
Reprints:Benhur Lee, Room 806, or Robert W. Doms, Room 807A, Department of Pathology and Laboratory Medicine, Abramson Bldg, 34th Street & Civic Center Blvd, Philadelphia, PA, 19104; e-mail: benhur@mail.med.upenn.edu ordoms@mail.med.upenn.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 6. SIV infection of homozygous ccr5▵32 PBLs. / We stimulated ccr5Δ32 PBLs with OKT3/IL-2. (A) One week after stimulation, the indicated pseudotyped GFP reporter viruses were used to infect the stimulated cells in the presence or absence of the indicated antibodies (10 μg/mL anti-CD4 [leu3A], 20 μg/mL anti-CCR52D7, and 20 μg/mL anti-STRL33 [mAb 699]). The cells were fixed and analyzed by FACS for GFP expression 3 days later. (B) Pseudotyped GFP reporter viruses were also used to infect CD4- and CCR5- or STRL33-transfected 293T cells in the presence or absence of 20 μg/mL of the indicated antibodies (anti-CD4 [leu3A], 2D7 [R5 mAb], and anti-STRL33 [mAb 699]). The cells were harvested, fixed, and analyzed for GFP expression 2 days later. The results are presented as the percent of infected cells (GFP+) normalized against the negative control (percent of infected cells in the presence of 20 μg/mL mouse IgG). (C) Infected ccr5Δ32 PBLs from panel A were costained with anti-STRL33. GFP+ cells can clearly be seen in the STRL33+ gate (compare with uninfected/isotype control, at far right dot-plot). Data shown is representative of results from 2 donors.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/1/10.1182_blood.v96.1.41/4/m_bloo01353006x.jpeg?Expires=1769089129&Signature=c3UjrR6dVAsYocfVXEGZe3Dke2VA9v4rFNsL~M-tutsPf1xA~cN6XQYbEMdXCGFjKcN7oxV1qKe67J9-9wCNEMaJnfEOmoOoBWIiRHliZsMlw4Dg8zaUxCbX5OdEV9VWrQNYKctXxSRR-j3AgGvSd47AtdNZDPblv3uGVRTW5eLOFFP~FWYvKNF4dElAr5PPbUFTu5h8nefUBlYajhfxC341fExcCyuq2WpYjSVz0PHIJAyQ4zWRuWFGVDWKsbLM2TJafgCtRFOoxJDhqE4uDiJqM7pht-E5kBxoazvgDzJoVsuj32saI1alJ2LD4DoNACuMRjsyhZvEEFXCKzhcug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal