Abstract
The Kell blood group protein is a zinc endopeptidase that yields endothelin-3, a potent bioactive peptide, by cleavage of big endothelin-3, a larger intermediate precursor. On red cells, Kell protein is linked by a single disulfide bond to XK, a protein that traverses the membrane 10 times and whose absence, as occurs in the McLeod phenotype, is associated with a set of clinical symptoms that include nerve and muscle disorders and red cell acanthocytosis. Previous studies indicated that Kell is primarily expressed in erythroid tissues, whereas XK has a wider tissue distribution. The tissue distribution of Kell protein has been further investigated by Northern blot analysis, PCR-screening of tissue complementary DNAs (cDNAs), and Western immunoblots. Screening of an RNA dot-blot panel confirmed that Kell is primarily expressed in erythroid tissues but is also expressed in a near equal amount in testis, with weaker expression in a large number of other tissues. PCR-screening of cDNAs from different tissues and DNA sequencing of the products gave similar results. In 2 of the nonerythroid tissues tested, testis and skeletal muscle, Kell protein was detected by Western immunoblotting. In skeletal muscle, isolation of XK with a specific antibody coisolated Kell protein. These studies demonstrate that Kell is expressed in both erythroid and nonerythroid tissues and is associated with XK.
Kell is a polymorphic 93 kDa type II membrane glycoprotein1 that on human red cells carries the majority of the Kell blood group antigens.2,3 Kell protein is a member of the neprilysin (M13) family of zinc endopeptidases, and it preferentially cleaves big endothelin-3 at a Trp21-Ile22 bond, yielding the 21-amino acid bioactive peptide, endothelin-3.4Previous Northern blot studies indicated that Kell is expressed primarily in bone marrow and fetal liver, with little or no Kell transcripts noted in brain, kidney, adult liver, and lung. In the erythroid tissues, the major Kell transcript was 2.5 kilobase (kb), but smaller amounts of larger transcripts, 6.6, 11.5, and 13.2 kb, were also observed.5
Early serological studies had predicted an association of Kell with another red cell membrane protein, XK, which expresses a surface antigen, termed Kx.2 XK is a 444-amino acid protein that is predicted to traverse the membrane 10 times.6 Although the function of XK is not known, its absence from red cells, the McLeod phenotype, is associated with a set of clinical symptoms that include red cell acanthocytosis, elevated levels of serum creatine phosphokinase, and late onset forms of muscular dystrophy and nerve abnormalities, characterized by areflexia and chorea. A covalent linkage between Kell and XK was shown, in that Kell and XK were coisolated by immunoprecipitation in nonreduced conditions.7,8 Site-directed mutagenesis of specific cysteine residues in Kell and XK proteins and coexpression in transfected cells demonstrated that Kell Cys72 is disulfide-linked to XK Cys347.9 Because XK had been shown to be expressed not only in erythroid tissues but also in skeletal muscle, pancreas, heart, brain, placenta, and several other tissues,6 we have reinvestigated the tissue distribution of Kell with the use of Northern blot analysis, PCR-screening of human tissue complementary cDNAs (cDNAs), and Western immunoblots. We find that Kell is also expressed in many nonerythroid tissues, primarily in testis, and that in the nonerythroid tissues, as exemplified by skeletal muscle, Kell is disulfide-linked to XK.
Materials and methods
RNA dot-blot analysis
A dot-blot panel, containing 89-514 ng of 50 different human tissue poly A+ RNAs, normalized for equal amounts of actin messenger RNA (mRNA), was purchased from Clontech (Palo Alto, CA). The panel, which also contained several RNA and DNA controls, was hybridized to a 32P-labeled 0.44-kb antisense RNA coveringKEL exons 9 to 13.10 The riboprobe was synthesized by SP6-directed reverse transcription of a fragment of the Kell cDNA, using the Strip-EZ RNA kit (Ambion, Austin, TX).
Northern blot analysis
Poly A+ RNA from various tissues, separated by electrophoresis and transferred to membranes, was purchased from Clontech. The membranes were hybridized at 68°C with32P-labeled Kell antisense mRNA. The riboprobe was synthesized by T7-directed reverse transcription of 2.4-kb Kell cDNA,1 10 using the Strip-EZ RNA kit purchased from Ambion. A random primed actin cDNA probe was synthesized, using the Strip-EZ DNA kit (Ambion).
PCR screening of KEL in human tissue cDNAs
First, strand cDNA preparations, normalized for equal amounts of 6 different housekeeping cDNAs, from 20 different human tissues were purchased from Clontech. About 1 ng of cDNA from each of the different tissues was used as a template for 25 cycles of PCR amplification as recommended by the manufacturer. The following primers, derived from exons 14 and 18 of the KEL gene,10 were used: 5′-TTG CAG CCT CAC CCC CAA CAC AGG TGG A-3′ and 5′-AGC CCC CCA ACG TCT GCA GCA TTC TCT A-3′.
The cDNA preparations were also used for detection of glycophorin A (GPA), using the following primers derived from the GPA gene11: 5′-CAG ACA AAT GAT ACG CAC AAA CGG GAC-3′ and 5′-CTT TAT CAG TCG GCG AAT ACC GTA AGA-3′. One of the primers contained sequences spanning exons 2 and 3, and the other was derived from exon 5.
PCR amplification of a skeletal muscle cDNA containing the Kell coding region
A Kell cDNA from a muscle cDNA library (Human Skeletal Muscle, Marathon-Ready, cDNA library, Clontech) was PCR-amplified by the nested 3′ end RACE procedure.
The following forward primers, specific for Kell sequences, were used: 5′-ATG GAA GGT GGG GAC CAA AGT GAG GAA G-3′ and 5′-GCC GAG GGA ACG CAG CCA GGC AGG TGG A-3′.
The first primer, nt 121-149, which includes the initiation codon inKEL exon 1, was used with adapter primer 1 (AP1), present at the 3′ end, in a first round of PCR amplification. The second primer, nt 150-177, from KEL exon 2, was used with adapter primer 2 (AP2), which is internal to AP1. A 2.4-kb PCR product was obtained, covering from nt 150 to the 3′ end of Kell cDNA.1 10
The 5′ transcription start site was obtained by 5′ RACE nested polymerase chain reaction (PCR), using the followingKEL-specific reverse primers: 5′-CTC CTC CCC AGA GCC TGG GTG CCA GGA ATT-3′ and 5′-CTG GCC ACT GCC CAT GGC CTG CTC CCT TCC-3′.
The first primer, derived from KEL exon 5, was used with AP1 present at the 5′ end, and the second primer, representing sequences from KEL exon 3, was used with AP2 that is internal to AP1. The PCR-amplified products were sequenced, using AP2 and the primer from exon 2.
DNA sequencing of PCR products
PCR-amplified products were separated by agarose gel electrophoresis, the bands excised and eluted for direct sequencing, using the Genclean kit, glassmilk-based method (Bio 101, Vista, CA). The PCR-amplified fragments from the various tissue cDNAs were sequenced, using the amplification primers.
The following Kell primers were used for sequencing the 2.4-kb PCR-derived Kell skeletal muscle cDNA: 5′-GCC GAG GGA ACG CAG CCA GGC AGG TGG A-3′ derived from KEL exon 2; 5′-GGT CCA GAA TTC CTG GCA CCC AGG C-3′ from exon 5 and 5′-CTC CTC CCC AGA GCC TGG GTG CCA GGA ATT-3′ from exon 5; 5′-GCT TGC CCT GTG CCC GCC GCT GCT C-3′ from exon 8; 5′-TGG CTG AGC TTT CTG CGT GCC TCC TGG A-3′ from exon 10 and 5′-GGA CTT TCT GCA GAG CCA CAT GAT C-3′ from exon 10; 5′-GGG TTG GAG GAG TCC AGC TGG AAA G-3′ from exon 15; 5′-ATG GCC CAC GAG CTG TTG CA-3′ from exon 16; 5′-AGC CCC CCA ACG TCT GCA GCA TTC TCT A-3′ from exons 17-18; and 5′-GGT GTT GGT CGA TAT TTC TGT GCT GTG GC-3′ from exon 19.
The Microchemistry Laboratory of the New York Blood Center performed DNA sequencing with the use of an ABI 373XL sequencer (Perkin-Elmer, Applied Biosystems, Foster City, CA), procedures from the manual, and ABI Big Dye reagents with BD Half-Term (Gen Pak).
Antibodies
Four antibodies were used in this study. (1) An antibody to the second extracellular loop of XK was used in immunoprecipitation studies, as previously described.9 This antibody was tagged with biotin, using sulfosuccinimidyl 6-(biotinoamido) hexanoate (sulfo-NHS-LC-biotin; Pierce, Rockford, IL), according to the manufacturer's directions. (2) Kell protein, isolated from red blood cells, was used to generate a polyclonal antibody in rabbits. (3) An antibody to the C-terminal intracellular domain of XK was raised in rabbits by treatment with a 39 mer synthetic peptide, corresponding to XK amino acids 406 to 444. The Microchemistry Laboratory of the New York Blood Center prepared the synthetic peptide. This peptide was also used to affinity purify the antibody. By Western blot assay, this antibody reacted with XK and XK/Kell complex from red cells of common Kell phenotype but not with proteins of the same size from McLeod red cells. The antibody did, however, have some cross-reaction with band 3 proteins from both McLeod and normal red cells. (4) A monoclonal antibody to glycophorin A and B was purchased from Sigma (St Louis, MO).
Preparation of an integral membrane fraction
Treatment of cell membranes with sodium carbonate at alkaline pH removes peripheral proteins and retains integral membrane proteins.12 This procedure was used to prepare cell membranes from various tissues. Frozen human tissues (testis and skeletal muscle) were obtained from the National Disease Research Interchange (NDRI; Philadelphia, PA). While still frozen (−70°C), about 2 grams of tissue was minced using a nickel scalpel, placed in 20 volumes of ice-cold 100 mmol/L sodium carbonate buffer, pH 11.5, and homogenized using a glass tissue grinder. The suspension was filtered through cheesecloth, and centrifuged at 4000 rpm for 10 minutes at 4°C in a Sorvall model RC-5C plus to remove unbroken cells and nuclei. The supernatant fractions were then centrifuged at 40 000 rpm for 30 minutes at 4°C with the use of a Beckman 60Ti rotor, and the pellet, composed mostly of cellular membranes, was rinsed twice with phosphate-buffered saline (PBS). The membranes were then solubilized by homogenization in 5 volumes of lysis buffer composed of PBS, containing 1% n-dodecyl α-d-Maltoside (Sigma), 0.5% of deoxycholic acid (sodium salt, Sigma), and protease inhibitors. The following protease inhibitors were used: 0.1 mmol/L L-1-tosylamido-2-phenylethyl chloromethyl ketone, 0.1 mmol/L phenylmethylsulfonyl fluoride, and 10 units/mL aprotinin (Sigma). The solution was than cleared by centrifugation in a Beckmann 50Ti rotor at 27 000 rpm for 20 minutes.
Isolation of XK complexes from nonerythroid tissues
The detergent-solubilized membrane preparation, described above, was incubated with biotinylated antibody to XK at 4°C overnight. The immunocomplex was recovered by adding 0.02 volumes of immobilized streptavidin suspension (Pierce), at 4°C for 3 hours with constant rocking. The streptavidin gel was then packed in a 5-mL column and washed 4 times with lysis buffer, and the XK complex was eluted with 100 mmol/L glycine buffer pH 2.8. The eluted material was collected in tubes containing 1 mol/L Tris to neutralize acidity.
SDS-PAGE and Western blotting
Membrane preparations, or isolated XK complexes, were dissolved in SDS-buffer (0.125 mol/L Tris-HCl, pH 6.8, 1% SDS, 5% glycerol), separated on precasted 4%-12% Tris-glycine gradient gels (Novex, San Diego, CA) and transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, NH) with the use of a Bio Rad (Hercules, CA) blotting apparatus. After treatment with rabbit antibodies, 2 different detection procedures were employed. The second antibody was conjugated either with alkaline phosphatase or horseradish peroxidase. Both second antibodies were purchased from Pierce. The alkaline phosphatase-conjugated antibody was detected with a chromogenic substrate (NBT/BCIP; purchased from BIORAD) and the horseradish peroxidase–conjugated antibody with a chemoluminescent substrate (Supersignal; purchased from Pierce).
Results
Detection of Kell RNA transcripts in nonerythroid tissues
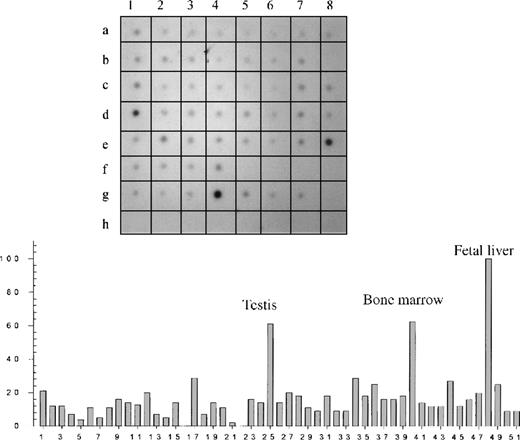
Hybridization of a RNA dot-blot panel with radioactive Kell antisense RNA followed by densitometry confirmed that Kell is expressed mainly in fetal liver and bone marrow (Figure1; Table 1). However, significant hybridization also occurred in many other tissues with the highest in testis. Weaker signals were observed throughout the dot-blot panel, including different parts of the brain and many other tissues. No hybridization occurred in the controls that contained nonhuman RNA or human DNA (Figure 1).
Dot-blot analysis of different tissues.
(Top) An autoradiography of a dot blot containing 51 different human tissue RNAs hybridized with a Kell radioactive riboprobe. (Bottom) Relative concentration values obtained by densitometry analysis. Tissue RNAs can be identified according to their position on the blot and their number as summarized in Table 1.
Dot-blot analysis of different tissues.
(Top) An autoradiography of a dot blot containing 51 different human tissue RNAs hybridized with a Kell radioactive riboprobe. (Bottom) Relative concentration values obtained by densitometry analysis. Tissue RNAs can be identified according to their position on the blot and their number as summarized in Table 1.
Tissue RNAs identified according to their position on the blot and their number
| Position . | Number . | Tissue sample* . |
|---|---|---|
| A1 | 1 | Whole brain |
| A2 | 2 | Amygdala |
| A3 | 3 | Caudates nucleus |
| A4 | 4 | Cerebellum |
| A5 | 5 | Cerebral cortex |
| A6 | 6 | Frontal lobe |
| A7 | 7 | Hippocampus |
| A8 | 8 | Medulla oblongata |
| B1 | 9 | Occipital lobe |
| B2 | 10 | Putamen |
| B3 | 11 | Substantia nigra |
| B4 | 12 | Temporal lobe |
| B5 | 13 | Thalamus |
| B6 | 14 | Subthalamic nucleus |
| B7 | 15 | Spinal cord |
| B8 | 16 | — |
| C1 | 17 | Heart |
| C2 | 18 | Aorta |
| C3 | 19 | Skeletal muscle |
| C4 | 20 | Colon |
| C5 | 21 | Bladder |
| C6 | 22 | Uterus |
| C7 | 23 | Prostate |
| C8 | 24 | Stomach |
| D1 | 25 | Testis |
| D2 | 26 | Ovary |
| D3 | 27 | Pancreas |
| D4 | 28 | Pituitary gland |
| D5 | 29 | Adrenal gland |
| D6 | 30 | Thyroid gland |
| D7 | 31 | Salivary gland |
| D8 | 32 | Mammary gland |
| E1 | 33 | Kidney |
| E2 | 34 | Liver |
| E3 | 35 | Small intestine |
| E4 | 36 | Spleen |
| E5 | 37 | Thymus |
| E6 | 38 | Peripheral leukocyte |
| E7 | 39 | Lymph node |
| E8 | 40 | Bone marrow |
| F1 | 41 | Appendix |
| F2 | 42 | Lung |
| F3 | 43 | Trachea |
| F4 | 44 | Placenta |
| F5 | — | |
| F6 | — | |
| F7 | — | |
| F8 | — | |
| G1 | 45 | Fetal brain |
| G2 | 46 | Fetal heart |
| G3 | 47 | Fetal kidney |
| G4 | 48 | Fetal liver |
| G5 | 49 | Fetal spleen |
| G6 | 50 | Fetal thymus |
| G7 | 51 | Fetal lung |
| G8 | — | |
| H1 | Yeast total RNA 100 ng | |
| H2 | Yeast tRNA 100 ng | |
| H3 | E colirRNA 100 ng | |
| H4 | E coli DNA 100 ng | |
| H5 | Poly r(A) 100 ng | |
| H6 | Human Cot1 DNA 100 ng | |
| H7 | Human DNA 100 ng | |
| H8 | Human DNA 500 ng |
| Position . | Number . | Tissue sample* . |
|---|---|---|
| A1 | 1 | Whole brain |
| A2 | 2 | Amygdala |
| A3 | 3 | Caudates nucleus |
| A4 | 4 | Cerebellum |
| A5 | 5 | Cerebral cortex |
| A6 | 6 | Frontal lobe |
| A7 | 7 | Hippocampus |
| A8 | 8 | Medulla oblongata |
| B1 | 9 | Occipital lobe |
| B2 | 10 | Putamen |
| B3 | 11 | Substantia nigra |
| B4 | 12 | Temporal lobe |
| B5 | 13 | Thalamus |
| B6 | 14 | Subthalamic nucleus |
| B7 | 15 | Spinal cord |
| B8 | 16 | — |
| C1 | 17 | Heart |
| C2 | 18 | Aorta |
| C3 | 19 | Skeletal muscle |
| C4 | 20 | Colon |
| C5 | 21 | Bladder |
| C6 | 22 | Uterus |
| C7 | 23 | Prostate |
| C8 | 24 | Stomach |
| D1 | 25 | Testis |
| D2 | 26 | Ovary |
| D3 | 27 | Pancreas |
| D4 | 28 | Pituitary gland |
| D5 | 29 | Adrenal gland |
| D6 | 30 | Thyroid gland |
| D7 | 31 | Salivary gland |
| D8 | 32 | Mammary gland |
| E1 | 33 | Kidney |
| E2 | 34 | Liver |
| E3 | 35 | Small intestine |
| E4 | 36 | Spleen |
| E5 | 37 | Thymus |
| E6 | 38 | Peripheral leukocyte |
| E7 | 39 | Lymph node |
| E8 | 40 | Bone marrow |
| F1 | 41 | Appendix |
| F2 | 42 | Lung |
| F3 | 43 | Trachea |
| F4 | 44 | Placenta |
| F5 | — | |
| F6 | — | |
| F7 | — | |
| F8 | — | |
| G1 | 45 | Fetal brain |
| G2 | 46 | Fetal heart |
| G3 | 47 | Fetal kidney |
| G4 | 48 | Fetal liver |
| G5 | 49 | Fetal spleen |
| G6 | 50 | Fetal thymus |
| G7 | 51 | Fetal lung |
| G8 | — | |
| H1 | Yeast total RNA 100 ng | |
| H2 | Yeast tRNA 100 ng | |
| H3 | E colirRNA 100 ng | |
| H4 | E coli DNA 100 ng | |
| H5 | Poly r(A) 100 ng | |
| H6 | Human Cot1 DNA 100 ng | |
| H7 | Human DNA 100 ng | |
| H8 | Human DNA 500 ng |
A dash denotes an empty sector. The empty sectors and sectors containing control nucleic acids (H1 to H8) were all zero. The empty sector B8 with assigned number 16, which also had a density value of 0, is shown in the histogram.
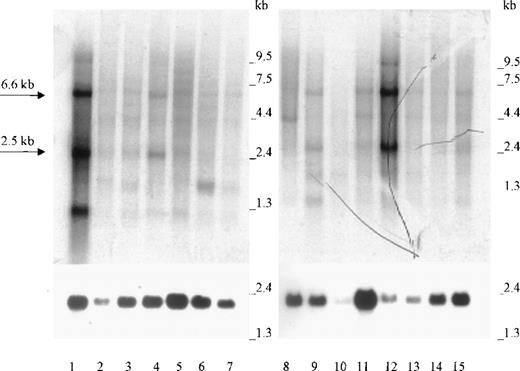
Northern blot analysis of 2 panels containing a total of 15 different human tissue RNAs showed that the 2.5-kb and 6.6-kb Kell transcripts present in bone marrow (Figure 2, lane 1) were also abundant in testis (Figure 2, lane 12). The same size transcripts were also detected in lesser amounts in lymph node, colon, and spleen (lanes 4, 9, and 15). Weaker signals for the 2.5-kb transcripts were detectable in several other tissues (Figure 2), including skeletal muscle (data not shown). Because of different amounts of mRNA, as evidenced by the actin signals, the relative amounts of Kell transcripts cannot be easily compared. Some tissues, such as small intestine (Figure 2, lane 10), had less actin mRNA than the other tissues. Other tissues, such as thyroid and peripheral blood lymphocytes (PBLs), showed transcripts of a different size than normally obtained for Kell transcripts. Thyroid had a smaller transcript of about 1.3 kb, and PBLs had a band at 3.5 kb. On occasion (as shown in Figure 2, lane 1), bone marrow also has a small, approximately 1 kb, transcript that may be caused by mRNA degradation. However, relative values were obtained by comparing the densities of the main 2.5-kb transcripts and correcting them for the different amounts of actin (Figure 1). The results presented in Table2 show that, unlike the results in Figure1, KEL appears to be expressed more in testis than in bone marrow.
Northern blot analysis.
Lanes 1 to 15 contain approximately 2 μg poly A+ RNA from (in the following order): bone marrow, adrenal gland, trachea, lymph node, spinal cord, thyroid, stomach, peripheral blood leukocytes, colon, small intestine, uterus, testis, prostate, thymus, and spleen. The 2.5-kb and 6.6-kb Kell transcripts are marked by arrows. Hybridization was performed with radioactive Kell antisense RNA (upper panel) or with radioactive β-actin cDNA (lower panel).
Northern blot analysis.
Lanes 1 to 15 contain approximately 2 μg poly A+ RNA from (in the following order): bone marrow, adrenal gland, trachea, lymph node, spinal cord, thyroid, stomach, peripheral blood leukocytes, colon, small intestine, uterus, testis, prostate, thymus, and spleen. The 2.5-kb and 6.6-kb Kell transcripts are marked by arrows. Hybridization was performed with radioactive Kell antisense RNA (upper panel) or with radioactive β-actin cDNA (lower panel).
Relative quantities of KEL 2.5-kb mRNA transcripts in different tissues
| Tissue . | Relative amount (%) . |
|---|---|
| Bone marrow | 100 |
| Testis | 218 |
| Colon | 45 |
| Lymph node | 43 |
| Spinal cord | 35 |
| Spleen | 31 |
| Adrenal gland | 30 |
| PBL | 29 |
| Small intestine | 29 |
| Thymus | 29 |
| Trachea | 26 |
| Thyroid | 26 |
| Prostate | 22 |
| Uterus | 21 |
| Stomach | 15 |
| Tissue . | Relative amount (%) . |
|---|---|
| Bone marrow | 100 |
| Testis | 218 |
| Colon | 45 |
| Lymph node | 43 |
| Spinal cord | 35 |
| Spleen | 31 |
| Adrenal gland | 30 |
| PBL | 29 |
| Small intestine | 29 |
| Thymus | 29 |
| Trachea | 26 |
| Thyroid | 26 |
| Prostate | 22 |
| Uterus | 21 |
| Stomach | 15 |
The relative values were obtained by measuring the densities of the 2.5 kb KEL transcript from Northern blots. All values were standardized for equal amounts of actin transcript. For ease of comparison, the amount of 2.5-kb KEL transcript in bone marrow was assigned a 100% value.
PBL, peripheral blood lymphocyte.
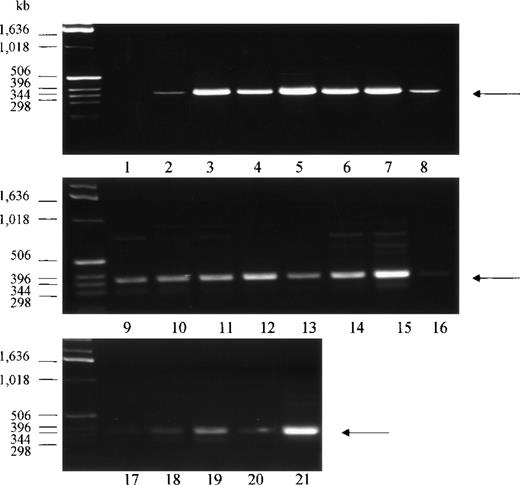
PCR analysis of human tissue first strand cDNAs, followed by DNA sequencing of the amplified DNA fragments, was conducted to confirm the presence of the Kell transcript in various non-erythroid tissues. A total of 20 tissue cDNAs of erythroid and nonerythroid origins were analyzed for the presence of Kell. Primers for PCR were designed to anneal to sequences derived from exon 14 and exon 18 of the KELgene to amplify a 386-base pair (bp) DNA fragment. A PCR product with the expected size was detected in all tissues, with the highest amounts in bone marrow, fetal liver, tonsils, spleen, placenta, and testis (Figure 3, lanes 3, 5, 6, 7, 15, and 21). No PCR product was detected in the negative control (Figure 3, lane 1), which had water instead of cDNA template. DNA sequencing analysis of the 386-bp DNA fragment from 4 of the samples, testis, adult liver, PBLs, and skeletal muscle, confirmed that the PCR-amplified products had identical sequences to that obtained from bone marrow or fetal liver. These tissues were chosen because there is abundant expression of Kell in testis, because there is very weak expression in skeletal muscle, and because previous studies had not detected Kell transcripts in adult liver and PBLs.
PCR analysis of Kell cDNA.
Normalized first strand cDNA preparations from different human tissues as indicated below were screened by PCR. Primers were designed to amplify a 386-bp segment (arrow). (1) Control (no template cDNA), (2) peripheral blood leukocytes, (3) bone marrow, (4) lymph node, (5) fetal liver, (6) tonsils, (7) spleen, (8) thymus, (9) brain, (10) heart, (11) kidney, (12) liver, (13) lung, (14) pancreas, (15) placenta, (16) skeletal muscle, (17) colon, (18) ovary, (19) prostate, (20) small intestine, and (21) testis.
PCR analysis of Kell cDNA.
Normalized first strand cDNA preparations from different human tissues as indicated below were screened by PCR. Primers were designed to amplify a 386-bp segment (arrow). (1) Control (no template cDNA), (2) peripheral blood leukocytes, (3) bone marrow, (4) lymph node, (5) fetal liver, (6) tonsils, (7) spleen, (8) thymus, (9) brain, (10) heart, (11) kidney, (12) liver, (13) lung, (14) pancreas, (15) placenta, (16) skeletal muscle, (17) colon, (18) ovary, (19) prostate, (20) small intestine, and (21) testis.
The complete DNA sequence of Kell cDNA obtained from skeletal muscle was further investigated by 3′ and 5′ RACE nested PCR and DNA sequencing. A 3′ RACE DNA fragment, of about 2.4 kb, was generated, using primers derived from KEL exon 1 and adapter primers at the 3′ end, thus containing the whole coding region. DNA sequence analysis showed that the coding region of the skeletal muscle Kell cDNA was nearly identical to that previously described from bone marrow Kell cDNA, with only one difference: a G to A mutation at nt 2197. This base change would encode an asparagine at Kell residue 692 instead of aspartic acid.
The 5′ untranslated region of skeletal muscle cDNA was obtained by 5′ RACE, sequenced, and found to be identical to Kell fetal liver and bone marrow cDNA.
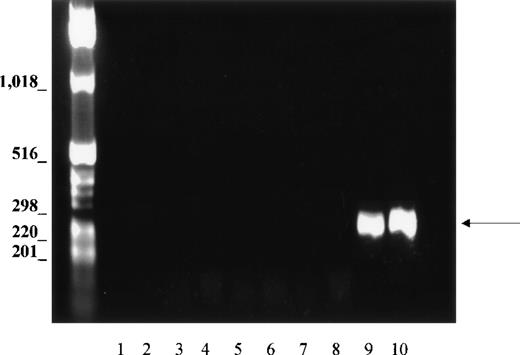
Because the tissues could be contaminated with erythroid cells during isolation, and because PCR is a sensitive technique capable of amplifying small amounts of cDNA, we evaluated this possibility with the use of primers specific for GPA, which is expressed only in erythroid tissues. If blood cells contaminated the non-erythroid tissues, GPA should be amplified, as well as Kell. Nine different tissue cDNAs, including bone marrow and fetal liver, were analyzed for GPA, using the same PCR conditions as for amplification of Kell transcripts. In bone marrow and fetal liver, primers derived from sequences of exons 2-3 and exon 5 of the GPA gene amplified a DNA fragment with the expected size of 330 bp (Figure4, lanes 9 and 10). By contrast, no GPA products were observed in any of the nonerythroid tissues (Figure 4, lanes 2 to 7), thus showing that erythroid contamination was not relevant and could not account for the amplification of Kell cDNA in non-erythroid cDNA libraries.
PCR analysis of GPA cDNA.
Normalized first strand cDNA preparations from different human tissues as indicated below. Primers were designed to amplify a 330-bp segment (arrow). (1) Control (no template cDNA), (2) brain, (3) skeletal muscle, (4) lung, (5) liver, (6) spleen, (7) lymph node, (8) peripheral blood leukocytes, (9) bone marrow, and (10) fetal liver.
PCR analysis of GPA cDNA.
Normalized first strand cDNA preparations from different human tissues as indicated below. Primers were designed to amplify a 330-bp segment (arrow). (1) Control (no template cDNA), (2) brain, (3) skeletal muscle, (4) lung, (5) liver, (6) spleen, (7) lymph node, (8) peripheral blood leukocytes, (9) bone marrow, and (10) fetal liver.
Kell protein is present in testis and skeletal muscle
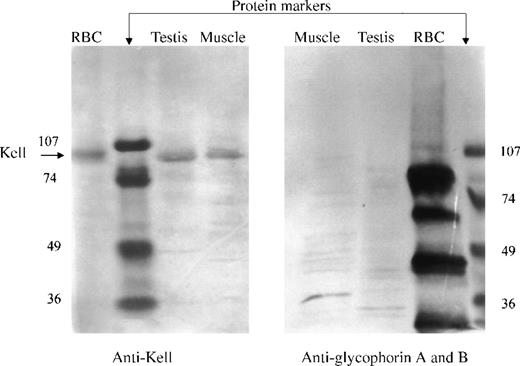
The presence of Kell protein in nonerythroid tissues was determined by direct Western immunoblotting of integral membrane proteins in testis and skeletal muscle. Membrane preparations, containing integral membrane proteins, were prepared by treatment of the membranes with Na2CO3 at pH 11.5. The proteins were separated by SDS-PAGE in reducing conditions and detected by Western immunoblotting, using a polyclonal antibody to Kell protein isolated from red cells. Kell protein, having the same electrophoretic mobility as that from red cell membranes, was detected in both testis and skeletal muscle (Figure 5, left panel). Unequal amounts of integral membrane proteins were loaded for the different tissues. The lanes containing skeletal muscle had about 35 μg protein, testis 15 μg protein, and red cells 5 μg protein.
Western blot analysis.
Integral membrane proteins (pH 11.5) from red blood cells, testis, or skeletal muscle were reduced, separated by SDS-PAGE, blotted, and detected with polyclonal antibody to Kell (left panel) or with a monoclonal antibody that recognizes glycophorins A and B (right panel). (Left panel) A single 93-kD band (arrow), the molecular size of Kell, was detected in red blood cells, testis, and skeletal muscle. (Right panel) Multiple bands corresponding to glycophorins A and B were detected in red blood cells but not in testis or skeletal muscle.
Western blot analysis.
Integral membrane proteins (pH 11.5) from red blood cells, testis, or skeletal muscle were reduced, separated by SDS-PAGE, blotted, and detected with polyclonal antibody to Kell (left panel) or with a monoclonal antibody that recognizes glycophorins A and B (right panel). (Left panel) A single 93-kD band (arrow), the molecular size of Kell, was detected in red blood cells, testis, and skeletal muscle. (Right panel) Multiple bands corresponding to glycophorins A and B were detected in red blood cells but not in testis or skeletal muscle.
A duplicate Western immunoblot, using monoclonal antibody to both glycophorin A and B, was performed to evaluate the possible contamination of testis and skeletal muscle with red cells. The antibody to the glycophorins reacted with multiple red cell membrane proteins but did not react with proteins from testis or skeletal muscle (Figure 5, right panel).
Association of Kell and XK in nonerythroid tissues
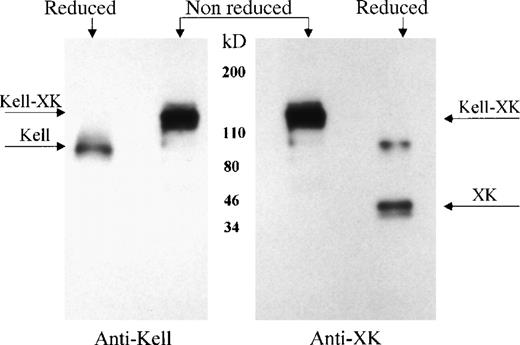
Previous studies7-9 demonstrated that Kell and XK are covalently linked on the red cell membrane. To determine whether Kell is also bound to XK in nonerythroid tissues, XK and its complexes were isolated from skeletal muscle membranes, using a biotinylated antibody to XK and characterized by Western immunoblotting, with the use of antibodies to Kell or to XK. Under reducing conditions, the antibody to the C-terminal domain of XK reacted with XK and with a higher molecular weight protein with the expected size of XK dimer (Figure6, second lane right panel). In reducing conditions, the presence of Kell protein was shown by reaction of antibody to Kell with a 93-kDa protein (Figure6, first lane left panel). Under nonreducing conditions, both antibodies recognized the Kell/XK protein complex, with a molecular weight of about 130 kDa (Figure 6, lane 2 left panel and lane 1 right panel), demonstrating the presence of a Kell/XK complex in skeletal muscle.
Kell/XK complex in skeletal muscle.
Skeletal muscle integral membrane proteins were isolated with antibody to XK, separated in reduced and nonreduced conditions by SDS-PAGE, and detected with antibody to Kell (left panel) or to XK (right panel). The location of Kell/XK complex, Kell, and XK are marked by arrows.
Kell/XK complex in skeletal muscle.
Skeletal muscle integral membrane proteins were isolated with antibody to XK, separated in reduced and nonreduced conditions by SDS-PAGE, and detected with antibody to Kell (left panel) or to XK (right panel). The location of Kell/XK complex, Kell, and XK are marked by arrows.
Discussion
Our studies show that expression of Kell protein, like that of its partner, XK, is not limited to erythroid tissues but is widely distributed. This finding is in keeping with the previous observation that Kell and XK are covalently linked on red cells.7-9 Earlier studies, in which expression of Kell and XK were determined only by Northern blot analysis, did not find that these 2 proteins were expressed in the same tissues, because expression of Kell was noted only in erythroid tissues and not in brain, adult liver, kidney, and lung,5 whereas expression of XK was detected in erythroid and nonerythroid tissues, primarily skeletal muscle, pancreas, heart, and brain.6 Our studies now indicate that Kell, like XK, is also expressed in nonerythroid tissues, primarily in testis, but with significant amounts in other tissues.
Kell expression was determined by 2 different Northern blot assays. One of the procedures measured a large number of tissues as part of a dot-blot panel in which the amount of poly A+ RNA in each well was varied to reflect equal amounts of actin poly A+RNA. The Kell probe used was a riboprobe covering the middle segment of Kell mRNA. Although this segment of Kell, representing exons 9 to 13 ofKEL, is not highly homologous with other members of the M13 family of zinc endopeptidases,1 there could be cross-reaction with other M13 RNAs. Because the other M13 members have larger mRNA transcripts,13-19 Northern blot analysis, detecting the major 2.5-kb and 6.6-kb Kell transcripts, was also performed. In general, the 2 procedures matched, with abundant Kell expression detected in erythroid tissues and testis and with weaker expression in other tissues. However, not all of the tissues represented in the dot-blot panel were tested by Northern blot analysis for the presence of 2.5-kb and 6.6-kb transcripts, and lengthy exposures of the film were necessary to detect Kell transcripts in tissues other than testis and the erythroid tissues. In the dot-blot experiments (Figure 1), there was near-equal expression of KELin both bone marrow and testis, whereas, by Northern blot analysis (Table 2), there was twice as much expressed in testis. The differences in the 2 procedures may be due to the fact that in the dot-blot assays total KEL transcripts were measured, whereas in the Northern blots only the main 2.5 kb was taken into account. Thus, any degradation of RNA that would lead to smaller transcripts, for example the 1-kb transcript in bone marrow (Figure 2), is not measured and will contribute to a lower value. Nevertheless, all procedures indicate that erythroid tissues and testis are the major sites of Kell expression.
PCR screening of cDNA from many tissues also supports that Kell is widely distributed. The 2 primers used to amplify the Kell transcripts are in areas that are homologous with sequences for NEP and ECE-1, and some amplification of these transcripts may have occurred. Sequencing of several of the transcripts showed, however, that only Kell transcripts were amplified. The PCR procedure employed is not quantitative, although it does give a rough indication of the relative distribution of Kell transcripts. The PCR method is limited by variations in the preparation of poly A+ RNA from different tissues and by the fact that the PCR procedure used is not quantitative. Thus, some minor discrepancies are found between the Northern blot analysis and the PCR determinations. For example, Kell appears to be moderately expressed in colon by Northern blot analysis (Figure 2, lane 9), but very little was detected by PCR screening (Figure 3, lane 17).
Because Kell is abundantly expressed in erythroid tissues, and the presence of erythroid cells may easily contaminate human tissues, it was important to rule out the possibility that the detection of Kell was due to contamination of tissues with blood. GPA is much more abundant in erythroid tissues than Kell, but PCR screening did not detect GPA in nonerythroid tissues, indicating that contamination with erythroid tissues is not a concern.
The presence of mRNA, as measured by Northern blot or RT-PCR, may not accurately reflect the presence of protein because other cellular mechanisms, such as protein stability or turnover, can affect protein content. Therefore, we determined, by Western immunoblot, the presence of Kell protein in 2 different tissues. We chose testis because both Northern blot and PCR procedures indicate that Kell is abundantly expressed in this tissue. For contrast, we also examined skeletal muscle that abundantly expresses XK as determined by Northern blot but shows weak expression of Kell by Northern blot and PCR (Figures 1 and3). Kell protein was present in both testis and skeletal muscle. According to different amounts of total integral membrane proteins loaded on the gels, we estimate that testis has twice as much Kell protein as skeletal muscle and that red cell membranes may have 7 times more than skeletal muscle. In skeletal muscle, most of XK isolated was complexed to Kell and very little appeared as free XK. However, this procedure may not account for all of the free XK protein because we have noticed that the antibody to XK preferentially reacts with XK when it is complexed to Kell and that free XK is more difficult to detect.
The Kell/XK complex may be more stable than the individual component proteins. This view is based on studies with 2 rare Kell red cell phenotypes in which either Kell or XK is absent or deficient.2 In the McLeod red cell phenotype, which lacks or is deficient in XK, there is also diminished amounts of Kell protein; whereas in the Ko (null) phenotype, which lacks Kell protein, there is less XK protein on the membrane, although an antigen Kx, carried by XK, appears to be enhanced.8 Other studies have shown that cellular transport and placement of Kell and XK on the plasma membrane does not require coexpression of both Kell and XK. In transfected COS cells, expression of Kell or XK by themselves is sufficient to allow the individual proteins to be expressed on the cell surface.20 In normal adult red cells, however, there is little free Kell or XK, and the majority of these proteins exists as a 130-kDa Kell/XK complex.
We do not know, however, whether the Kell/XK complex in nonerythroid tissues is similar to that in red cells. Red cells have a single membrane representing the plasma membrane and, by contrast, nonerythroid tissues may have multiple intracellular membranes. The enzymatic activity of Kell protein is optimal at an acidic pH,4 suggesting that Kell may play an intracellular role, possibly in transport and processing of endothelin precursors or in the endocytic pathway. In this regard, it should be noted that endothelin-converting enzymes are not only ectoenzymes but are also present intracellularly.21-24 Future immuno-histochemical studies are necessary to determine the cellular and intracellular locations of Kell and XK. Whether the full cellular function of Kell requires XK, or whether Kell is complementary to the function of XK, also remains to be determined. Thus far, big endothelin-3 is the preferred substrate for Kell, as determined by in vitro studies with recombinant Kell and by measuring enzymatic activity of red cells.4 We do not know, however, if big endothelin-3 is the only substrate for Kell and whether Kell, which may be present intracellularly, has a different function to Kell present on cell surfaces.
DNA sequencing of a Kell cDNA obtained by PCR amplification from a skeletal muscle cDNA library showed that DNA sequence was identical to that obtained from erythroid tissues with the single exception of a possible Asp 692 Asn substitution because of a single base change. This single base change may, however, be due to the PCR procedure and will have to be confirmed by other methods. The sequence identity between erythroid and non-erythroid Kell suggests similar enzymatic functions in erythroid and nonerythroid tissues. The fact that Kell is disulfide-linked to XK in nonerythroid tissues also indicates complementary functions for XK and Kell proteins.
Acknowledgments
We thank Dr James D. Farmar and Andrea Molinaro of the Laboratory of Microchemistry for their work in DNA sequencing.
Supported in part by a National Institutes of Health Specialized Center of Research (SCOR) grant in Transfusion Biology and Medicine, HL54459.
Reprints:Colvin M. Redman, The New York Blood Center, 310 East 67 St, New York, NY 10021; e-mail credman@nybc.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal