Abstract
An attractive hypothesis is that in utero exposure of hematopoietic cells to oncogenic agents can induce molecular changes leading to overt acute lymphoblastic leukemia (ALL) in infants and perhaps older children as well. Although supported by studies of identical infant twins with concordant leukemia, and of nontwined patients withMLL gene rearrangements, this concept has not been extended to the larger population of B-lineage ALL patients who lack unique nonconstitutive mutations or abnormally rearranged genes. We therefore sought to demonstrate a prenatal origin for 7 cases of B-cell precursor ALL (either CD10+ or CD10−) that had been diagnosed in infants and children 14 days to 9 years of age. Using a polymerase chain reaction–based assay, we identified the same clonotypic immunoglobulin heavy-chain complementarity determining region or T-cell receptor VD2-DD3 sequences in the neonatal blood spots (Guthrie card) and leukemic cell DNAs of 2 infants with CD10− ALL and 2 of the 5 older patients with CD10+ ALL. Nucleotide sequencing showed a paucity of N or P regions and shortened D germ line and conserved J sequences, indicative of cells arising from fetal hematopoiesis. Our findings strongly suggest a prenatal origin for some cases of B-cell precursor ALL lacking specific clonotypic abnormalities.
B-cell precursor acute lymphoblastic leukemia (ALL) is by far the most common form of leukemia in children. Its incidence is highest between 1 and 5 years of age, with a peak between 2 and 4 years.1,2 Recent studies of identical twins with concordant ALL established that leukemic transformation in these cases was initiated during the fetal period,3-6 which would help to explain the common phenotypes, karyotypes, and clonal gene rearrangements that typify the leukemic cells of such patients. This finding was interpreted as evidence for in utero leukemic conversion in one twin, with the transformed cells migrating to the other twin through the placental circulation.5 In 1997, Gale et al7 demonstrated MLL-AF4 gene fusion in neonatal blood spots (Guthrie card) of nontwined patients who developed acute leukemia at the ages of 5 months to 2 years. Taken together, these observations provide compelling support for the in utero origin of some cases of childhood ALL. However, approximately 50% of childhood ALL patients, most with leukemic B-cell precursors, lack abnormal gene rearrangements that could be used to backtrack their leukemias to the fetal stage of development.
We therefore analyzed the Guthrie card blood spots of infants and children who were diagnosed with either CD10+ or CD10− ALL at 14 days to 9 years of age (median, 2 years), using a combination of nonpathologic immunoglobulin heavy chain (IGH) and T-cell receptor delta (TCRD) gene rearrangements to identify individual leukemic clones. This polymerase chain reaction (PCR) strategy was based on the high frequency of such changes in B-lineage ALL (95% IGH, 60% to 70% TCRD)8-14 and the uniqueness of somatic mutations within complementarity determining region 3 (CDR3) of the IGH gene.15 16 CDR3 comprises the 3′ end of VH, all of D, and the 5′ end of JH; it also contains randomly inserted nucleotides. The TCRD gene contains a limited number of functional V and J segments, but rearrangements affecting VD2 and DD3 are relatively common in B-lineage ALL, prompting us to include primers for these 2 regions in our PCR analysis. Here we report clonotypic IGH and TCRD gene rearrangements in the Guthrie card blood spots of 4 patients with either CD10+ or CD10− B-lineage ALL.
Patients, materials, and methods
Patients and controls
We studied the leukemic bone marrow cells and Guthrie cards of 7 pediatric patients with B-cell precursor ALL after obtaining informed consent from the patients' parents or guardians. The clinical and laboratory features of these patients are summarized in Table1. Two of the cases developed in infants (CD10−, CD19+, and HLA-DR+) and 5 in children (CD10+, CD19+, and HLA-DR+). The median age at diagnosis in the latter group was 2 years (range, 1.5 to 9 years). The leukocyte counts on admission ranged from 2300 to 388 000/μL (median, 121 600/μL). Both of the infants hadMLL gene rearrangement by Southern blot analysis (data not shown). It was confirmed that the 5 CD10+ patients had no infectious symptoms at the time of neonatal blood spot collection. Bone marrow (n = 2) and peripheral blood (n = 15) samples from healthy infants, children, and adults and Guthrie cards of the same-aged patients (n = 8) who did not develop leukemia were obtained with informed consent and served as normal controls.
Clinical data on patients with CD10+ or CD10− B-precursor ALL
| Case no. . | Sex . | Age at onset* . | Leukocyte count (per μL) . | Percentage of blasts in peripheral blood† . | Karyotype . | Positive immunological markers‡ . |
|---|---|---|---|---|---|---|
| 1 | F | 14 d | 121 600 | 68 | 46XX,t(4;11;15) | 19, 34, 45, 79a, HLA-DR |
| 2 | M | 2 mo | 257 000 | 93 | 46XY,t(4;11)(q21;q23) | 15, 19, 34, 45, 79a, HLA-DR |
| 3 | M | 2 y | 67 900 | 87 | 46XY | 10, 19, 33, 34, HLA-DR |
| 4 | M | 2.2 y | 159 000 | 90 | 63XXY | 10, 19, 20, 34, 45, HLA-DR |
| 5 | M | 1.5 y | 388 000 | 88 | 46XY,t(11;12)(q13;q24) | 10, 19, 34, 45, HLA-DR |
| 6 | M | 9 y | 6 500 | 30 | 46XY | 10, 19, 20, 34, HLA-DR |
| 7 | M | 2 y | 2 300 | 0 | 46XY | 10, 19, 20, 22, 34, HLA-DR |
| Case no. . | Sex . | Age at onset* . | Leukocyte count (per μL) . | Percentage of blasts in peripheral blood† . | Karyotype . | Positive immunological markers‡ . |
|---|---|---|---|---|---|---|
| 1 | F | 14 d | 121 600 | 68 | 46XX,t(4;11;15) | 19, 34, 45, 79a, HLA-DR |
| 2 | M | 2 mo | 257 000 | 93 | 46XY,t(4;11)(q21;q23) | 15, 19, 34, 45, 79a, HLA-DR |
| 3 | M | 2 y | 67 900 | 87 | 46XY | 10, 19, 33, 34, HLA-DR |
| 4 | M | 2.2 y | 159 000 | 90 | 63XXY | 10, 19, 20, 34, 45, HLA-DR |
| 5 | M | 1.5 y | 388 000 | 88 | 46XY,t(11;12)(q13;q24) | 10, 19, 34, 45, HLA-DR |
| 6 | M | 9 y | 6 500 | 30 | 46XY | 10, 19, 20, 34, HLA-DR |
| 7 | M | 2 y | 2 300 | 0 | 46XY | 10, 19, 20, 22, 34, HLA-DR |
d indicates days; mo, months; y, years.
Case 7 had 98% lymphoblasts in 2 successive bone marrow aspirates.
An MLL fusion gene was demonstrated in cases 1 and 2 by Southern blot analysis (not shown).
Specimens
DNA was extracted from the leukemic bone marrow cells that had been stored in liquid nitrogen since diagnosis and from control peripheral blood and bone marrow cells (DnaQuick DNA extraction kit; Dainippon Pharmaceutical Co, Osaka, Japan). The Guthrie cards had been stored either at room temperature or at −80°C at several different laboratories. Samples cut from each Guthrie card (3 mm × 3-mm squares containing the equivalent of about 1 × 104mononuclear cells) were extracted in 30μL of double-distilled water for 2 hours at 55°C. The resultant eluates were directly amplified with the use of the Ampdirect PCR kit (Shimadzu Co, Tokyo, Japan). Successful DNA extraction was confirmed by demonstration of the amplified beta-globin gene in all experiments.
In preliminary sensitivity experiments, the starting aliquot containing 1 × 105 leukemic cells added to 1 mL blood (cell count, 1 × 107) obtained from a healthy adult was sequentially diluted at concentrations ranging from 10−1 to 10−6/mL. These samples were then used to determine the limit of detection of the PCR products amplified from extracted leukemic cell DNA as well as material eluted from the Guthrie cards. In the latter situation, the analyses were performed with cards on which a 50-μL droplet of blood containing the diluted leukemic cells had been stored for at least 2 months prior to extraction.
PCR analysis using consensus primers
Samples of DNA (100 ng) from leukemic or nonleukemic peripheral blood and bone marrow cells were subjected to PCR with the use of consensus primers for rearranged IGH and TCRD regions.11,17 18 The primers for the IGH CDR3 region were FR3A (5′-ACACGGC(C/T)(G/C) TGTATTACTGT-3′), LJH (5′TGAGGAGACGGTG-ACC-3′), and VLJH (5′-GTGACCAGGGTNCCTTGGCCCCAG-3′). The primers for the TCRD region were VD2E (5′-GAGTCATGTCAGCC-ATTGAG-3′) and DD3E (5′-AGGAAATGGCACTTTTGCC-3′) for the first PCR reaction, and VD2I (5′-GCACCATCAGAGAGAGATGA-3′) and DD3I (5′-TTGTAGCACTGTGCGTATCC-3′) for nested PCR. The amplification conditions for each targeted segment were as follows: 1 × buffer (25 mmol/L Taps buffer, pH 9.3 containing 50 mmol/L KCl, 2 mmol/L MgCl2, 1 mmol/L 2-mercaptoethanol), 200μmol/L dNTP, 0.5 μmol/L primers, and 2.5 units of ExTaq (Takara Shuzo Co, Shiga, Japan) in a final reaction volume of 50 μL. The cycling conditions were 1 minute at 92°C, 45 seconds at 62°C (annealing temperature), and 1 minute at 72°C for a total of 35 cycles followed by a final incubation period at 72°C for 5 minutes. (Perkin-Elmer Gene Amp. 2400; Perkin-Elmer Cetus, Norwalk, CT). The annealing temperature was reduced by 0.2°C per cycle from 62°C to 55°C. These amplifications were performed on either a Perkin-Elmer Gene Amp 2400 or a Perkin-Elmer 9700 instrument.
PCR analysis using allele-specific primers
For cases 5, 6, and 7, in which leukemic clone with same sequence was not amplified in Guthrie cards, we set up the allele-specific primers for the rearranged IGH CDR3 regions of leukemic cells for each case. The primer sequences used were 5′-CCT TATTACTATGGTTCGGGGAGTTATTA-3′ for case 5, 5′-CCGGATTCAGCAGCTCGTCC-3′ for case 6, and 5′-GAGATCTGGTAGTACTACTTTGTTGA-3′ for case 7, respectively, and nested PCR was performed by using FR3A, LJH, and each allele-specific primer. The cycle conditions were the same as previously described, except for case 7, in which the annealing temperature was reduced from 60°C to 52°C. Similarly, we attempted to set up the allele-specific primers in the rearranged TCRD regions for these cases, but this was not possible because of a paucity of N region.
Cloning and sequencing
Amplified PCR products of the rearranged IGH CDR3 or TCRD region were cloned by ligation with the use of a pCR2.1 TA cloning kit (Invitrogen, Carlsbad, CA). At least 15 independent colonies were picked and expanded (Table 2). The plasmids were purified with a DNA purification kit (Promega, Madison, WI), and sequencing was performed on a mixture containing 8 μg of DNA, 2 μL each of dNTP reagents from the Thermo Sequenase fluorescent labeled primer cycle sequencing kit containing 7-deazo–deoxyguanosine triphosphate (dGTP) (Amersham Pharmacia Biotech AB, Uppsala, Sweden) and 2 μL of 1.5 pmol of upstream or understream primers. Sequencing was performed under the following conditions: 92°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds for 30 cycles in an ALF DNA sequencer (Pharmacia, LKB Biotech, Piscataway, NJ). The sequence data were assigned on the basis of their similarity to BLAST sequences and confirmed by homology to each germ-line sequence.19-22
Ratio of colony numbers showing identical nucleotide sequences per picked-up colonies
| Case . | IGH . | TCRD . | ||
|---|---|---|---|---|
| Guthrie card . | Leukemic cell . | Guthrie card . | Leukemic cell . | |
| 1 | 14/15 (93.3) | 15/16 (93.8) | 13/15 (86.7) | 12/15 (80.0) |
| 2 | 11/17 (64.7) | 13/15 (86.7) | NA | NA |
| 3 | 13/18 (72.2) | 15/21 (71.4) | 8/19 (42.1) | 11/16 (68.8) |
| 4a | 14/21 (66.7) | 13/16 (81.3) | 6/15 (40.0) | 10/18 (55.6) |
| 4b | 9/16 (56.3) | NA | ||
| 5 | NA | 12/16 (75.0) | NA | NA |
| 6 | NA | 8/15 (53.3) | NA | 10/17 (58.8) |
| 7 | NA | 13/16 (81.3) | NA | 12/18 (66.7) |
| Case . | IGH . | TCRD . | ||
|---|---|---|---|---|
| Guthrie card . | Leukemic cell . | Guthrie card . | Leukemic cell . | |
| 1 | 14/15 (93.3) | 15/16 (93.8) | 13/15 (86.7) | 12/15 (80.0) |
| 2 | 11/17 (64.7) | 13/15 (86.7) | NA | NA |
| 3 | 13/18 (72.2) | 15/21 (71.4) | 8/19 (42.1) | 11/16 (68.8) |
| 4a | 14/21 (66.7) | 13/16 (81.3) | 6/15 (40.0) | 10/18 (55.6) |
| 4b | 9/16 (56.3) | NA | ||
| 5 | NA | 12/16 (75.0) | NA | NA |
| 6 | NA | 8/15 (53.3) | NA | 10/17 (58.8) |
| 7 | NA | 13/16 (81.3) | NA | 12/18 (66.7) |
Results
Limiting dilution experiments
The sensitivity of our PCR analysis was studied both with DNA extracted from serially diluted leukemic bone marrow cells and with material directly eluted from experimentally prepared blood spots on Guthrie cards, following previously described methods.7Analysis of leukemic cell DNA for rearrangements in the IGH CDR3 region indicated a detection limit of 1 leukemic cell per 1000 normal cells (10−3) (Figure 1A). A similar limit was established for DNA eluted from Guthrie cards (Figure 1B). In the PCR assay for rearrangements in the TCRD region, using serial dilutions of leukemic bone marrow cells, we found a detection limit of 1 leukemic cell per 10 000 normal cells, which was 1 log more sensitive than the result for Guthrie card blood spots (10−3) (Figure 1C,D).
Limiting dilution experiments for rearranged IGH and TCRD.
Rearranged IGH and TCRD amplified segments are shown by ethidium bromide gel staining. (A) IGH, leukemic cell DNA. (B) TCRD, leukemic cell DNA. (C) IGH, Guthrie card DNA. (D) TCRD, Guthrie card DNA. Standard limiting dilution (10−1 through 10−6) was prepared with positive control cells. The limit for detecting the IGH rearrangement was approximately 10−3 compared with 10−4 for TCRD in leukemic cells DNA. M indicates DNA size marker; P, amplified product from 100 ng positive control DNA; and N, amplified product from negative control DNA.
Limiting dilution experiments for rearranged IGH and TCRD.
Rearranged IGH and TCRD amplified segments are shown by ethidium bromide gel staining. (A) IGH, leukemic cell DNA. (B) TCRD, leukemic cell DNA. (C) IGH, Guthrie card DNA. (D) TCRD, Guthrie card DNA. Standard limiting dilution (10−1 through 10−6) was prepared with positive control cells. The limit for detecting the IGH rearrangement was approximately 10−3 compared with 10−4 for TCRD in leukemic cells DNA. M indicates DNA size marker; P, amplified product from 100 ng positive control DNA; and N, amplified product from negative control DNA.
PCR analysis of bone marrow, peripheral blood cell, and Guthrie card DNA from normal controls
In the normal controls, DNA obtained from bone marrow, peripheral blood cells, and Guthrie cards did not produce the clonally amplified products in a diffuse smear of IGH and TCRD amplification (data not shown).
PCR analysis of leukemic cell and Guthrie card DNA.
In the 2 infants and in all 5 of the remaining patients, the amplified bands of rearranged IGH CDR3 DNA from leukemic cells varied in size from 90 to 120 base pairs (bp), while those of the TCRD region ranged from 70 to 100 bp by electrophoresis. All amplified segments showed a single rearranged allele, which was subcloned and sequenced. In cases 2 and 5, it was not possible to amplify the targeted TCRD region (Tables3 and 4).
Comparison of nucleotide sequences in rearranged IGH CDR3 leukemic cell DNA (L) and corresponding Guthrie card DNA (G)3-150
| Case . | V . | N . | D . | N . | J . | DH . | JH . |
|---|---|---|---|---|---|---|---|
| 1(G) | TGTGCAAGAGATCCC | c | TAGTAGTACCAGCT | cgtcc | TTACTACTACTACATGGAC | Dlr-4b | J6 |
| 1(L) | TGTGCAAGAGATCCC | c | TAGTAGTACCAGCT | cgtcc | TTACTACTACTACATGGAC | ||
| 2(G) | TGTGCAAGAGATCC | GTATAGCAGCTCGTCC | ccaa | TACTACTACATGGAC | DN4 | J6 | |
| 2(L) | TGTGCAAGAGATCC | GTATAGCAGCTCGTCC | ccaa | TACTACTACATGGAC | |||
| 3(G) | CAGGGGTCCGTATTTT | TATGGTTCG | ccg | CTGGTTCGACCCCTGG | Dxp′-1 | J5 | |
| 3(L) | CAGGGGTCCGTATTTT | TATGGTTCG | ccg | CTGGTTCGACCCCTGG | |||
| 4a(G)3-151 | TGTGCAAGAGATCCC | aaggat | TAGCAGCTCG | cccct | TTACTACTACATGGAC | DN4 | J6 |
| 4a(L) | TGTGCAAGAGATCCC | aaggat | TAGCAGCTCG | cccct | TTACTACTACATGGAC | ||
| 4b(G) | TGTGCGAGAGATC | ATTACGATATTTTGACTGGTTATTA | ACTGGTTCGACCCCTG | Dxp-1 | J5 | ||
| 4b(L) | not amplified | ||||||
| 5(G) | not amplified | ||||||
| 5(L) | TGTGCAAGAGA TCCC | cct | TATTACTATGGTTCGGGGAGTTATTATTAC | gttatta | CTACTTTGACTACTGG | Dxp′-1 | J4 |
| 6(G) | not amplified | ||||||
| 6(L) | TGTGCAAGAGATCCC | ccggattc | AGCAGCTCGTCC | cc | TTACTACTACTATGGAC | DN4 | J6 |
| 7(G) | not amplified | ||||||
| 7(L) | TGTGCGAGATC | tgg | TAGTACTACTTT | g | TTGACTACTGGGGCCAA | Dlr-5 | J6 |
| Case . | V . | N . | D . | N . | J . | DH . | JH . |
|---|---|---|---|---|---|---|---|
| 1(G) | TGTGCAAGAGATCCC | c | TAGTAGTACCAGCT | cgtcc | TTACTACTACTACATGGAC | Dlr-4b | J6 |
| 1(L) | TGTGCAAGAGATCCC | c | TAGTAGTACCAGCT | cgtcc | TTACTACTACTACATGGAC | ||
| 2(G) | TGTGCAAGAGATCC | GTATAGCAGCTCGTCC | ccaa | TACTACTACATGGAC | DN4 | J6 | |
| 2(L) | TGTGCAAGAGATCC | GTATAGCAGCTCGTCC | ccaa | TACTACTACATGGAC | |||
| 3(G) | CAGGGGTCCGTATTTT | TATGGTTCG | ccg | CTGGTTCGACCCCTGG | Dxp′-1 | J5 | |
| 3(L) | CAGGGGTCCGTATTTT | TATGGTTCG | ccg | CTGGTTCGACCCCTGG | |||
| 4a(G)3-151 | TGTGCAAGAGATCCC | aaggat | TAGCAGCTCG | cccct | TTACTACTACATGGAC | DN4 | J6 |
| 4a(L) | TGTGCAAGAGATCCC | aaggat | TAGCAGCTCG | cccct | TTACTACTACATGGAC | ||
| 4b(G) | TGTGCGAGAGATC | ATTACGATATTTTGACTGGTTATTA | ACTGGTTCGACCCCTG | Dxp-1 | J5 | ||
| 4b(L) | not amplified | ||||||
| 5(G) | not amplified | ||||||
| 5(L) | TGTGCAAGAGA TCCC | cct | TATTACTATGGTTCGGGGAGTTATTATTAC | gttatta | CTACTTTGACTACTGG | Dxp′-1 | J4 |
| 6(G) | not amplified | ||||||
| 6(L) | TGTGCAAGAGATCCC | ccggattc | AGCAGCTCGTCC | cc | TTACTACTACTATGGAC | DN4 | J6 |
| 7(G) | not amplified | ||||||
| 7(L) | TGTGCGAGATC | tgg | TAGTACTACTTT | g | TTGACTACTGGGGCCAA | Dlr-5 | J6 |
The findings are categorized by IGH region (V, N, D, and J). N-region sequences are given in lowercase letters. Germ-line sequences are identical to those reported in the literature.19-22
Biallelic amplification (4a, 4b) for Guthrie card DNA, but not for leukemic cell DNA (4a only).
Comparison of nucleotide sequences of the VD2-DD3 rearrangements in leukemic cell DNA (L) and corresponding Guthrie card DNA (G)4-150
| Case . | VD2 . | P N P . | DD3 . |
|---|---|---|---|
| GL | GGT CTT ACT GTG CCT GTG ACA CC | ACT GGG GGA TAC G | |
| 1(G) | GGT CTT ACT GTG CCT GTG ACA CC | gt | ACT GGG GGA TAC G |
| 1(L) | GGT CTT ACT GTG CCT GTG ACA CC | gt | ACT GGG GGA TAC G |
| 2(G) | not amplified | ||
| 2(L) | not amplified | ||
| 3(G) | GGT CTT ACT GTG CCT GTG ACA CC | ta | ACT GGG GGA TAC G |
| 3(L) | GGT CTT ACT GTG CCT GTG ACA CC | ta | ACT GGG GGA TAC G |
| 4(G) | GGT CTT ACT GTG CCT GTG ACA | a | GGA TAC G |
| 4(L) | GGT CTT ACT GTG CCT GTG ACA | a | GGA TAC G |
| 5(G) | not amplified | ||
| 5(L) | not amplified | ||
| 6(G) | not amplified | ||
| 6(L) | GGT CTT ACT GTG CCT GTG ACA CC | g | T GGG GGA TAC G |
| 7(G) | not amplified | ||
| 7(L) | GGT CTT ACT GTG CCT GTG A | gt | ACT GGG GGA TAC G |
| Case . | VD2 . | P N P . | DD3 . |
|---|---|---|---|
| GL | GGT CTT ACT GTG CCT GTG ACA CC | ACT GGG GGA TAC G | |
| 1(G) | GGT CTT ACT GTG CCT GTG ACA CC | gt | ACT GGG GGA TAC G |
| 1(L) | GGT CTT ACT GTG CCT GTG ACA CC | gt | ACT GGG GGA TAC G |
| 2(G) | not amplified | ||
| 2(L) | not amplified | ||
| 3(G) | GGT CTT ACT GTG CCT GTG ACA CC | ta | ACT GGG GGA TAC G |
| 3(L) | GGT CTT ACT GTG CCT GTG ACA CC | ta | ACT GGG GGA TAC G |
| 4(G) | GGT CTT ACT GTG CCT GTG ACA | a | GGA TAC G |
| 4(L) | GGT CTT ACT GTG CCT GTG ACA | a | GGA TAC G |
| 5(G) | not amplified | ||
| 5(L) | not amplified | ||
| 6(G) | not amplified | ||
| 6(L) | GGT CTT ACT GTG CCT GTG ACA CC | g | T GGG GGA TAC G |
| 7(G) | not amplified | ||
| 7(L) | GGT CTT ACT GTG CCT GTG A | gt | ACT GGG GGA TAC G |
See footnotes to Table 3 for additional information. P indicates P region; N, N region.
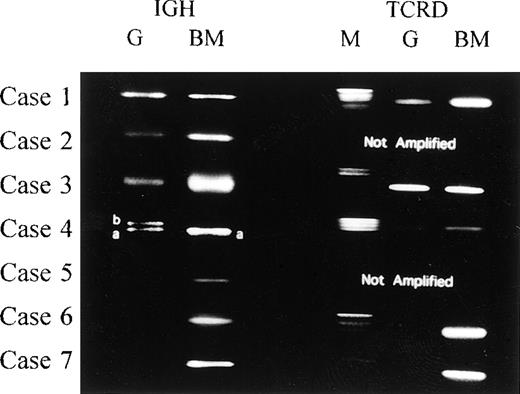
Using consensus primers for the IGH CDR3 region, we succeeded in amplifying Guthrie card DNA both from infant cases and from 2 of the 5 remaining cases. A single band of rearranged TCRD DNA was visible by agarose gel electrophoresis in cases 1, 3, and 4 (Figure2). Case 4 also had biallelic bands of amplified IGH CDR3 DNA (Figure 2). As with the DNA from leukemic bone marrow cells in cases 2 and 5, we were unable to amplify the targeted TCRD segments in Guthrie card eluates representing cases 2, 5, 6, and 7. In cases 5, 6, and 7, even with the allele-specific primers in nested PCR analysis, none of the cases demonstrated the clonal amplification.
PCR analysis for rearranged IGH and TCRD.
Amplification of rearranged IGH and TCRD from 7 cases in our study. Comparison of the PCR products from cases 1, 2, and 3 showed no size differences between Guthrie card and leukemic cell DNAs representing the IGH. We could not detect the rearranged TCRD from leukemic cell DNA and corresponding Guthrie card DNA in case 2. For case 4, the rearranged IGH product from Guthrie card DNA consisted of 2 different-sized fragments (4a and 4b), 1 of which (4a) was identical to the fragment amplified from leukemic cell DNA. Sequencing of the (a) and (b) DNAs revealed the same V-D-J or V-D rearrangements (Tables 3and 4). M indicates DNA size marker; G, PCR products from Guthrie card DNA; and BM, PCR products from leukemic bone marrow cell DNA.
PCR analysis for rearranged IGH and TCRD.
Amplification of rearranged IGH and TCRD from 7 cases in our study. Comparison of the PCR products from cases 1, 2, and 3 showed no size differences between Guthrie card and leukemic cell DNAs representing the IGH. We could not detect the rearranged TCRD from leukemic cell DNA and corresponding Guthrie card DNA in case 2. For case 4, the rearranged IGH product from Guthrie card DNA consisted of 2 different-sized fragments (4a and 4b), 1 of which (4a) was identical to the fragment amplified from leukemic cell DNA. Sequencing of the (a) and (b) DNAs revealed the same V-D-J or V-D rearrangements (Tables 3and 4). M indicates DNA size marker; G, PCR products from Guthrie card DNA; and BM, PCR products from leukemic bone marrow cell DNA.
Summary of cloning sequencing results.
As summarized in Table 2, the ratio of colony numbers showing identical nucleotide sequences per picked-up colonies in leukemic cells was in a range of IGH (53.3% to 93.8%) and TCRD (55.6% to 80.0%). In Guthrie cards, we identified identical nucleotide sequences as that of leukemic cells in frequencies of IGH (56.3% to 93.3%) and TCRD (40.0% to 86.7%). Tables 3 and 4 also summarize the sequencing data for the 7 leukemia patients. In 4 cases, the IGH CDR3 findings in Gurthrie card and leukemic bone marrow cell DNAs were identical, as were the TCRD results in 3 cases. In case 4, biallelic IGH rearrangements (4a and 4b) were amplified from Guthrie card DNA, but 1 of these bands, 4b, was not detectable in leukemic cells, even though the 4a sequences were identical. Overall, the N regions of IGH sequences consisted of 0 to 6 nucleotide insertions in 5 of the 7 cases. By contrast, the TCRD N regions possessed only 1 or 2 nucleotide insertions in the 5 cases analyzed, in which 3 cases had identical TCRD sequences in leukemic bone marrow cell and Guthrie card DNAs.
Discussion
Demonstrating the clonal origin of leukemic cells can be difficult. One accepted method is to trace specific karyotypic changes to a suspected precursor cell; however, this option is eliminated when Guthrie cards are the only source of prediagnostic genetic material. Moreover, as in the present study, it may not be possible to extract RNA from Guthrie card blood spots, obviating the use of reverse transcriptase–PCR to detect leukemia-specific chimeric gene expression.
Identification of antigen-receptor gene rearrangements affords an attractive alternative for backtracking the clonal evolution of leukemic cells. The IGH and TCRD regions, for example, undergo rearrangements that are unique for an individual cell and its progeny. Since these changes occur in 95% and 54% of the cases, respectively, we reasoned that a combination of these markers would allow amplification of rearranged gene segments in all but a small proportion of cases. Indeed, in recent studies, this strategy was successfully used to detect minimal residual disease in ALL patients who were receiving chemotherapy or had undergone bone marrow transplantation.23-27
Here we report identically rearranged IGH or TCRD segments in Guthrie card and leukemia cell DNA from 4 patients with ALL (ages 14 days, 2 months, 2.2 years, and 1.5 years). In case 4, we detected the biallelic rearranged IGH products (4a and 4b) from Guthrie card DNA. On the other hand, we amplified the single allele region (4a). This result suggested that 2 clonal populations may have existed at birth or that continuing rearrangement or the deletion of the allele may have occurred in the process of developing leukemia. However, the precise mechanism remains to be determined. In cases 5, 6, and 7, we attempted to demonstrate the clonal band by preparing allele-specific primers, which are more sensitive than the consensus primers, but no amplification was obtained with this procedure. Failure to demonstrate leukemic progenitor cells in the neonatal blood spots of 3 patients could indicate the absence of leukemia or too few leukemic cells for detection with our PCR methods. The probability that identical V-D-J or V-D sequences were detected by chance alone is vanishingly small—occurring in 1 in 106 to 1 in 108 cells under our study conditions. Thus, taken together, our data suggest the presence of leukemic progenitor cells in the neonatal blood spots in at least 4 of the 7 patients studied. The fetal characteristics of the leukemic cells were indicated by the small numbers of nucleotides inserted into the N region, the shortened D germ line and conserved J germ-line segments (IGH), and the conserved germ-line sequences of VD2 and DD3 (TCRD).28-30
It may be important that each of the patients with positive findings in Guthrie card blood spots developed overt leukemia within 2.5 years after birth. This result agrees with previous molecular studies demonstrating the presence of leukemic cells in neonatal blood specimens and suggests a relatively rapid onset of disease in patients with Guthrie card evidence of fetally transformed lymphoid cells. However, a recent account of identical twins who developed T-cell leukemia at 9 or 10 years of age provides a strong argument for protracted latency in isolated cases originating from fetal cells.
In infants, B-cell precursor ALL is characterized by hyperleukocytosis, the absence of surface CD10 expression, rearrangements of theMLL gene on chromosome 11q23, and a generally poor response to chemotherapy. By contrast, in children 2 to 4 years of age, the disease tends to be associated with a low presenting leukocyte count, positive CD10 expression, and good response to standard chemotherapy.31 Since both types of ALL originate, and probably begin clonal proliferation, during the fetal period, the reason for their dissimilar clinical features is unclear. Continued study of Guthrie card neonatal blood using immunologic markers to detect clonotypic changes should help answer this question and others regarding the natural history of childhood ALL.
Acknowledgment
The authors are grateful to Yasuko Hashimoto for her assistance in the preparation of this manuscript.
Reprints:Tomohito Yagi, Department of Pediatrics, Kyoto Prefectural University of Medicine, 602-8566, Kajiicho 465 Hirokoji Kamigyoku, Kyoto, Japan; e-mail: yagi@ped.kpu-m.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal