Abstract
Congenital afibrinogenemia is a rare, autosomal, recessive disorder characterized by the complete absence of detectable fibrinogen. We previously identified the first causative mutations in a nonconsanguineous Swiss family; the 4 affected persons have homozygous deletions of approximately 11 kb of the fibrinogen alpha (FGA) gene. Haplotype data implied that these deletions occurred on distinct ancestral chromosomes, suggesting that this region may be susceptible to deletion by a common mechanism. We subsequently showed that all the deletions were identical to the base pair and probably resulted from a nonhomologous recombination mediated by 7-bp direct repeats. In this study, we have collected data on 13 additional unrelated patients to identify the causative mutations and to determine the prevalence of the 11-kb deletion. A common recurrent mutation, at the donor splice site of FGA intron 4 (IVS4 + 1 G > T), accounted for 14 of the 26 (54%) alleles. One patient was heterozygous for the previously identified deletion. Three more frameshift mutations, 2 nonsense mutations, and a second splice site mutation were also identified. Consequently, 86% of afibrinogenemia alleles analyzed to date have truncating mutations of FGA, though mutations in all 3 fibrinogen genes, FGG, FGA, and FGB, might be predicted to cause congenital afibrinogenemia.
Congenital afibrinogenemia (Mendelian Inheritance in Man #202,400) was originally described in 1920.1 To date some 150 families with this disorder have been reported,2 with approximately 50% of instances occurring in consanguineous pedigrees.3 Although functional assays of clot formation are infinitely prolonged in affected persons, the coagulation defect is surprisingly no more severe than in severe hemophilias A and B, varying from severe to moderate.4Umbilical cord hemorrhage is often the first sign of the disorder; gum bleeding, epistaxis, menorrhagia, gastrointestinal bleeding, and hemarthrosis occur with different degrees of intensity, and spontaneous intracerebral bleeding and splenic rupture can occur throughout life. Patients generally respond well to fibrinogen replacement therapy. The genetic defect was assumed to be at the level of fibrinogen synthesis because the half-life of infused fibrinogen is essentially normal.2 The fibrinogen locus is comprised of 3 genes coding for fibrinogen gamma (FGG), fibrinogen alpha (FGA), and fibrinogen beta (FGB), clustered in a region of approximately 50 kb on chromosome 4q28-q31.5 We previously studied this region in a Swiss family with 2 pairs of affected brothers by using microsatellite analysis, polymerase chain reaction (PCR) amplification, and Southern blotting, identifying the first causative mutations for the disorder.6 We found that the genetic defect in this family was an apparently recurrent deletion of approximately 11 kb of DNA that eliminates most of the FGAgene, leading to an absence of functional fibrinogen. We subsequently identified the molecular mechanism involved in the generation of the deletions responsible for congenital afibrinogenemia by cloning and sequencing the deletion junctions for all mutated chromosomes. The deletions were all identical to the base pair and probably resulted from nonhomologous (illegitimate) recombination, mediated by a direct 7-bp repeat, AACTTTT, and perhaps also by indirect repeats in the breakpoint region.7
In this study, we performed mutation analysis in the FGA gene in 13 additional unrelated patients with congenital afibrinogenemia from Puerto Rico, France, Pakistan, Belgium, and the United States. One patient, from the United States, was a heterozygous carrier of the originally described 11 kb deletion, as revealed by Southern blot analysis. In addition, we identified the most common mutation, in the invariant GT dinucleotide of the donor splice site of FGAintron 4 (IVS4 + 1 G > T), which accounted for 14 of the 26 (54%) afibrinogenemia alleles analyzed in this study. Five new mutations leading to premature protein truncation were also identified: 34insC (exon 1), 3121del AA (exon 4), and 4329del C, G316X, and W334X (all exon 5). Finally, a potential IVS1 + 3 donor splice site mutation was identified. All the mutations in patients of Caucasian origin were identified; in contrast, no mutations were found in FGAin the 2 non-Caucasian patients studied.
Patients, materials, and methods
Patients
Clottable fibrinogen levels were measured by the method of Clauss.8 Fibrinogen concentrations were determined either by radial immunodiffusion, by rocket immunoelectrophoresis, or by nephelometry.9 10 Prothrombin time, activated partial thromboplastin time, thrombin time, and clotting factors were measured locally using standard methods. Blood was collected from 13 unrelated patients with congenital afibrinogenemia, as diagnosed by functional or antigenic assays. There was known consanguinity for 3 of the families (Table 1). Informed consent was obtained from all patients participating in the study.
Patients with afibrinogenemia investigated in this study
| Patient . | Origin (consanguinity) . | Age at Diagnosis . | Mutation . | Haplotypes . |
|---|---|---|---|---|
| A2 | France (NC) | Birth | IVS4 + 1 G > T | 1-5-2 |
| IVS4 + 1 G > T | 2-5-2 | |||
| A4 | Puerto Rico (C) | 7 months | Not found | 1-1-3 |
| 1-1-3 | ||||
| A5 | France (?) | Birth | IVS4 + 1 G > T | 1-3-2 |
| IVS4 + 1 G > T | 1-4-1 | |||
| A6 | France (NC) | 1 year | IVS4 + 1 G > T | 1-5-2 |
| IVS4 + 1 G > T | 2-4-2 | |||
| A7 | France (NC) | 10 months | IVS4 + 1 G > T | 2-5-2 |
| G316X (exon 5) | 2-6-2 | |||
| A8 | Belgium (NC) | 5 years | 34insC (exon 1) | 2-5-2 |
| 3121delAA (exon 4) | 2-2-2 | |||
| A9 | Belgium (C) | 2 years | IVS4 + 1 G > T | 1-4-1 |
| IVS4 + 1 G > T | 1-4-1 | |||
| A10 | France (NC) | 17 years* | IVS4 + 1 G > T | 1-5-1 |
| IVS1 + 3 A > G | 3-3-1 | |||
| A12 | France (NC) | 5 years | 3121delAA (exon 4) | 2-2-? |
| W334X (exon 5) | 2-5-? | |||
| B1 | France (?) | Birth | IVS4 + 1 G > T | 2-3-1 |
| IVS4 + 1 G > T | 2-3-1 | |||
| B2 | Pakistan (C) | 17 months | Not found | 1-3-1 |
| 1-3-1 | ||||
| B3 | United States (?) | Birth | IVS4 + 1 G > T | 2-4-2 |
| 4329delC (exon 5) | 2-4-1 | |||
| B4 | United States (?) | Birth | IVS4 + 1 G > T | 2-5-2 |
| 11-kb deletion | 3-del-1 |
| Patient . | Origin (consanguinity) . | Age at Diagnosis . | Mutation . | Haplotypes . |
|---|---|---|---|---|
| A2 | France (NC) | Birth | IVS4 + 1 G > T | 1-5-2 |
| IVS4 + 1 G > T | 2-5-2 | |||
| A4 | Puerto Rico (C) | 7 months | Not found | 1-1-3 |
| 1-1-3 | ||||
| A5 | France (?) | Birth | IVS4 + 1 G > T | 1-3-2 |
| IVS4 + 1 G > T | 1-4-1 | |||
| A6 | France (NC) | 1 year | IVS4 + 1 G > T | 1-5-2 |
| IVS4 + 1 G > T | 2-4-2 | |||
| A7 | France (NC) | 10 months | IVS4 + 1 G > T | 2-5-2 |
| G316X (exon 5) | 2-6-2 | |||
| A8 | Belgium (NC) | 5 years | 34insC (exon 1) | 2-5-2 |
| 3121delAA (exon 4) | 2-2-2 | |||
| A9 | Belgium (C) | 2 years | IVS4 + 1 G > T | 1-4-1 |
| IVS4 + 1 G > T | 1-4-1 | |||
| A10 | France (NC) | 17 years* | IVS4 + 1 G > T | 1-5-1 |
| IVS1 + 3 A > G | 3-3-1 | |||
| A12 | France (NC) | 5 years | 3121delAA (exon 4) | 2-2-? |
| W334X (exon 5) | 2-5-? | |||
| B1 | France (?) | Birth | IVS4 + 1 G > T | 2-3-1 |
| IVS4 + 1 G > T | 2-3-1 | |||
| B2 | Pakistan (C) | 17 months | Not found | 1-3-1 |
| 1-3-1 | ||||
| B3 | United States (?) | Birth | IVS4 + 1 G > T | 2-4-2 |
| 4329delC (exon 5) | 2-4-1 | |||
| B4 | United States (?) | Birth | IVS4 + 1 G > T | 2-5-2 |
| 11-kb deletion | 3-del-1 |
This female patient was considered to have severe hemophilia from an early age; the formal diagnosis of afibrinogenemia was not made until she was 17 years old. Haplotypes were defined with the 3 closely linked markers, D4S3021, FGAi3, and D4S2999. Because analysis of the parental chromosomes was not possible, allele phases were inferred to determine the minimum number of different chromosomes in the patient sample. C, consanguineous; NC, not consanguineous; ?, unknown status.
Mutation screening
Genomic DNA was purified from blood leukocytes according to standard protocols. The 5 exons encoding the fibrinogen α isoform were amplified from genomic DNA by PCR, under standard conditions. The alternatively spliced exon 6 was excluded from this study because it persists only in the mRNA encoding the longer αE isoform, which represents just 1% of adult fibrinogen.11 Exons 2 and 3 were coamplified in a single fragment, whereas the larger exon 5 with part of the 3′UTR was amplified in 3 overlapping fragments (the first fragment was coamplified with exon 4). The intron–exon boundaries and flanking sequences were included in the regions analyzed. Primers were designed from the published FGA sequence (Genbank M64982) and are listed as shown in Table2.
Primers used for PCR amplifications (5′ >> 3′)
| Target . | Forward . | Reverse . |
|---|---|---|
| Exon 1 | CAGCCCCACCCTTAGAAAAG | CCTGGGGTCATAAAGCTAAG |
| Exons 2-3 | CCTCTTCTGGCTAACATTGC | CAGGGATATTATGAAGGTATG |
| Exon 4 | CAGCAGCTACTTCAATAACC | GTGCATAACTATCGCCTTCC |
| Exon 5, 5′ | (same as exon 4) | GCGGCATGTCTGTTAATGCC |
| Exon 5, mid | GATCTTGTCGAGGGTCATGC | CTCCTAACATAGGTGAGAAG |
| Exon 5, 3′ | CTGGACCTCTGAGAGCTCTG | ATGGCTCTGTACTGTTAGGC |
| Target . | Forward . | Reverse . |
|---|---|---|
| Exon 1 | CAGCCCCACCCTTAGAAAAG | CCTGGGGTCATAAAGCTAAG |
| Exons 2-3 | CCTCTTCTGGCTAACATTGC | CAGGGATATTATGAAGGTATG |
| Exon 4 | CAGCAGCTACTTCAATAACC | GTGCATAACTATCGCCTTCC |
| Exon 5, 5′ | (same as exon 4) | GCGGCATGTCTGTTAATGCC |
| Exon 5, mid | GATCTTGTCGAGGGTCATGC | CTCCTAACATAGGTGAGAAG |
| Exon 5, 3′ | CTGGACCTCTGAGAGCTCTG | ATGGCTCTGTACTGTTAGGC |
PCR products were directly sequenced with the primers used for the amplification on a semiautomated sequencer, using standard dye-terminator protocols (PE Biosystems, Foster City, CA). For all patients with apparently heterozygous changes, the 2 alleles were cloned (TOPO-TA; Invitrogen, Groningen, The Netherlands) and sequenced as above.
Southern blotting
Genomic DNA was digested with BamHI and electrophoresed and blotted by standard techniques. Filters were hybridized with the pAFS1 probe as previously described.6
Haplotype analysis
Microsatellite analyses were performed as previously described.7 The locus order and genetic distances in centimorgans were: D4S3021–(0.0)–fibrinogen(FGAi3/FGB)–(0.6)–D4S2999 (according to the Marshfield sex-averaged genetic map, http://www.marshmed.org). Because parental chromosomes were unavailable for analysis, allele phases were inferred to determine the minimal number of different chromosomes in the patient collection; the real number was possibly higher.
Screening of normal alleles
Ninety-five DNA samples from anonymous European control subjects were screened for the IVS1 + 3 A > G mutation by PCR amplification ofFGA exon 1 followed by nonradioactive, single-stranded conformation analysis using GeneGel Excel gels (Pharmacia Biotech, St Albans, UK) as previously described.12
Results
Data from 13 unrelated patients with afibrinogenemia were collected for this study. For all these patients, plasma fibrinogen was undetectable by functional and immunologic assays, and the prothrombin, activated partial thromboplastin, and thrombin time tests were unclottable in all patients. Factor V, factor VII, and factor X were measured in all patients and were within normal ranges. In the 7 patients tested, factors VIII, IX, and XI were within normal ranges. Furthermore, for some patients, inhibitors (antithrombin, protein C, and protein S), factor V Leiden, and prothrombin G20210A mutations were assayed, and all results were normal.
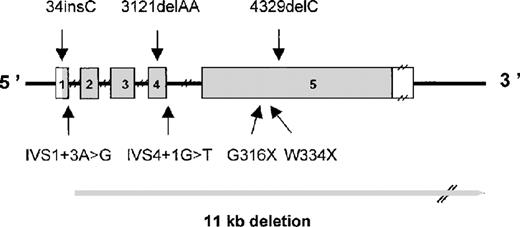
The FGA gene was screened for mutations by PCR amplification of the entire coding region of the α isoform, and this was followed by DNA sequencing. Heterozygous PCR products were cloned, and the 2 alleles were sequenced. Causative mutations were identified in 22 of 26 disease alleles. Interestingly, the mutations were identified in all patients of Caucasian origin but not in the 2 non-Caucasian patients (Table 1 and Figure 1).
Schematic representation of the FGA gene showing mutations causing congenital afibrinogenemia.
Exons are numbered and are drawn to scale; the shaded portions represent the coding region. Introns are indicated by narrow lines and are not to scale.
Schematic representation of the FGA gene showing mutations causing congenital afibrinogenemia.
Exons are numbered and are drawn to scale; the shaded portions represent the coding region. Introns are indicated by narrow lines and are not to scale.
Deletion of 11 kb
We had previously predicted that the originally reported 11-kbFGA deletion would be a relatively common recurrent mutation, and we therefore wanted to determine its prevalence in this patient group. The deletion was excluded in patients A5, A6, A8, and A12 because of heterozygosity for the FGAi3 polymorphism, which is situated within the deletion in intron 3, and in patient B3, a compound heterozygote for 2 different mutations (Table 1). For the remaining patients, Southern blot analysis was performed as previously described. The patient from the United States (B4) was identified as a carrier of the deletion (Figure 2), confirming its recurrent nature.7
Southern blot diagnosis of the FGA 11-kb deletion.
The normal fragment detected by probe pAFS1 measures 13 kb, the deleted allele is detected as a 17-kb fragment because 2 BamHI sites are eliminated. The results for 3 patients are shown. C, homozygous deleted control; N, normal control.
Southern blot diagnosis of the FGA 11-kb deletion.
The normal fragment detected by probe pAFS1 measures 13 kb, the deleted allele is detected as a 17-kb fragment because 2 BamHI sites are eliminated. The results for 3 patients are shown. C, homozygous deleted control; N, normal control.
Splice site mutations
Five patients were homozygous for a previously undescribed splice site mutation, IVS4 + 1 G > T (A2, A5, A6, A9, and B1), and 4 were compound heterozygotes (A7, A10, B3, and B4; Table 1, Figures 1 and 2).
The IVS4 + 1 G > T represents 54% (14 of 26) of the alleles in this study or 48% of all the afibrinogenemia mutations described to date (including our previous study6).
Haplotypes were obtained for 3 microsatellite markers within or closely flanking the fibrinogen gene cluster. Because the parental DNA was unavailable, allele phases could not be determined; remarkably, however, the IVS4 + 1 mutation was found on at least 8 discrete haplotypes (for the 3 markers used; Table 1). Significantly, the mutation was associated with 3 different alleles just for the intragenic FGA intron 3 microsatellite marker (arbitrarily numbered 3, 4, and 5; Table 1).
In 1 patient (A10) who was heterozygous for the IVS4 + 1 mutation, the only other change identified by sequencing of the FGA gene was an A > G mutation in the intron 1 donor site (IVS1 + 3 A > G). Although the donor site +3 position is not highly conserved in splice sites, this mutation was not found in 190 normal alleles (or in any of the other patients in this study). We predicted that the IVS1 + 3 mutation was the second causative mutation in this patient (see “Discussion”).
Deletions and insertions
One patient (A8) was found to be a compound heterozygote for 2 frameshift mutations in FGA. The first mutation, 34insC (numbered according to Genbank M64982), was a 1-bp insertion in the second codon of exon 1 that led to an in-frame TAG stop codon in exon 2. The second mutation, 3121delAA, was a 2-bp deletion in exon 4 that led to an in-frame TAG stop codon 15 codons downstream, in the same exon. Another patient (A12) was also a compound heterozygote for 3121delAA. A third frameshift mutation in exon 5, 4329delC, was identified in patient B3. This deletion generated an in-frame TAA stop codon 69 codons downstream.
Nonsense mutations
Two nonsense mutations were identified in exon 5, both in compound heterozygosity: G316X (patient A7, GGA > TGA) and W334X (patient A12, TGG > TGA).
Discussion
In this study, we analyzed the FGA gene in 13 unrelated patients with congenital afibrinogenemia from Puerto Rico, France, Pakistan, Belgium, and the United States to identify the causative mutations in affected persons and to determine the prevalence of the 11-kb FGA deletion. Including the 3 alleles in the 4 affected family members described in our original report,6 we have now studied a total of 29 alleles in 17 patients and have identified 8 different FGA mutations that account for 86% (25 of 29) of the afibrinogenemia alleles.
In our previous studies, we described 4 affected persons, all homozygous for an 11-kb deletion that was identical to the base pair.6 7 Surprisingly, haplotype analysis suggested that the deletion recurred on 2 if not 3 discrete ancestral chromosomes, perhaps as a result of an illegitimate recombination mediated by direct and indirect repeats in the breakpoint region. The identification here of 1 additional allele bearing the 11-kb deletion, in an American patient unrelated to the Swiss family previously described, confirms that the FGA 11-kb deletion is indeed recurrent.
The novel IVS4 + 1 G > T mutation we report here, at the invariant GT dinucleotide of the intron 4 donor splice site, also appeared to be recurrent and is the most common known cause of congenital afibrinogenemia, accounting for 14 of the 29 (48%) mutant alleles we studied. Alternatively, it remains possible that these 2 mutations were not recurrent but were inherited from common ancestors; it could be predicted that they are very ancient mutations because of their association with numerous haplotypes.
Although it proved impossible to determine the effect of this mutation in patients' leukocyte RNA (the levels of illegitimate expression were too low to detect), the fact that these patients have clinical afibrinogenemia and not severe hypofibrinogenemia implies that essentially no normal splicing occurs at this site. Among mutations of the consensus dinucleotide splice sites, those at the +1 position of the donor 5′) site are the most frequent.13 In most patients, they lead to skipping of the exon immediately 5′ of the mutation.
We performed computer splice prediction analysis of the region around the IVS4 donor site with the program Spliceview,14 which predicts splice-site efficiencies taking into account consensus sequence homology and the mutual dependency of nucleotides (http://www.itba.mi.cnr.it/webgene/). In the normal sequence, 2 distinct predicted donor sites were detected, the first “physiological,” and the second 5 bases downstream that resulted in a frameshift mutation. In the presence of the IVS4 + 1 G > T mutation, the physiological site is no longer detected by the analysis. We therefore predict that the common mutation leads to aberrant usage of the alternative site and to a consequent frameshift mutation in 100% of the transcripts.
The second donor splice site mutation, IVS1 + 3 A > G, was present in only 1 patient, in compound heterozygosity with the IVS4 + 1 G > T mutation. It was the only other sequence variant identified in this patient. We excluded the possibility that this was a common polymorphism in the human FGA gene by screening 190 alleles from normal persons; no other carrier was found. Spliceview analysis of the normal sequence around the IVS1 donor site predicts a single site; however, in the mutant IVS1 + 3 G sequence, no donor site is identified. Furthermore, although the consensus donor splice site has either A or G at this position,15 11 disease-causing splice site + 3 A > G mutations have been reported to date; in 10 of these, aberrant splicing was proven.16 Consequently, it is probable that this represents the second pathogenic mutation in patient A10, though the undetectable levels of FGA mRNA in blood leukocytes made it impossible to evaluate its in vivo effect.
We have also identified 3 novel frameshift mutations, all of which are predicted to result in premature termination of translation because of the creation of in-frame stop codons and 2 nonsense mutations.
Although all the mutations were identified in Caucasian patients, we did not find any FGA mutations in patients A4 and B2 (of Puerto Rican and Pakistani origins, respectively). Both these patients are consanguineous and homozygous for the 3 polymorphic markers used. It is likely that they have mutations in the closely linked FGG orFGB genes, which were not analyzed in this study. Missense mutations in FGB have recently been identified in 2 patients with afibrinogenemia.17
We have now identified 8 different mutations in the fibrinogen Aα gene, accounting for the majority of afibrinogenemia alleles investigated, though intuitively it could be expected that all the fibrinogen genes would be equally implicated. All the mutations were predicted to lead to premature termination (Figure 1). Four such mutations have been previously described in FGA exon 5, in association with either dysfibrinogenemia or severe hypofibrinogenemia. Three of these are situated much closer to the carboxy-terminus than the mutations we describe here, leading to termination after amino acid 451 or higher.18-20 Fibrinogen Otago,21however, was associated with a homozygous frameshift mutation, leading to the truncation of fibrinogen Aα at amino acid 270, which is proximal to 3 of the afibrinogenemia mutations we describe. In our patients, these mutations were present in compound heterozygosity associated with more amino-terminal truncation mutations (Table 1). It is possible that the more distal truncation mutations are functionally less severe and permit the secretion of minute quantities of fibrinogen, which, in homozygosity or in compound heterozygosity with less severe FGA mutations, result in hypofibrinogenemia rather than afibrinogenemia. If this is the case, we expect to find some of the mutations we described here in patients with less severe fibrinogen disorders in association with less severe mutations. The characterization of these new mutations will facilitate diagnosis of these disorders and permit prenatal diagnosis for families who so desire.
Acknowledgments
We thank all the families for their cooperation and for donating blood samples. We thank Hewlett-Packard (Switzerland) for their kind support through their donation of computer equipment.
Supported by a Marie Heim-Voegtlin grant (M.N.-A.), Swiss National Science Foundation grant 31-55848.98, and the Roche Research Foundation.
Reprints:Marguerite Neerman-Arbez, Centre Médical Universitaire, 1 rue Michel Servet, CH-1211 Geneva, Switzerland; e-mail: marguerite.arbez@medecine.unige.ch.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal