Abstract
We studied a large kindred with nonsyndromic autosomal dominant thrombocytopenia to define the phenotype and used genomic linkage analysis to determine the locus of the abnormal gene. Affected family members are characterized by lifelong moderate thrombocytopenia (mean = 42.7 × 109/L) with moderate propensity toward easy bruising and minor bleeding. Megakaryocytes are present in bone marrow with reduced frequency, and there are no apparent abnormalities of myeloid or erythroid cells. This type of inherited thrombocytopenia has no evident association with hematopoietic malignancy or progression to aplastic anemia. In the past, members of this family have failed therapeutic trials of immunosuppression and splenectomy. In our investigation, we found that affected individuals had normal platelet size compared with unaffected family members and modestly increased thrombopoietin levels. Hematopoietic colony assays from bone marrow and peripheral blood demonstrated that megakaryocyte precursors (CFU-Mk) were dramatically increased in both number and size in affected individuals. Bone marrow cells grown in liquid culture with thrombopoietin failed to develop polyploid cells greater than 8N. Also, electron microscopy demonstrated that megakaryocytes from an affected individual had markedly delayed nuclear and cytoplasmic differentiation. Genome-wide linkage analysis established a single locus for the disease gene on the short arm of chromosome 10 with a maximum 2-point lod score of 5.68 (at θ = 0). By recruiting additional family members, the genomic region was narrowed to 17 centimorgans. We conclude that a gene in this locus plays an important role in megakaryocyte endomitosis and terminal maturation.
Inherited thrombocytopenia is a rare human condition that is frequently mistaken for an acquired platelet disorder. A careful medical and family history can generally help categorize congenital thrombocytopenia based on the duration of symptoms and onset immediately after birth. A detailed pedigree can often help identify the inheritance pattern (ie, autosomal recessive, autosomal dominant, or sex-linked). Furthermore, in some cases, the coexpression of additional phenotypic anomalies defines clinically recognized syndromes associated with thrombocytopenia (ie, deafness and nephritis in Alport syndrome, eczema and immunodeficiency in Wiskott-Aldrich syndrome). The known causes of inherited thrombocytopenia have been recently reviewed.1 It is important to distinguish disorders that are primarily of platelet number from those that are predominantly of platelet dysfunction (often with normal numbers). Glanzmann thrombasthenia (GpIIb/IIIa deficiency) and type I von Willebrand disease (vWF deficiency) are examples of such thrombocytopathies.1 2
In this report, we present new information regarding a family with autosomal dominant thrombocytopenia that was first described by Bithell and colleagues in 1965.3 This large family has kept meticulous records, which allow the inheritance pattern to be traced over 6 generations from a single founder.3 The recent identification of thrombopoietin (TPO) and its receptor, c-Mpl, has provided the means to study megakaryocyte and platelet development in vitro.4 5 Studies of rare familial human conditions provide the opportunity to understand normal physiology and identify novel gene functions in vivo.
We studied 30 members of a single extended family to establish linkage of inherited thrombocytopenia to a locus of the human genome. This led to the identification of a 17-centimorgan (cM) region of chromosome 10 as the only site of statistically significant linkage (maximum lod score = 5.68 by 2-point linkage, θ = 0). Moreover, we found that bone marrow and peripheral blood from thrombocytopenic patients were highly enriched for megakaryocyte colony-forming units (CFU-Mk) compared with unaffected individuals. The megakaryocytes were unable to achieve normal polyploidy or ultrastructural maturation after 11 days in liquid culture. These findings suggest a late block in terminal differentiation of megakaryocytes.
Patients, materials, and methods
Patient recruitment
Members of the family with hereditary thrombocytopenia were enrolled as study subjects in a protocol approved by the University of Washington Institutional Review Board. After obtaining informed consent, peripheral venous blood was collected in Vacutainer tubes (Becton Dickenson, San Jose, CA) containing ethylenediaminetetraacetic acid (EDTA), and complete blood counts were measured using an automated blood analyzer (Serono Diagnostics Systems 9018CP, Norwell, MA) or were provided by personal physicians. Blood smears were prepared for Wright-Giemsa stain. In all cases, blood was processed within 24 hours of collection.
Preparation of platelet-poor plasma, genomic DNA, and platelets
Erythrocytes and mononuclear cells were separated from platelet-rich plasma via low-speed centrifugation (360g for 10 minutes). The plasma was carefully transferred into a new tube and subjected to high-speed centrifugation (1550g for 20 minutes). The resulting supernatant was then frozen in aliquots as platelet-poor plasma and was used for TPO enzyme-linked immunosorbent assays (ELISAs). The platelet pellet was resuspended in approximately 1 mL of RPMI media and was used as a source of platelets for flow cytometry and signaling studies. From the original low-speed separation, the buffy coat layer was carefully transferred to separate tubes. Four aliquots of 200 μL were used to extract genomic DNA (QIAamp Blood Kit, Qiagen Corp, Valencia, CA). Isolated DNA was measured by spectrophotometry and agarose gel electrophoresis to ensure that sufficient quantity and purity was achieved.
Flow cytometry
Platelets were resuspended in Tris-buffered saline with 10-mmol/L EDTA (TBSE), counted (Coulter platelet analyzer, Santa Ana, CA), and adjusted to approximately 200 × 109/L. Two hundred microliters of platelets were incubated with monoclonal antibodies to glycoproteins (Gp) IIb-IIIa, GpIV, or Gp Ia-IIa (Immunotech, Marseille, France) at a final concentration of 10 μg/mL (30 minutes at 37°C). Platelets were washed twice with TBSE plus 1% human albumin (Alpine Biologics, Blauvelt, NY) by centrifugation at 1500g for 10 minutes. All supernatant was removed, and the pellet was resuspended in 10 μL of fluorescein isothionate (FITC)-conjugated goat antimouse immunoglobulin (Becton Dickenson). The platelets were incubated for 15 minutes in the dark and washed by centrifugation in TBSE plus 1% human albumin. The pellet was resuspended in Hema-Line 2 (ABX Diagnostic, Allentown, PA) and analyzed by flow cytometry.
Thrombopoietin ELISA
TPO levels were measured from platelet-poor plasma for each study participant using a sandwich ELISA (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. Samples were measured in duplicate and were repeated in 2 separate experiments to ensure that results were internally consistent; 96-well plates were read using a microplate reader (Bio-Rad 550, Bio-Rad, Hercules, CA), 450- to 570-nm dual wavelength protocol.
Preparation of bone marrow and CD34+ peripheral blood
One unaffected and 2 affected family members consented to donate bone marrow (5-6 mL) from the iliac crest and a large volume of peripheral blood (50 mL). Bone marrow smears were prepared on cover slips and stained with Wright-Giemsa stain. Next, low-density cells were separated from both bone marrow and peripheral blood cells by overlayering the cells onto Ficoll (1.077-1.08 g/mL; ICN Biomedicals Inc, Costa Mesa, CA) and by centrifugation (400g for 30 minutes). Cells were collected from the interface, washed 2 times in Iscove's modified Dulbecco's medium (IMDM) with 2% fetal calf serum (Stem Cell Technologies, Vancouver, Canada), and counted using a hemocytometer. No further manipulations of bone marrow cells were carried out. Peripheral blood cells were subjected to immunomagnetic enrichment of CD34+ cells using the MidiMACS system (CD34 Select Kit, Miltenyi Biotec, Auburn, CA). Selected cells were washed in IMDM with 2% fetal calf serum (Stem Cell Technologies) and counted.
Colony assays
Low-density bone marrow cells were mixed with 1% methylcellulose in IMDM containing 30% fetal bovine serum, 1% bovine serum albumin, 0.1-mmol/L β-mercaptoethanol, and the following recombinant human cytokines: 50-ng/mL stem cell factor, 10-ng/mL interleukin-3, 3-U/mL erythropoietin, and 10-ng/mL granulocyte-monocyte colony-stimulating factor (MethoCult Assay, Stem Cell Technologies). The final concentration of cells was adjusted to 7.5 × 104cells/plate (1.1 mL) for each sample. Five replicate plates were made for each of the affected individuals and the unaffected family member. After 12 days at 37°C and 5% CO2 in a humidified incubator, burst-forming units, erythroid (BFU-E), CFU-GM (granulocyte, monocyte), and mixed-lineage CFU-GEMM (granulocyte, erythroid, monocyte, megakaryocyte) colonies were counted from each of the plates, and averages were determined for each individual. CFU-Mk colonies were determined separately using the MegaCult System (Stem Cell Technologies), in which cells were cultured on collagen-based slide chambers in the presence of recombinant human TPO, interleukin-3, and interleukin-6. Low-density bone marrow cells were plated at a density of either 5.0 × 104 or 2.0 × 105 cells per chamber, and CD34-selected peripheral blood cells were plated at either 4.0 × 103 or 1.2 × 104 cells per chamber. After 14 days in culture, the chamber slides were dehydrated and stained for GpIIb/IIIa according to the manufacturer's guidelines. All colony numbers were quantified using an inverted phase light microscope.
DNA ploidy evaluation
Ficoll-purified bone marrow cells were cultured under serum-free conditions in IMDM with 1% Nutridoma (Sigma Corp, St Louis, MO) and supplemental antibiotics (penicillin and streptomycin, BioWhittaker, Walkersville, MD) at a final concentration of 1 × 106 cells/mL. Recombinant TPO was added at a final concentration of 10 ng/mL (generous gift from Don Foster, Zymogenetics, Seattle, WA), and the cultures were incubated at 37°C in a humidified incubator containing 5% CO2. On day 11, cells were removed from liquid culture and incubated with FITC-labeled CD41a-specific antibody (Pharmingen Corp, San Diego, CA) as recommended (20 μL/106 cells). After gentle washing, the cells were resuspended in buffer containing 50-μg/mL propidium iodide, 0.1% Triton X-100, 0.1% sodium citrate, and 30-μg/mL RNAse (Sigma Corp). Flow cytometry was performed (Becton Dickinson FACScalibers), and ploidy classes for megakaryocytes were determined by plotting the propidium iodide fluorescence of the cells with highest CD41 expression (brightest 10% to 15%) using a semilogarithmic scale.
Signal transduction studies
Platelets were obtained by centrifugation of platelet-rich plasma (see “Preparation of platelet-poor plasma, genomic DNA, and platelets”). Platelets were prepared by incubation with aspirin, prostaglandin E1, and apyrase as previously described.6 Final cell pellets were resuspended in a modified HEPES-Tyrodes buffer (10-mmol/L HEPES, pH 7.4; 129-mmol/L NaCl; 8.9-mmol/L NaHCO3; 0.8-mmol/L KH2PO4; 0.8-mmol/L MgCl2; 5.6-mmol/L dextrose; 2-U/mL apyrase; 1-mmol/L CaCl2) and were incubated at 37°C in the absence of serum and growth factors for 2 hours. One half of each sample was then stimulated with recombinant human TPO (10 ng/mL) for 10 minutes while the other half remained unstimulated. Protein extracts were prepared using a modified RIPA buffer as previously described.5 7 Samples of unstimulated and stimulated lysates were analyzed by Western blot and probed with a phospho-specific STAT3 (signal transducers and activators of transcription) antibody generously provided by David Frank, Boston, MA (1:3000 in Tris-buffered saline solution plus 0.05% Tween 20 and 3% bovine serum albumin [TBST + BSA]). After extensive washing in TBST, tyrosine-phosphorylated STAT3 protein was visualized using a secondary goat antirabbit immunoglobulin G conjugated to horseradish peroxidase (1:5000 in TBST + BSA; Bio-Rad) and chemiluminescent reagents (New England Nuclear Corp, Boston, MA). The blot was later stripped (50°C for 30 minutes in 62.5-mmol/L Tris, pH 6.8; 2% sodium dodecyl sulfate; 100-mmol/L β-mercaptoethanol) and reprobed to demonstrate equal loading of STAT3 in all lanes (STAT3 polyclonal antibody K-18, 0.5 μg/mL in TBST + BSA; Santa Cruz Biotechnology, Santa Cruz, CA).
Electron microscopy
After 10 days in liquid culture (10-ng/mL TPO, serum-free), a portion of the marrow culture was recovered by low-speed centrifugation (200g for 5 minutes) and was enriched for megakaryocytes by discontinuous BSA density gradient sedimentation as previously described.5 7 The fractions with highest megakaryocyte numbers were pooled and collected by low-speed centrifugation. The cell pellet was preserved by gently adding fixative containing 1% glutaraldehyde and 4% formaldehyde. The pellet was minced to provide fragments about 1 mm in maximum diameter. These fragments were postfixed in 2% osmium tetroxide and processed into Eponate 12. Each plastic block was cut to provide sections 0.5 μm thick, which were stained (methylene blue-azure II dye) and examined by light microscopy to select the block with maximum cell density. This block was cut into thin sections (60-90 nm), stained with lead acetate and uranyl acetate, and examined by electron microscopy.
Genome-wide linkage analysis
Genomic localization was performed using the Weber-9 human linkage set, consisting of 388 microsatellite markers with an average distance of 10 cM between markers. Polymerase chain reaction of genomic DNA and allelic assignments for the initial 16 participants (Figure 1) were done by Research Genetics Corp (Birmingham, AL). This service reports a genotyping error rate of less than 1%.8 Using that data and allelic frequencies obtained from the web sites of CEPH (Centre d'Etude du Polymorphisme Humain; www.cephb.fr/cgi-bin/wdb/ceph/systeme/form) and CHLC (Cooperative Human Linkage Center;http://cgap.nci.nih.gov/CHLC), 2-point linkage analysis was performed for each individual marker using the LINKAGE analysis software.9 10 An autosomal dominant, fully penetrant gene with an allele frequency of 10−4 and no phenocopies was modeled (see “Autosomal dominant inheritance pattern”). The initial genomic scan had a maximum potential lod score of 3.31. A lod score is the logarithm of ratio of the likelihood of the data given linkage at a certain recombination fraction divided by the likelihood of the data assuming no linkage. A lod score of 3 or more is generally considered significant evidence of genetic linkage (ie, cosegregation of the marker and disease). A chromosome 10 region that achieved a significant lod score was studied further by adding additional markers from the PE Biosystems high-density microsatellite set (Foster City, CA) and accrual of additional family members for genotyping. After the initial genomic scan, all subsequent genotyping was performed in our laboratory using an ABI 310 genetic analyzer. Linkage analysis was then used to calculate lod scores for the extended pedigree, delimit the range of linkage, and identify recombination events.
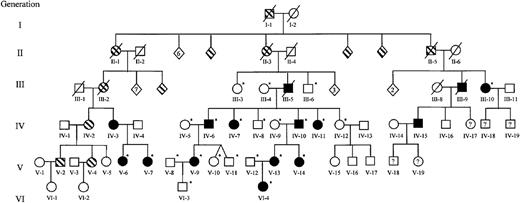
Extended family pedigree.
Squares represent males; circles, females; diamonds, unspecified gender. The founder and partial pedigrees of 3 of his 12 children are depicted. Documented thrombocytopenia is indicated by solid figures; presumed or reported thrombocytopenia, striped figures; unaffected, open figures; unknown platelet status, question mark. Individuals who were studied during the genomic scan and those recruited to further delimit the area of linkage are indicated by an asterisk. Numbers inside diamonds indicate the number of offspring. Three unaffected children of unaffected individuals (not depicted) were included in the complete blood count and TPO data but were not used for genotyping. The generation and person numbers for each individual are used in Figures2-7.
Extended family pedigree.
Squares represent males; circles, females; diamonds, unspecified gender. The founder and partial pedigrees of 3 of his 12 children are depicted. Documented thrombocytopenia is indicated by solid figures; presumed or reported thrombocytopenia, striped figures; unaffected, open figures; unknown platelet status, question mark. Individuals who were studied during the genomic scan and those recruited to further delimit the area of linkage are indicated by an asterisk. Numbers inside diamonds indicate the number of offspring. Three unaffected children of unaffected individuals (not depicted) were included in the complete blood count and TPO data but were not used for genotyping. The generation and person numbers for each individual are used in Figures2-7.
Results
Autosomal dominant inheritance pattern
We obtained peripheral venous blood from 28 members of a single pedigree and classified them as thrombocytopenic (less than 125 × 109/L) or normal (150 × 109/L to 400 × 109/L). We were able to assign affectation status to all individuals without ambiguity. There were no instances of transmission of thrombocytopenia from an asymptomatic individual (ie, carrier) that might suggest incomplete penetrance. Males and females were equally likely to transmit the thrombocytopenic trait and to inherit it, suggesting that there is no sex-linked component. There was no consanguinity within the family, and accurate records have been maintained for 6 generations. These data were completely consistent with an autosomal dominant inheritance of thrombocytopenia in this family (Figure 1).
Medical history
Individual members of this family have come to medical attention at various times over the past 30 years and have been extensively evaluated to determine the nature of their thrombocytopenia. At least 3 individuals were treated for presumed immune-mediated thrombocytopenia with immunosuppression followed by splenectomy without any improvement in platelet levels. There were no major bleeding diatheses even with minor surgery and childbirth, suggesting that the residual platelets have normal hemostatic function. None of the affected individuals developed additional hematopoietic problems, such as myelodysplasia and leukemia, which have been described as occurring in another familial thrombocytopenia syndrome.11 Autologous platelet survival studies done elsewhere in 3 individuals showed moderately decreased survival—likely insufficient to account for the degree of thrombocytopenia.
Thrombocytopenia associated with increased white blood cells and polymorphonuclear granulocytes
We analyzed peripheral blood to determine the platelet count, mean platelet volume (MPV), hematocrit, mean corpuscular volume (MCV), total white blood cells, and complete differential using an automated blood analyzer (Table 1). Most notably, the affected individuals were thrombocytopenic, with a mean platelet count of 42.7 × 109/L (range, 18 × 109/L to 106 × 109/L) compared with a mean of 241.8 × 109/L for unaffected family members (range, 162 × 109/L to 342 × 109/L). Morphologically, the platelets of affected individuals appeared normal without clumping or variation in size (Figure 2). Moreover, the MPV was equivalent between the affected and unaffected family members, distinguishing this form of inherited thrombocytopenia from others that are associated with either large platelets (ie, macrothrombocytes, observed in Alport syndrome, May-Hegglin anomaly, and grey platelet syndrome) or small platelets (ie, microthrombocytes, seen in Wiskott-Aldrich syndrome). The total white blood cell count was significantly higher in individuals with thrombocytopenia than without thrombocytopenia (10.8 × 109/L vs 6.2 × 109/L, respectively, P = .002). This was primarily due to an increased number of neutrophils for thrombocytopenic individuals. The lymphocyte and mononuclear cell numbers were not statistically different for these populations. Average hematocrits were similar in the thrombocytopenic and unaffected groups (47.5% vs 47.1%, respectively), but the MCV was significantly lower in affected family members than in unaffected individuals (P = .02). Because the percentage of women was higher among thrombocytopenic (11 of 14; 79%) than among unaffected (6 of 16; 38%) individuals, we stratified our data by sex. Although the number of observations is relatively small, a lower MCV was observed in affected males and females than in unaffected males and females (data not shown). Mean corpuscular hemoglobin concentration and red cell distribution width showed no statistically significant differences between affected and unaffected family members.
Peripheral blood analysis
| Mean values ± standard deviation . | |||
|---|---|---|---|
| Measured parameter . | Affected (n = 13) . | Unaffected (n = 17) . | P value . |
| Platelets (×109/L) | 42.7 ± 24.2 | 241.8 ± 62.1 | <.001 |
| MPV (fL) | 9.7 ± 1.4* | 9.6 ± 1.2 | .85 |
| White blood count (×109/L) | 10.8 ± 4.3 | 6.2 ± 1.6 | .002 |
| Neutrophils | 7.9 ± 4.0* | 3.5 ± 1.4 | .003 |
| Lymphocytes | 2.3 ± 0.4* | 2.1 ± 0.9 | .32 |
| Monocytes | 0.8 ± 0.8* | 0.6 ± 0.2 | .37 |
| Hematocrit (%) | 47.5 ± 3.8 | 47.1 ± 4.5 | .76 |
| MCV (fL) | 88.3 ± 5.0 | 92.9 ± 5.0 | .02 |
| Mean corpuscular hemoglobin concentration | 32.3 ± 0.9 | 32.3 ± 1.2 | .97 |
| Red cell distribution width | 12.7 ± 0.9† | 13.9 ± 2.3 | .082 |
| Mean values ± standard deviation . | |||
|---|---|---|---|
| Measured parameter . | Affected (n = 13) . | Unaffected (n = 17) . | P value . |
| Platelets (×109/L) | 42.7 ± 24.2 | 241.8 ± 62.1 | <.001 |
| MPV (fL) | 9.7 ± 1.4* | 9.6 ± 1.2 | .85 |
| White blood count (×109/L) | 10.8 ± 4.3 | 6.2 ± 1.6 | .002 |
| Neutrophils | 7.9 ± 4.0* | 3.5 ± 1.4 | .003 |
| Lymphocytes | 2.3 ± 0.4* | 2.1 ± 0.9 | .32 |
| Monocytes | 0.8 ± 0.8* | 0.6 ± 0.2 | .37 |
| Hematocrit (%) | 47.5 ± 3.8 | 47.1 ± 4.5 | .76 |
| MCV (fL) | 88.3 ± 5.0 | 92.9 ± 5.0 | .02 |
| Mean corpuscular hemoglobin concentration | 32.3 ± 0.9 | 32.3 ± 1.2 | .97 |
| Red cell distribution width | 12.7 ± 0.9† | 13.9 ± 2.3 | .082 |
P value for 2-tailed Student t test assuming unequal variances.
n = 12.
n = 11.
Platelets from thrombocytopenic individuals.
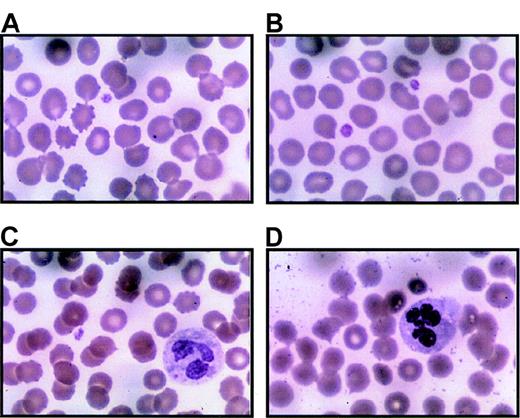
Peripheral blood smears were prepared immediately upon receipt (within 24 hours of collection) with Wright-Giemsa stain. Representative regions are shown for 3 separate thrombocytopenic individuals (A and B are from IV-10; C from V-14; D from III-10). Panel A includes 1 normal-sized platelet and a red blood cell containing a Howell-Jolly body due to prior splenectomy. Panel B is unusual because 2 platelets can be seen in 1 field. Panel C demonstrates a Döhle body in the cytoplasm of a neutrophil. Panel D includes hypersegmented neutrophils. Both Döhle bodies and hypersegmented neutrophils appear to be common in thrombocytopenic individuals. Magnification, 1000 ×.
Platelets from thrombocytopenic individuals.
Peripheral blood smears were prepared immediately upon receipt (within 24 hours of collection) with Wright-Giemsa stain. Representative regions are shown for 3 separate thrombocytopenic individuals (A and B are from IV-10; C from V-14; D from III-10). Panel A includes 1 normal-sized platelet and a red blood cell containing a Howell-Jolly body due to prior splenectomy. Panel B is unusual because 2 platelets can be seen in 1 field. Panel C demonstrates a Döhle body in the cytoplasm of a neutrophil. Panel D includes hypersegmented neutrophils. Both Döhle bodies and hypersegmented neutrophils appear to be common in thrombocytopenic individuals. Magnification, 1000 ×.
Similar platelet-specific proteins found on the surface platelets from thrombocytopenic and nonthrombocytopenic individuals
Flow cytometry of affected versus unaffected family members showed no major differences in expression of GpIIb/IIIa, GpIV, and GpIa/IIa (data not shown). These assays were difficult to compare because they were run as samples became available, and the number of platelets in thrombocytopenic samples was low. Nonetheless, these data demonstrate that there are no major differences in cell surface expression for several well-characterized platelet proteins.
Examination of the bone marrow
We examined bone marrow from 2 affected patients and 1 unaffected family member. Megakaryocytes were evident in all 3 samples, although the number of mature, multilobulated megakaryocytes was reduced in the 2 thrombocytopenic individuals and the size of the average megakaryocyte was decreased (Figure3). Erythroid and myeloid maturation appeared to be unaffected.
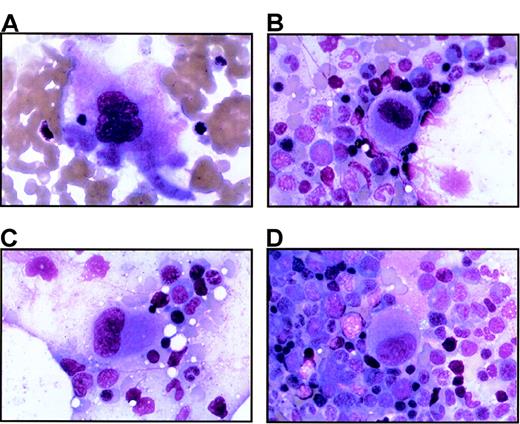
Bone marrow from thrombocytopenic individuals contains small megakaryocytes with decreased nuclear complexity.
Photomicrographs of bone marrow aspirates are shown after Wright-Giemsa staining (magnification, 750 ×). (A) A representative megakaryocyte from an unaffected individual (IV-8). (B-D) The characteristic morphology of megakaryocytes seen in 2 thrombocytopenic individuals (IV-10 and V-14).
Bone marrow from thrombocytopenic individuals contains small megakaryocytes with decreased nuclear complexity.
Photomicrographs of bone marrow aspirates are shown after Wright-Giemsa staining (magnification, 750 ×). (A) A representative megakaryocyte from an unaffected individual (IV-8). (B-D) The characteristic morphology of megakaryocytes seen in 2 thrombocytopenic individuals (IV-10 and V-14).
Intact tyrosine phosphorylation of STAT3 in platelets from thrombocytopenic individuals
Fifty milliliters of blood was obtained from 1 unaffected and 2 affected individuals, and platelets were isolated for signaling studies. Platelets were divided into 2 aliquots, and half were stimulated with TPO (10 ng/mL) for 10 minutes before lysis. Whole-cell extracts from each patient were then analyzed by Western blot and probed with an antibody specific for tyrosine-phosphorylated STAT3 (Figure 4). We found that both the thrombocytopenic and unaffected platelets were capable of mediating TPO-induced STAT phosphorylation. This demonstrates that early signaling events in response to TPO stimulation are similar in affected and unaffected platelets. This finding is in marked contrast to patients with the TAR syndrome (thrombocytopenia with absent radii syndrome), in which a failure of TPO-induced signaling has been demonstrated.12
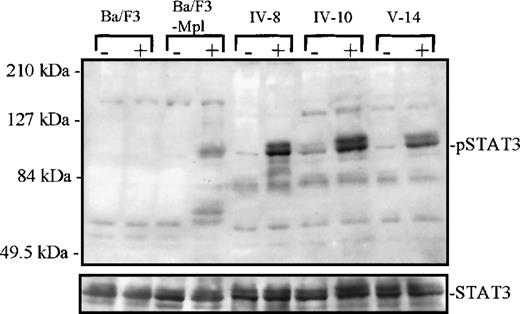
Tyrosine phosphorylation of STAT3.
Platelets from 1 unaffected (IV-8) and 2 thrombocytopenic family members (IV-10 and V-14) were either unstimulated (−) or stimulated (+) with recombinant human TPO for 10 minutes, 10 ng/mL. Cellular extracts were analyzed by Western blot (7.5% acrylamide gel) and probed with a phospho-STAT3–specific antibody. Below, the blot was stripped and reprobed with a STAT3 antibody to confirm equal protein in each lane. At left, extracts from parental Ba/F3 and Ba/F3-mMpl cells (a cell line engineered to express the murine Mpl receptor) were used as negative and positive control, respectively, for TPO-dependent STAT3 phosphorylation.
Tyrosine phosphorylation of STAT3.
Platelets from 1 unaffected (IV-8) and 2 thrombocytopenic family members (IV-10 and V-14) were either unstimulated (−) or stimulated (+) with recombinant human TPO for 10 minutes, 10 ng/mL. Cellular extracts were analyzed by Western blot (7.5% acrylamide gel) and probed with a phospho-STAT3–specific antibody. Below, the blot was stripped and reprobed with a STAT3 antibody to confirm equal protein in each lane. At left, extracts from parental Ba/F3 and Ba/F3-mMpl cells (a cell line engineered to express the murine Mpl receptor) were used as negative and positive control, respectively, for TPO-dependent STAT3 phosphorylation.
TPO levels are elevated in affected versus unaffected individuals
Thrombocytopenic individuals had an average TPO value of 94.1 ± 63.9 pg/mL (range, 26.6-224.7 pg/mL) compared with 48.7 ± 42.4 pg/mL (range, 8.1-155 pg/mL) for unaffected family members (P = .04, 2-tailed Student t test). TPO levels in plasma are primarily regulated by platelets and megakaryocytes, which internalize and degrade TPO bound to Mpl receptors.13,14Therefore, the minimal elevation in TPO levels, despite marked reduction in circulating platelets, suggests that a compensatory increase in TPO uptake and degradation is occurring in the marrow.
Megakaryocytes lack full polyploidization
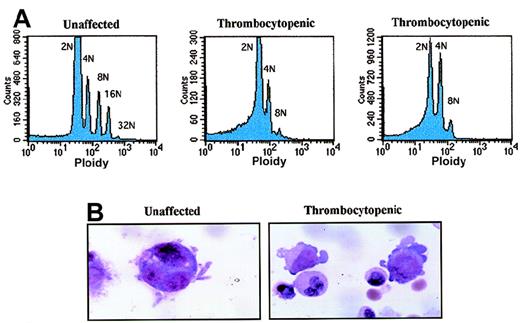
Low-density cells from bone marrow were cultured for 11 days under serum-free conditions in the presence of recombinant human TPO (10 ng/mL). Then, 2 × 106 to 3 × 106 cells were double-stained with FITC-conjugated CD41 antibody and propidium iodide. Cells were analyzed by flow cytometry to determine ploidy classes for the CD41+ cells (Figure 5A). We found that megakaryocytes from an unaffected family member contained megakaryocytes with ploidy classes of 2N, 4N, 8N, 16N, and 32N. In contrast, the 2 affected thrombocytopenic individuals had predominantly 2N and 4N megakaryocytes with a few 8N cells but no higher ploidy classes. The CD41 staining was similar in all 3 samples, and the highest 10% to 15% of all cells were analyzed. Representative megakaryocytes from an unaffected and a thrombocytopenic family member are shown in Figure 5B (grown in vitro with 10-ng/mL TPO for 7 days).
Abnormal polyploidy formation in thrombocytopenic individuals.
(A) Megakaryocytes from 1 unaffected family member (IV-8) and 2 thrombocytopenic individuals (IV-10 and V-14) were tested for the ability to become polyploid after 10 to 11 days in culture. The cells with highest CD41a expression were selected and plotted as cell number (y-axis; linear scale) versus propidium iodide content (x-axis; logarithmic scale). (B) Photomicrographs demonstrate representative megakaryocytes derived by liquid culture in 10-ng/mL TPO for 7 days (magnification, 1500 ×). At left is a single megakaryocyte from an unaffected individual (IV-8); right, 2 immature megakaryocytes with cytoplasmic blebs around the periphery (IV-10).
Abnormal polyploidy formation in thrombocytopenic individuals.
(A) Megakaryocytes from 1 unaffected family member (IV-8) and 2 thrombocytopenic individuals (IV-10 and V-14) were tested for the ability to become polyploid after 10 to 11 days in culture. The cells with highest CD41a expression were selected and plotted as cell number (y-axis; linear scale) versus propidium iodide content (x-axis; logarithmic scale). (B) Photomicrographs demonstrate representative megakaryocytes derived by liquid culture in 10-ng/mL TPO for 7 days (magnification, 1500 ×). At left is a single megakaryocyte from an unaffected individual (IV-8); right, 2 immature megakaryocytes with cytoplasmic blebs around the periphery (IV-10).
CFU-Mk increased in affected individuals
Colony assays were performed to determine the number of erythroid, myeloid, and megakaryocytic progenitors in this form of hereditary thrombocytopenia. An equal number of low-density bone marrow cells or peripheral blood cells were plated in semisolid methylcellulose medium in the presence of multiple growth factors as described in the “Materials and methods.” After 12 days, BFU-E, CFU-GM, and CFU-GEMM were counted. As shown in Table2 and Table3, thrombocytopenic patients had moderately increased BFU-E and normal CFU-GM and CFU-GEMM in the bone marrow. In contrast, peripheral blood from an affected individual had markedly higher BFU-E, CFU-GM, and CFU-GEMM, indicating a greater number of progenitors in circulation. Qualitatively, the erythroid colonies from thrombocytopenic patients were less strongly hemaglobinized and had less discrete borders than those from the unaffected individual. Also, there were significantly more scattered cells and diffuse colonies of unclear lineage in the cultures from thrombocytopenic patients.
Bone marrow colonies
| . | Marrow colonies/104low-density cells . | Colonies/0.5 × 105 low-density cells CFU-Mk . | ||
|---|---|---|---|---|
| BFU-E . | CFU-GM . | CFU-GEMM . | ||
| Unaffected (IV-8)* | 39.5 + 4.7 | 18.8 + 5.0 | 13.1 + 6.3 | 58 |
| Affected (IV-10)* | 75.7 + 11.2 | 27.1 + 7.6 | 22.9 + 9.1 | 155† |
| Affected (V-14)* | 64.5 + 15.6 | 17.3 + 3.9 | 16.0 + 5.2 | 123† |
| . | Marrow colonies/104low-density cells . | Colonies/0.5 × 105 low-density cells CFU-Mk . | ||
|---|---|---|---|---|
| BFU-E . | CFU-GM . | CFU-GEMM . | ||
| Unaffected (IV-8)* | 39.5 + 4.7 | 18.8 + 5.0 | 13.1 + 6.3 | 58 |
| Affected (IV-10)* | 75.7 + 11.2 | 27.1 + 7.6 | 22.9 + 9.1 | 155† |
| Affected (V-14)* | 64.5 + 15.6 | 17.3 + 3.9 | 16.0 + 5.2 | 123† |
Mean ± standard deviation of 5 plates for BFU-E, CFU-GM, and CFU-GEMM. CFU-Mk data is the average of 2 separate slide chambers.
Numbers in parentheses indicate patient identifier (Figure 1).
Numbers for affected individuals are likely to be underestimates because confluent colonies were difficult to distinguish.
Peripheral blood colonies
| . | Peripheral blood colonies/ 105 low-density cells . | Colonies/104 CD34+ cells CFU-Mk . | ||
|---|---|---|---|---|
| BFU-E . | CFU-GM . | CFU-GEMM . | ||
| Normal control | 22.3 ± 2.1 | 5.7 ± 2.0 | 0.5 ± 0.4 | 1.9 ± 2.4 |
| Unaffected (IV-8)3-150 | 68.1 ± 2.2 | 7.9 ± 2.4 | 1.7 ± 0.4 | 3.8 ± 2.6 |
| Affected (V-14)3-150 | 108.0 ± 18.0 | 18.5 ± 0 | 10.5 ± 2.8 | 74.5 ± 26.3 |
| . | Peripheral blood colonies/ 105 low-density cells . | Colonies/104 CD34+ cells CFU-Mk . | ||
|---|---|---|---|---|
| BFU-E . | CFU-GM . | CFU-GEMM . | ||
| Normal control | 22.3 ± 2.1 | 5.7 ± 2.0 | 0.5 ± 0.4 | 1.9 ± 2.4 |
| Unaffected (IV-8)3-150 | 68.1 ± 2.2 | 7.9 ± 2.4 | 1.7 ± 0.4 | 3.8 ± 2.6 |
| Affected (V-14)3-150 | 108.0 ± 18.0 | 18.5 ± 0 | 10.5 ± 2.8 | 74.5 ± 26.3 |
Mean ± standard deviation of 3 plates for BFU-E, CFU-GM, and CFU-GEMM and 4 chamber slides for CFU-Mk.
Numbers in parentheses indicate patient identifier (Figure 1).
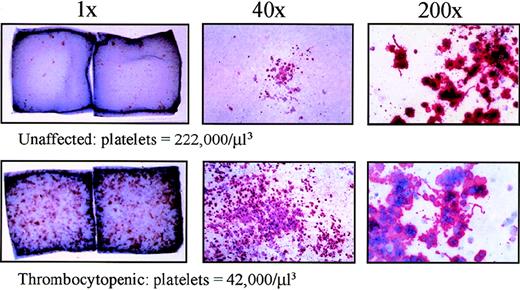
Ficoll-separated bone marrow progenitors as well as Ficoll-separated and CD34-selected peripheral blood cells were plated in a collagen-based CFU-Mk assay with recombinant TPO, interleukin-3, and interleukin-6. After 14 days in culture, the megakaryocyte colonies were dehydrated, fixed, and immunostained with a GpIIb/IIIa antibody conjugated to alkaline phosphatase. Upon incubation with an appropriate substrate (naphthol), the megakaryocyte colonies were stained red or pink, and the nuclei were counterstained with Evan's blue dye. The thrombocytopenic individuals had greatly increased numbers of CFU-Mk to the extent that they were confluent in some areas (Table 2, Figure 6). Using CD34-selected peripheral blood, very few CFU-Mk were detected from 1 unaffected family member and 1 normal donor. However, 1 thrombocytopenic individual had 20-fold increased level of CFU-Mk in the peripheral blood (approximately 1 per 133 CD34+ cells; Table 3). Furthermore, the megakaryocyte colonies from affected family members were much larger (most above 21 cells) than those from an unaffected individual and normal control. These data demonstrate a tremendous increase in CFU-Mk from individuals with autosomal dominant thrombocytopenia.
Increased CFU-Mk in thrombocytopenic individuals.
Megakaryocyte colonies were grown on collagen matrix as described in “Materials and methods.” Cells were fixed and stained for GpIIb/IIIa expression. On the left, double-chamber slides are shown (1 ×, unmagnified) for 1 unaffected (IV-8) and 1 thrombocytopenic individual (IV-10). Dark spots (pink or red) represent individual megakaryocyte colonies; only the larger colonies are visible without magnification. In the center panels, representative megakaryocyte colonies are shown under low-power magnification (40 ×). On the right, higher power views of megakaryocytes (200 ×) demonstrate the presence of proplatelet processes and platelet fragmentation in unaffected and thrombocytopenic samples. Similar data were obtained for a second thrombocytopenic individual (V-14).
Increased CFU-Mk in thrombocytopenic individuals.
Megakaryocyte colonies were grown on collagen matrix as described in “Materials and methods.” Cells were fixed and stained for GpIIb/IIIa expression. On the left, double-chamber slides are shown (1 ×, unmagnified) for 1 unaffected (IV-8) and 1 thrombocytopenic individual (IV-10). Dark spots (pink or red) represent individual megakaryocyte colonies; only the larger colonies are visible without magnification. In the center panels, representative megakaryocyte colonies are shown under low-power magnification (40 ×). On the right, higher power views of megakaryocytes (200 ×) demonstrate the presence of proplatelet processes and platelet fragmentation in unaffected and thrombocytopenic samples. Similar data were obtained for a second thrombocytopenic individual (V-14).
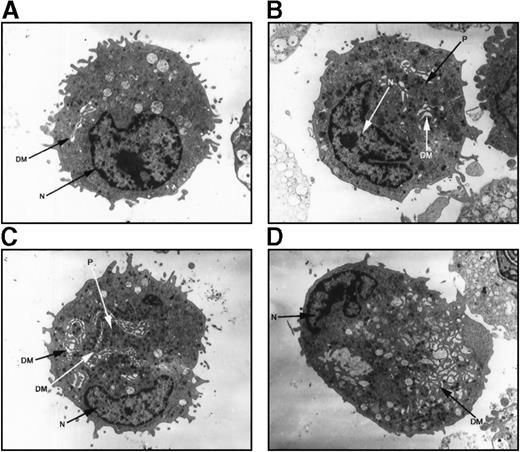
Electron microscopy evaluation of megakaryocytes in hereditary thrombocytopenia
Megakaryocytes from a family member with inherited thrombocytopenia were unusually small, contained relatively hypolobulated nuclei, and had a paucity of demarcation membranes and platelet fields (Figure7). The photomicrographs in Figure 7support a model in which the affected megakaryocytes are capable of many of the elements of terminal differentiation and platelet production but are unable to generate the normal nuclear or cytoplasmic mass found in mature megakaryocytes.
Ultrastructure of megakaryocytes from a thrombocytopenic family member.
Electron micrographs of 4 characteristic megakaryocytes from 1 affected individual (IV-10) are shown, arranged from least to most differentiated. (A) Very early megakaryocyte with a minimal demarcation membrane (DM) system; the diameter is 11 μm. (B and C) Most common megakaryocytic cells include a more obvious demarcation membrane system and occasional developing platelets (P); the diameters are 12 and 15 μm, respectively. (D) Most fully differentiated megakaryocyte observed with increased demarcation membranes; the maximum diameter is 14 μm. Nuclei (N) are indicated. Magnification, 2900 × for panels A, B, and D; 2300 × for panel C.
Ultrastructure of megakaryocytes from a thrombocytopenic family member.
Electron micrographs of 4 characteristic megakaryocytes from 1 affected individual (IV-10) are shown, arranged from least to most differentiated. (A) Very early megakaryocyte with a minimal demarcation membrane (DM) system; the diameter is 11 μm. (B and C) Most common megakaryocytic cells include a more obvious demarcation membrane system and occasional developing platelets (P); the diameters are 12 and 15 μm, respectively. (D) Most fully differentiated megakaryocyte observed with increased demarcation membranes; the maximum diameter is 14 μm. Nuclei (N) are indicated. Magnification, 2900 × for panels A, B, and D; 2300 × for panel C.
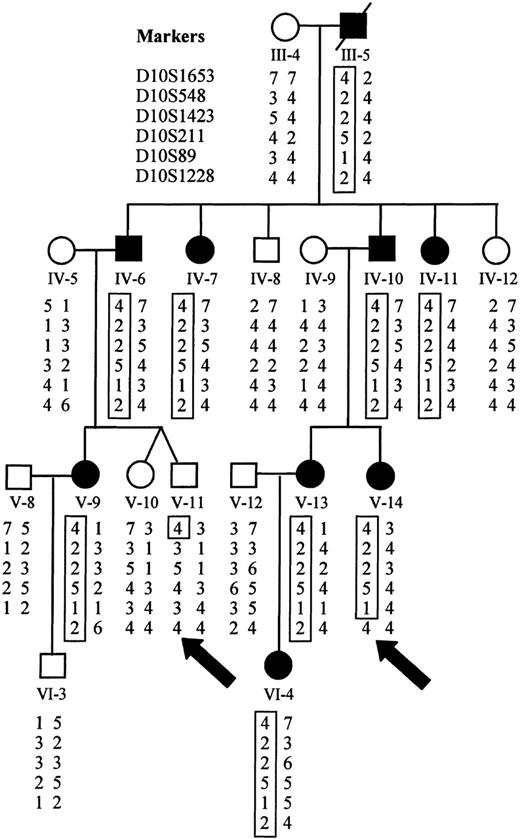
Genomic linkage analysis
Initially, 16 family members were analyzed using the Weber-9 microsatellite marker set. We calculated a maximum possible 2-point lod score of 3.31, assuming that all meioses were informative for a given marker. Of the 360 autosomal markers that were analyzed, 3 achieved a lod score of 3.31 at a recombination fraction of zero, all located on chromosome 10 between 48.4 cM and 63.3 cM using sex-averaged recombination rates (D10S211, D10S197, D10S1208). Evidence of genetic recombination was observed at 33.2 cM (D10S1430) and 85.6 cM (GATA121A08), establishing a maximal linkage interval of 52 cM. Only 1 marker at a different locus achieved a lod score above 2.0 (lod = 2.7 on chromosome 17; D17S1294). However, each of the flanking markers on chromosome 17 had maximal lod scores of less than 1. By recruiting additional family members and using additional markers for finer mapping of this region of chromosome 10, we were able to establish recombination at 40.4 cM (D10S1653) and 57.4 cM (D10S1228), limiting the area of study to 17 cM (Figure 8). Recalculation of lod scores with additional family members gave a maximum value of 5.68 at a recombination rate of zero for D10S89 (Table4).
Haplotype analysis demonstrates sites of recombination.
Haplotypes for markers on chromosome 10 were derived from genotyping data. The disease haplotype is boxed for each affected individual. Recombination is evident in affected individual V-14 at D10S1228 and in unaffected individual V-11 at D10S1653 (marked by arrows).
Haplotype analysis demonstrates sites of recombination.
Haplotypes for markers on chromosome 10 were derived from genotyping data. The disease haplotype is boxed for each affected individual. Recombination is evident in affected individual V-14 at D10S1228 and in unaffected individual V-11 at D10S1653 (marked by arrows).
Genomic linkage analysis
| Marker name . | Distance (cM) . | Maximum lod score . |
|---|---|---|
| D10S1653 | 40.4 | 1.07 (θ = 0.2) |
| D10S548 | 45.7 | 4.36 (θ = 0) |
| D10S1423 | 46.2 | 4.26 (θ = 0) |
| D10S211 | 48.4 | 4.06 (θ = 0) |
| D10S89 | 52.1 | 5.68 (θ = 0) |
| D10S1228 | 57.4 | 1.19 (θ = 0.1) |
| Marker name . | Distance (cM) . | Maximum lod score . |
|---|---|---|
| D10S1653 | 40.4 | 1.07 (θ = 0.2) |
| D10S548 | 45.7 | 4.36 (θ = 0) |
| D10S1423 | 46.2 | 4.26 (θ = 0) |
| D10S211 | 48.4 | 4.06 (θ = 0) |
| D10S89 | 52.1 | 5.68 (θ = 0) |
| D10S1228 | 57.4 | 1.19 (θ = 0.1) |
θ indicates recombination fraction.
Discussion
In this paper, we describe a single extended family with autosomal dominant thrombocytopenia that results in a phenotype of incomplete maturation of megakaryocytes. This syndrome is characterized by mild to moderate bruising throughout life, normal erythroid and myeloid cells, moderately elevated plasma TPO levels, expanded progenitors across all hematopoietic lineages, and dramatic expansion of immature megakaryocytes (CFU-Mk). Through a genomic scan with follow-up linkage analysis, we have established a locus of linkage encompassing a maximal region of below 20 cM on human chromosome 10. We propose that a mutation of 1 allele in this region inhibits terminal differentiation of megakaryocytes, either through a loss of function or dominant-negative mechanism of action.
Linkage to chromosome 10 excludes several proposed candidate genes, such as tpo (chromosome 3q26.3-27), mpl (chromosome 1p34), NF-E2 (chromosome 12q13), CBFA2 (formerlyAML1, chromosome 21q22), and GATA-1 (chromosome Xp11.23), each of which have been demonstrated to play a critical role in megakaryocyte development or platelet production.15-20Furthermore, the early Mpl signaling apparatus is intact as evidenced by phosphorylation of STAT3 in platelets from affected family members as well as the expansion of bone marrow cells and megakaryocytes in culture conditions that contain only TPO as an exogenous growth factor. These findings clearly distinguish the current phenotype from conditions such as TAR or congenital amegakaryocytic thrombocytopenia, in which response to TPO is absent or severely diminished.12 21-23
TPO plasma levels are primarily regulated by internalization and degradation of the growth factor after binding Mpl on platelets and megakaryocytes.13,24,25 Conditions in which platelets and megakaryocytes are both decreased (ie, chemotherapy, amegakaryocytic thrombocytopenia, aplastic anemia) result in very high plasma TPO levels.13,14,26 However, immune-mediated thrombocytopenia purpura (ITP) results in slightly elevated or normal TPO levels because of a compensatory increase in megakaryocyte mass in the bone marrow.13,14,26 27 Our results are similar to those seen in ITP. Although the increase in TPO levels is measurable (94 vs 54 pg/mL;P = .04), it is much lower than would be expected given the level of thrombocytopenia. The dramatic increase in megakaryocyte progenitors explains why these values were not higher.
The mechanism by which mutation of a single allele causes autosomal dominant thrombocytopenia in this family is not known. As illustrated by the recent description of autosomal dominant thrombocytopenia with propensity toward leukemia, deletion of a single allele ofCBFA2 results in hematopoietic defects through loss of function.19 It is also possible that the mutation underlying inherited thrombocytopenia in the present study operates through a dominant-negative mechanism. If this were the case, the abnormal protein may be deficient in dimerization or multimerization that occurs during normal function. The region of genetic linkage, on the short arm of chromosome 10, includes a number of potential candidate genes that may play an important role in megakaryocyte maturation but none that are known to be expressed predominantly in megakaryocytes or their precursors. We are currently investigating several known genes, including AF10, which encodes a zinc-finger protein that is involved in chromosomal rearrangements in some leukemias28,29; supervillin, an actin binding molecule with 3 potential actin binding sites30-32; and BMI-1,a protooncogene that has been implicated in human malignancy.33
It is of particular interest that the phenotype of autosomal dominant thrombocytopenia in this family demonstrates increased proliferation and decreased terminal maturation, similar to recent reports of cord blood and fetal megakaryocytes.34,35 Around the time of birth, megakaryocyte development changes to favor terminal differentiation, but the mechanism of this evolution is unknown. It is possible that the family we are studying expresses a perpetual fetal type of megakaryocytopoiesis. If correct, this may help explain the molecular basis for delayed platelet production in recipients of cord blood stem cell transplants36,37 and may lead to new strategies for improving platelet recovery in this setting. In the final stages of preparing this manuscript, a similar unrelated family from Italy was described with linkage of autosomal dominant thrombocytopenia to the same locus of human chromosome 10.38 39 It is likely that as other unrelated families with the same condition are discovered, the abnormal gene will be identified soon and insight will be gained into the molecular basis of megakaryocyte maturation.
Acknowledgments
We wish to acknowledge Nancy Lin for assistance with the hematopoietic colony assays, Jennifer Luthi for CD34+enrichment of peripheral blood cells, Gayle Teramura for flow cytometry of platelets, Don Foster for purified human TPO, David Frank for phospho-STAT3 antibody, David Thorning for electron microscopy, and Sarah Serafimidis for administrative assistance and editing. Most importantly, we wish to thank the family members who participated in this study. Without their enthusiastic support and assistance, this work would not have been possible.
Supported by the Doris Duke Charitable Foundation, Clinical Scientist Award (J.G.D.).
G.P.J. is an American Heart Association clinician-scientist awardee.
Reprints:Jonathan Drachman, Puget Sound Blood Center, 921 Terry Ave, Seattle, WA 98104; e-mail: drachman@u.washington.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal