Abstract
Because immunoglobulin (Ig)-β (CD79b) is required for immunoglobulin allelic exclusion, we examined the CD79b expressed by four chronic lymphocytic leukemia (CLL) samples that expressed more than one immunoglobulin heavy-chain allele and five samples that had normal immunoglobulin heavy-chain allelic exclusion. All leukemia cell samples stained poorly with monoclonal antibodies specific for extracellular epitopes of CD79b. However, all samples expressed functional CD79b genes, regardless of whether they did or did not express more than one immunoglobulin heavy-chain allele. We identified variant CD79b genes that had conservative base substitutions restricted to regions encoding the extracellular immunoglobulin-like domain of CD79b. However, these variants were not restricted to samples lacking immunoglobulin heavy-chain allelic exclusion and most likely reflect genetic polymorphism. Collectively, these data indicate that the unusual expression of more than one immunoglobulin heavy allele by CLL B cells is not associated with structural, nonconservative mutations in the signal-transduction domains of CD79b.
Immunoglobulin (Ig)-β (CD79b), together with Ig-α (CD79a), are components of the B-cell receptor complex that are critical for signal transduction following surface immunoglobulin cross-linking.1 It was reported that the chronic lymphocytic leukemia (CLL) B cells of some patients have somatic mutations in the cytoplasmic domain of CD79b that impair its ability to engage in effective signal transduction.2 Because CD79b also is necessary for immunoglobulin heavy-chain allelic exclusion during normal B-cell development,3,4 we examined whether mutations in CD79b account for the lack of immunoglobulin heavy-chain allelic exclusion that is noted for the leukemia B cells of approximately 6% of the patients with CLL.5 To test this hypothesis, we analyzed the CD79b molecule of four CLL samples that expressed more than 1 immunoglobulin heavy-chain allele and 5 CLL samples that had normal immunoglobulin heavy-chain allelic exclusion.
Study design
Patient samples
Flow cytometry
Direct immunofluorescence analyses and calculation of mean fluorescence intensity ratio were performed as described.5,6 CD79a expression was detected using an R-Phycoerythrin (R-PE)-conjugated mouse IgG1 anti-human monoclonal antibody (mAb) produced by clone HM47 (PharMingen, San Diego, CA) as described.7 CD79b was detected using a fluorescein isothiocyanate (FITC)-labeled mAb of clone SN88(DAKO, Glostrup, Denmark) and an R-PE–conjugated mouse IgG1 anti-human mAb derived from clone CB3-19(PharMingen). FITC-conjugated mAbs against CD19, IgM, IgD, and κ or λ immunoglobulin light chains, along with respective isotype control mouse IgG, were purchased from Becton Dickinson (San Jose, CA).10
Molecular analyses
Results and discussion
Characterization of the expressed immunoglobulin
We used the anchored reverse transcription-polymerase chain reaction (RT-PCR) ELISA technique to identify the Ig VH gene subgroups expressed by each of the CLL samples. CLL samples 6, 8, and 9 had leukemic B cells that expressed immunoglobulin heavy chains of the VH3 and VH4 subgroups, whereas sample 7 had B cells that expressed immunoglobulin heavy chains of the VH2 and VH3 subgroups. The other five CLL samples (1-5) expressed only one immunoglobulin heavy-chain allele.
Expression of CD79a and CD79b polypeptides
CD79a was detected in all 9 CLL cases using the HM47 mAb. CLL samples that expressed only 1 immunoglobulin heavy-chain allele had a mean fluorescence intensity ratio (MFIR) for CD79a ranging from 3.1 (for sample 4) to 16.3 (for sample 2) (mean = 8.7 ± 4.9, SD). Similarly, the CLL samples that lacked immunoglobulin heavy-chain allelic exclusion had MFIR values ranging from 8 (for sample 6) to 20.3 (for sample 8) when stained for CD79a (mean = 11.1 ± 6.0, SD). The MFIR values for cells stained with HM47 (MFIRHM47) between CLL samples that did or did not manifest immunoglobulin heavy-chain allelic exclusion were not significantly different (P > .5, Student paired t test).
We examined the relative expression of CD79b using SN8,8 an mAb that reacts with an extracellular epitope of CD79b.13We noted that B cells from human tonsils (n = 6) had MFIRSN8 values ranging from 8.3 to 25.5 (mean = 14.8 ± 7.2, SD). Similarly, the MFIRSN8values of blood B cells from eight healthy donors ranged from 12.3 to 27.6 (mean = 19.9 ± 5.0, SD). In contrast, the average MFIRSN8 of all CLL samples examined (mean = 3.8 ± 2.0, SD, n = 9) was significantly lower than that of tonsillar or blood B cells of healthy donors (P = .001, Student paired t test). MFIRSN8 values of the CLL samples with immunoglobulin heavy-chain allelic exclusion ranged from 2.6 (for sample 5) to 7.8 (for sample 2) (mean = 4.9 ± 2.5, SD). Similarly, the MFIRSN8 of samples expressing more than 1 immunoglobulin heavy-chain allele ranged from 1.5 (for sample 6) to 5.6 (for sample 8) (mean = 2.9 ± 1.9, SD). There was no significant difference between these groups (P > .2, Student pairedt test). Similar results were obtained using another CD79b-specific mAb, CB3-1.9
Nucleic acid sequence analyses of the expressed CD79b genes
Base substitutions were detected in all samples regardless of whether the leukemic cells expressed more than one immunoglobulin heavy-chain allele. However, in contrast to the mutations noted in the study by Thompson et al,2 the substitutions that we detected were only within the region encoding extracellular immunoglobulin-like domain of CD79b, or its leader sequence (Figure1 and Table1). We conclude that such mutations cannot account for the lack of immunoglobulin heavy-chain allelic exclusion that is noted for some CLL B-cell populations.
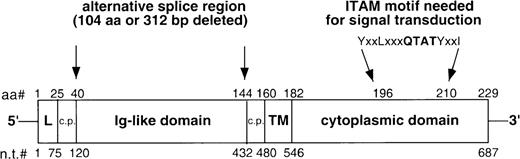
Schematic of the CD79b molecule.
Top numbers refer to the amino acid positions (aa#), bottom numbers refer to the nucleotide positions (n.t.#). L indicates the leader peptide, TM indicates the transmembrane domain, c.p. connotes the connecting peptide, and ITAM refers to the immunoreceptor-tyrosine-based activation motif.
Schematic of the CD79b molecule.
Top numbers refer to the amino acid positions (aa#), bottom numbers refer to the nucleotide positions (n.t.#). L indicates the leader peptide, TM indicates the transmembrane domain, c.p. connotes the connecting peptide, and ITAM refers to the immunoreceptor-tyrosine-based activation motif.
Summary of nucleotide changes identified in the CLL samples*
| AA Position . | Substitution . | AA Change . | CD79b . | CLL Samples† . |
|---|---|---|---|---|
| 16 | GCT → GCG | ala → ala | Leader | 1, 2, 3, 4, 5, 6, 7, 8, 9 |
| 23 | GCA insertion | ala insertion | Leader | 2, 3, 4, 5, 6, 7, 9 |
| 34 | CGG → CAG | arg → gln | Connecting peptide | 4 |
| 54 | GCC → CCC | ala → pro | Ig domain | 8 |
| 58 | GCC → GGC | ala → gly | Ig domain | 1, 2, 3, 4, 5, 6, 7, 8 |
| 73 | AAT → TAT | asn → tyr | Ig domain | 8 |
| 75 | AGC → ACC | ser → thr | Ig domain | 8 |
| 81 | GAG → GAT | glu → asp | Ig domain | 2 |
| 84 | GCG → GAG | ala → glu | Ig domain | 4, 5, 6, 7 |
| 100 | CAG → CAT | gln → his | Ig domain | 3 |
| 102 | GAA → GAT | glu → asp | Ig domain | 3 |
| 122 | TGT → TGC | cys → cys | Ig domain | 1, 2, 7, 9 |
| AA Position . | Substitution . | AA Change . | CD79b . | CLL Samples† . |
|---|---|---|---|---|
| 16 | GCT → GCG | ala → ala | Leader | 1, 2, 3, 4, 5, 6, 7, 8, 9 |
| 23 | GCA insertion | ala insertion | Leader | 2, 3, 4, 5, 6, 7, 9 |
| 34 | CGG → CAG | arg → gln | Connecting peptide | 4 |
| 54 | GCC → CCC | ala → pro | Ig domain | 8 |
| 58 | GCC → GGC | ala → gly | Ig domain | 1, 2, 3, 4, 5, 6, 7, 8 |
| 73 | AAT → TAT | asn → tyr | Ig domain | 8 |
| 75 | AGC → ACC | ser → thr | Ig domain | 8 |
| 81 | GAG → GAT | glu → asp | Ig domain | 2 |
| 84 | GCG → GAG | ala → glu | Ig domain | 4, 5, 6, 7 |
| 100 | CAG → CAT | gln → his | Ig domain | 3 |
| 102 | GAA → GAT | glu → asp | Ig domain | 3 |
| 122 | TGT → TGC | cys → cys | Ig domain | 1, 2, 7, 9 |
The numbering of the amino acid (AA) sequences of the CD79b molecule begins with the ATG methionine start codon. The leader peptide is from AA position 1 to 25, the connecting peptide is from AA 26 to 40, the immunoglobulin-like domain is between AA position 41 to 144, the second connecting peptide is between AA position 145 and 160, the transmembrane domain is between AA position 161 and 182, the cytoplasmic domain is between AA position 183 and 229. Ala = alanine; arg = arginine; asn = asparagine; asp = aspartic acid; CLL = chronic lymphocytic leukemia; cys = cysteine; gln = glutamine; glu = glutamic acid; gly = glycine; his = histidine; Ig = immunoglobulin; pro = proline; ser = serine; thr = threonine; try = tyrosine.
CLL samples 1, 2, 3, 4, and 5 express only one immunoglobulin heavy-chain allele; CLL samples 6, 7, 8, and 9 express both immunoglobulin heavy-chain alleles.
Instead, the cloned CD79b genes had >99% sequence homology to CD79b genes isolated from human B-cell lines.11,14 Four of the 5 samples that expressed only 1 immunoglobulin heavy-chain allele and 3 of the 4 samples that lacked immunoglobulin heavy-chain allelic exclusion had an insertion of an alanine amino acid (AA) at position 23 within the leader peptide sequence (Table 1 and Figure 1). Moreover, all samples had a conservative nucleotide change of GCT → GCG at position 16. The CLL B cells of 8 of the 9 patients had a point mutation (GCC → GGC) at AA position 58 located in the extracellular domain of the CD79b molecule as previously described,14 resulting in an alanine-to-glycine substitution. Another single base change was detected in the immunoglobulin-like domain at AA position 122 (TGT → TGC) in 2 of the 5 samples that expressed only 1 immunoglobulin heavy-chain allele, and in 2 of the 4 samples that lacked immunoglobulin heavy-chain allelic exclusion. A third base substitution was detected in the immunoglobulin-like domain at AA position 84 (GCG → GAG). This substitution resulted in an AA change of alanine to glutamic acid in 2 of the 5 samples that expressed only 1 immunoglobulin heavy-chain allele and in 2 of the 4 samples that lacked immunoglobulin heavy-chain allelic exclusion. Because we observed the same AA substitutions in the CD79b expressed by leukemia cells from different individuals, we reason that these changes reflect genetic polymorphism rather than somatic mutations, as noted by others.15 In any case, the identified changes in the extracellular domain cannot account for the lack of immunoglobulin heavy-chain allelic exclusion.
Expression of the splice variant of CD79b
The 2 CD79b RNA transcripts, corresponding to the full length (709 nucleotides) and the truncated form (397 nucleotides) of the CD79b molecule (ΔCD79b), were detected in all 9 CLL samples tested. Sequence analyses of the spliced variant forms were found to be >99% homologous to the previously published CD79bsplice sequences obtained from human B-cell lines.16 The CLL B cells examined in this study expressed relatively high ratios of ΔCD79b to CD79b messenger RNA, as noted recently by others.13However, the relative expression of ΔCD79b to CD79b did not vary, depending on the presence or absence of allelic exclusion.
Origin of CLL cells that lack immunoglobulin heavy-chain allelic exclusion
CLL cells that express more than 1 immunoglobulin heavy-chain allele also may be derived from a B cell that originally had immunoglobulin receptors with deleterious anti-self reactivity that should have resulted in clonal deletion. Successful rearrangement of a second immunoglobulin heavy-chain could change the specificity of the B-cell receptor even in the absence of silencing the originally expressed allele by generating mixed molecules that no longer have the binding activity conducive for clonal deletion. In a similar manner, approximately 1% of T cells that also have been found to disregard the conventional rule of T-cell receptor allelic exclusion.17-19 In such cases, expression of T-cell receptors containing combinations of two beta-chain alleles apparently changes the overall avidity of the expressed T-cell receptors to allow such cells to escape negative selection in the thymus or to permit selective expansion in the periphery. Similarly, the expression of two immunoglobulin heavy-chain alleles on the B cells may reduce the overall affinity of the expressed immunoglobulin for self-antigen, allowing for escape from clonal deletion.
Recent studies indicate that a subset of patients may have CLL cells that are derived from a postgerminal center memory B-cell compartment.20,21 Such CLL cells have somatic mutations in their expressed immunoglobulin heavy-chain genes. However, some CLL cells that lack immunoglobulin heavy-chain allelic exclusion may express one immunoglobulin heavy-chain allele without somatic mutations along with the other that apparently has undergone somatic mutation.5 Conceivably, such cells may arise from the germinal center compartment.
In any case, the prevalence of patients who have CLL cells that express 2 immunoglobulin heavy-chain alleles appears high. We examined 122 additional CLL samples since our initial report5 and found that 8 (6%) lacked immunoglobulin heavy-chain allelic exclusion (data not shown). As such, CLL cells that express more than 1 immunoglobulin heavy-chain allele may constitute a distinctive subset with a cytogenesis or clinical behavior that is different from that of conventional CLL. However, we conclude from this study that the expression of 2 immunoglobulin heavy-chain alleles does not require distinctive structural mutations in CD79b, indicating that other factors contribute to the cytogenesis of this subgroup of leukemia B cells.
Acknowledgments
We acknowledge the excellent technical assistance of Ester Avery and Todd Johnson.
Supported by grant R37-CA49870 from the National Institutes of Health.
Reprints:Thomas J. Kipps, Division of Hematology/Oncology, Department of Medicine, University of California-San Diego, La Jolla, CA 92093-0663.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal