Abstract
There is strong evidence that infant leukemias with a t(4;11) translocation originate in utero. To test whether other subtypes of childhood leukemias are also initiated during fetal life, we used clone-specific genetic markers for the analysis of neonatal blood spots from 5 children aged 6 months to 4 years 8 months at diagnosis of pro-B, common acute lymphoblastic leukemia (ALL), and T-ALL. In all children, the clonotypic antigen receptor gene rearrangements were already present at birth. The estimated amount of clonotypic cells was in the range of 10 to 100 cells per blood spot. In 2 infants with a t(4;11) positive ALL, we detected similar amounts of the fusion gene sequences compared with the clonal antigen receptor gene rearrangements, suggesting the presence of both markers in the same cells. Our data indicate that the first leukemogenic event of diverse types of childhood ALL may already occur in utero.
Childhood acute lymphoblastic leukemia (ALL) is a biologically heterogeneous disease. B-cell precursor (BCP) ALL is the most common type of childhood ALL, with a peak incidence between 2 to 5 years of age.1 A small biologically different subgroup of these BCP ALL has a t(4;11) translocation and occurs most frequently in infancy.2 ALLs with a T-cell immunophenotype account for only 15% of ALLs and occur most often in older children.3There is good evidence that at least some of these childhood leukemias originate in utero: children who develop B-lineage leukemias within the first 3 years of life have fetal type DJ rearrangements of the immunoglobulin heavy (IgH) chain genes, implying an in utero transforming event.4-6 Furthermore, an identical leukemia-specific translocation and/or an identical clonotypic antigen receptor gene rearrangement have been demonstrated in monozygotic twins with ALL.7-12 The proof for the prenatal origin of pro-B ALL with a t(4;11) translocation comes from the detection of MLL/AF4 gene fusions in the neonatal blood spots from 3 children.13The analysis of leukemia-specific fusion gene sequences is not applicable to determine most other kinds of leukemias, because only a third of all childhood ALLs have detectable chromosomal translocations,14 and methods for their investigation are intricate and therefore difficult to establish. The examination of clone-specific antigen receptor gene rearrangements is widely used for studies of clonality and the detection of minimal residual disease, and can be applied in most childhood ALL to investigate the evolution of preleukemic/leukemic clones.15
We therefore screened neonatal blood spots from 5 children with B- and T-lineage (T) ALL for the presence of clone-specific antigen receptor gene rearrangements.
Study design
Patients
Five patients with ALL were selected by their age at diagnosis, the immunophenotype of the leukemia, the detection of leukemia clone-specific antigen receptor gene rearrangements, the presence of nonconstitutive chromosomal translocations t(4;11), and the availability of their neonatal blood spots (Guthrie cards): 2 infants aged 6 months and 1 year with a t(4;11) positive pro-B ALL, 1 child aged 4 years and 8 months with common ALL, and 2 children aged 2 years and 1 month and 2 years and 2 months with an intermediate T-ALL with a normal karyotype. The study was approved by the local institutional ethics committee and informed consent was obtained from the parents.
Determination of leukemia clone-specific immunoglobulin heavy chain and T-cell receptor γ gene rearrangements, and of the clonotypic MLL-AF4 or AF4-MLL breakpoint
IgH and T-cell receptor γ (TCRG) rearrangements were identified as described previously.16-18 Cloning of chromosomal breakpoints between the human MLL and AF4 gene was performed as described previously.19 Amplified breakpoints were sequenced and data were deposited in public data bases (database accession numbers: AJ000172, AJ235350, and AJ235351). Optimal conditions for amplification of each rearrangement and fusion region using clone-specific oligonucleotides (Table1) were established on DNA from bone marrow mononuclear cells obtained at diagnosis (Figure1).
Sequences of clonotypic antigen receptor gene rearrangements
| Pts . | IgH . | |||
|---|---|---|---|---|
| D . | N . | J . | VDJ . | |
| 1 | gtattacgatattttgactggttattataac | cctcg | aactggttcgacc | VH4-D3-10-J5 |
| 2 | tagtagtaccagctgctat | tgaggggg | ttggtactactacggtat | VH6-D2-2-J6 |
| 3 | ataagttcgatagctgctcgc | gactactgggcc | VH1-(?D)-J4 | |
| TCRG | ||||
| rowV | N | J | VJ | |
| 4 | ccacctgggac | cgggg | tattataagaaactctttg | TCRGV1S3J1S3 |
| 5 | ccacctgggatg | agtcttc | ttataagaaactctttggcag | TCRGV1S4J1S3 |
| Pts . | IgH . | |||
|---|---|---|---|---|
| D . | N . | J . | VDJ . | |
| 1 | gtattacgatattttgactggttattataac | cctcg | aactggttcgacc | VH4-D3-10-J5 |
| 2 | tagtagtaccagctgctat | tgaggggg | ttggtactactacggtat | VH6-D2-2-J6 |
| 3 | ataagttcgatagctgctcgc | gactactgggcc | VH1-(?D)-J4 | |
| TCRG | ||||
| rowV | N | J | VJ | |
| 4 | ccacctgggac | cgggg | tattataagaaactctttg | TCRGV1S3J1S3 |
| 5 | ccacctgggatg | agtcttc | ttataagaaactctttggcag | TCRGV1S4J1S3 |
Pts: patients; patients 1 and 2 had a pro-B ALL; patient 3, a common ALL; patients 4 and 5, a T-lineage ALL. They were screened for the presence of preleukemic/leukemic cells in their neonatal blood spots by the use of ASO-PCR. The sequences are subdivided into V (variable), D (diversity), and J (joining) regions. N are the candidate nucleotides for template-independent insertion, D are the residual nucleotides of the D regions, and J the 5′ end of the J regions. The V(D)J combinations are shown. Underlined sequences indicate the position of the leukemia clone-specific primers. In patient 1, the primer was placed into the VD junction (gccagaggagggtagtattacg).
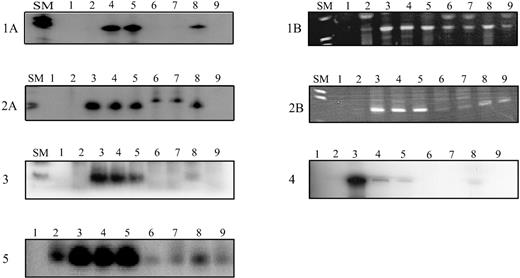
Amplification of antigen receptor gene rearrangements and fusion regions of MLL-AF4/AF4-MLL rearrangements in neonatal blood spots from children with ALL using radioactive nested (TCRG, fusion sequences) or seminested (IgH, pt 3) or a nonradioactive seminested (IgH, pts 1 and 2) ASO-PCR, respectively.
Radioactive PCR products were size fractionated on denaturing polyacrylamide gels and exposed overnight. Amplified IgH products were separated on 3% agarose gels. Patients 1 and 2: (A) Amplification of the leukemia clone-specific IgH rearrangements (size: 75 base pairs (bp) and 74 bp for patients 1 and 2, respectively). (B) Amplification of the leukemia-specific MLL-AF4 (patient 1) and AF4-MLL (patient 2) fusion sequence resulting in a band of 146 bp and 143 bp, respectively. Patients 3, 4, and 5: Amplification of the leukemia clone-specific IgH rearrangement (67 bp in patient 3) and TCRG rearrangement (148 bp and 124 bp for patients 4 and 5, respectively). SM: Size marker VIII (Boehringer Mannheim, Mannheim, Germany); controls: lane 1: no DNA; lane 2: peripheral blood DNA; lanes 3-7: dilutions of leukemic DNA in DNA from peripheral blood mononuclear cells from healthy donors from 10−3 to 10−7; a sensitivity of 10−5 was achieved for all clonotypic rearrangements; lane 8: Guthrie card DNA; lane 9: Guthrie card DNA from age-matched controls.
Amplification of antigen receptor gene rearrangements and fusion regions of MLL-AF4/AF4-MLL rearrangements in neonatal blood spots from children with ALL using radioactive nested (TCRG, fusion sequences) or seminested (IgH, pt 3) or a nonradioactive seminested (IgH, pts 1 and 2) ASO-PCR, respectively.
Radioactive PCR products were size fractionated on denaturing polyacrylamide gels and exposed overnight. Amplified IgH products were separated on 3% agarose gels. Patients 1 and 2: (A) Amplification of the leukemia clone-specific IgH rearrangements (size: 75 base pairs (bp) and 74 bp for patients 1 and 2, respectively). (B) Amplification of the leukemia-specific MLL-AF4 (patient 1) and AF4-MLL (patient 2) fusion sequence resulting in a band of 146 bp and 143 bp, respectively. Patients 3, 4, and 5: Amplification of the leukemia clone-specific IgH rearrangement (67 bp in patient 3) and TCRG rearrangement (148 bp and 124 bp for patients 4 and 5, respectively). SM: Size marker VIII (Boehringer Mannheim, Mannheim, Germany); controls: lane 1: no DNA; lane 2: peripheral blood DNA; lanes 3-7: dilutions of leukemic DNA in DNA from peripheral blood mononuclear cells from healthy donors from 10−3 to 10−7; a sensitivity of 10−5 was achieved for all clonotypic rearrangements; lane 8: Guthrie card DNA; lane 9: Guthrie card DNA from age-matched controls.
Polymerase chain reaction with DNA from Guthrie spots
Guthrie spots were routinely cut into 3 pieces and DNA was extracted from each piece separately at different occasions using the DNAzol kit (Vienna Labs, Vienna, Austria). Integrity of DNA was confirmed.20 A third of the DNA from 1 piece was used for 1 clonotypic polymerase chain reaction (PCR). The sequence of the PCR product obtained from Guthrie card DNA was confirmed by sequencing in both directions. PCR products of the expected size of the specific PCR product in the control samples were also sequenced. To avoid contamination with amplified material, precautions were taken as previously described,21 and experiments were performed on 2 to 3 independent occasions. Amplifications were performed on a PTC 300 Thermocycler (Techne, Cambridge, UK).
Estimation of clonotypic cells in neonatal blood spots was performed by comparing the intensity of the signals in all segments of the neonatal blood spot with those from the dilution series, assuming that a Guthrie spot contains approximately 30 μL blood.13
Results and discussion
The sequence of the clone-specific junctional region of the IgH and of the TCRG rearrangements was used to develop an ASO-PCR for their detection at low frequency (Table 1). By this approach, the clonotypic rearrangements were detected in the neonatal blood spots from all 5 patients (Figure 1). In addition to the clone-specific IgH rearrangements, we detected the leukemia-specific MLL-AF4 and the AF4-MLL fusion sequences in the Guthrie cards from the 2 infants with a t(4;11) positive leukemia (Figure 1). This observation is in line with a previous report by Gale et al,13 demonstrating that the MLL-AF4 rearrangements were present in the neonatal blood spots from three 6-month- to 2-year-old children. We estimated that about 10 clonotypic cells were present per Guthrie spots from patients 1, 3, and 5, and about 100 cells from patients 2 and 4. These numbers of preleukemic/leukemic cells are within the range reported by Gale et al.13 Of note, the estimated number of clonotypic cells as defined by the IgH rearrangements correspond to that as defined by the MLL-AF4/AF4-MLL fusion regions in patients 1 and 2. The similar quantity of the clonal cells suggests that the clonal IgH cells carried also the t(4;11) translocation.
The third patient with a common BCP ALL and a clone-specific IgH rearrangement had a normal karyotype and no exploitable leukemia-specific molecular genetic marker. He was already aged 4 years 8 months at diagnosis and is, so far, the oldest child in whom the presence of the preleukemic/leukemic cell clone was demonstrated at birth. Thus, the latency period in this case was probably more than 5 years. Our data and a recent report demonstrating the identification of the identical clonotypic TEL/AML fusion sequences in twins who developed leukemia 9 years apart from each other9 indicate, that diverse subtypes of ALLs may have variable latency periods.
The occurrence of an identical TCR beta rearrangement in twins with a T-NHL and a T-ALL at 9 and 11 years of age, respectively, provided first evidence that T-cell malignancies may also evolve from a clone that is already transformed in utero.10 We therefore investigated neonatal blood spots from 2 young children with T-ALL and detected the clonotypic TCRG rearrangements in both. Thus, we prove that T-cell leukemias can also be initiated in utero.
An array of molecular events within a cell is assumed to determine initiation, latency, and progression of leukemias. The detection of leukemia-specific fusion genes and clonotypic antigen receptor gene rearrangements may complement each other to define a time frame for the initiation and the duration of the dormant phase of the neoplastic clone, because the biologic meaning of the 2 rearrangement processes may be different in the evolution of the leukemia. We show in this study that clone-specific antigen receptor gene rearrangements are already present at birth in children who develop t(4;11) translocation positive ALL in infancy, and B- or T-lineage ALL during early childhood. We cannot exclude that the concordance of clonotypic sequences shared by the leukemic clone and a clone present at birth is due to an antigen receptor gene rearrangement predating the first oncogenic event. This is unlikely, however, because most of the IgH rearrangements in BCP ALL are nontranslatable.22 A normal cell with such a rearrangement undergoes apoptosis, but the survival of these cells indicates a rescue mechanism. Further, cells were even able to clonally expand. We therefore assume that a first leukemogenic event in utero is a common phenomenon in young children with ALL.
Note added after submission. After submission of this paper, a report by Wiemels et al23 described that in the majority of cases of childhood ALL with TEL-AML1 fusion, the clonotypic fusion was present in the patients' Guthrie spots, ie, did occur prenatally.
Acknowledgments
We would like to thank Dr Shai Israeli (NCI) for fruitful discussions. We would also like to thank Dr S. Stöckler-Ipsiroglu from the University of Vienna, for providing the Guthrie cards.
This work was supported in part by the Österreichische Kinderkrebshilfe and by private donations to the CCRI.
Reprint:E. Renate Panzer-Grümayer, CCRI, St. Anna Kinderspital, Kinderspitalg 6, A-1090 Vienna, Austria; e-mail:panzer@ccri.univie.ac.at.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal