Abstract
The weak D phenotype is caused by many different RHD alleles encoding aberrant RhD proteins, raising the possibility of distinct serologic phenotypes and of anti-D immunizations in weak D. We reported 6 new RHD alleles, D category III type IV, DIM, and the weak D types 4.1, 4.2.1, 4.2.2, and 17. The immunohematologic features of 18 weak D types were examined by agglutination and flow cytometry with more than 50 monoclonal anti-D. The agglutination patterns of the partial D phenotypes DIM, DIII type IV, and DIVtype III correlated well with the D epitope models, those of the weak D types showed no correlation. In flow cytometry, the weak D types displayed type-specific antigen densities between 70 and 4000 RhD antigens per cell and qualitatively distinct D antigens. A Rhesus D similarity index was devised to characterize the extent of qualitative changes in aberrant D antigens and discriminated normal D from all tested partial D, including D category III. In some rare weak D types, the extent of the alterations was comparable to that found in partial Ds that were prone to anti-D immunization. Four of 6 case reports with anti-D in weak D represented auto-anti-D. We concluded that, in contrast to previous assumptions, most weak D types, including prevalent ones, carry altered D antigens. These observations are suggestive of a clinically relevant potential for anti-D immunizations in some, but not in the prevalent weak D types, and were used to derive an improved transfusion strategy in weak D patients.
The Rhesus D antigen (ISBT 004.001; RH1) is expressed by the RhD protein. Anti-D is still the leading cause for the hemolytic disease of the newborn.1,2 Depending on the population, 3% to 25% of white individuals lack the antigen D. As anti-D immunizations can occur readily in D-negative recipients,3the antigen D is of critical importance for the blood transfusion strategy, and the most important blood group antigen determined by a protein.
About 0.2% to 1% of white individuals carry red cells with a reduced expression of the D antigen (weak D).4-6 For more than 45 years, anti-D immunizations are known to occur infrequently in D-positives,7-10 often in individuals with such a low antigen D density.7 Usually these cases could be traced to a few partial D phenotypes,11,12 especially D category VI.10 Most, but not all, partial D can be identified by the lack of reactivity with certain monoclonal anti-D antibodies, which is interpreted as lack of certain “D epitopes.”13 The increasingly elaborated D epitope schemes13-16 allowed the identification and classification of many new partial D.
However, it had been impossible to obtain unequivocal evidence for serologic differences in the majority of weak D. In such samples, the lack of reactivity with anti-D may be attributed to the reduced expression of the D antigen rather than the lack of any D epitope.11,17,18 As no definitive serologic variation could be established over the years, it became generally accepted that most weak Ds possess a normal D antigen.19-23 As a consequence, the possibility of anti-D immunization in weak D was often disregarded.20,22,23 It should be noted that a substantial number of anti-D immunizations in weak D24 and in unclassified samples25 remained unexplained in regard to a serologic and molecular classification.
We showed that most, if not all, weak D phenotypes carried altered RhD proteins.26 For example, in the weak D type 4, 2 transmembraneous amino acid residues are substituted, which was reminiscent of ARRO-1 reported as partial D.27 These findings raised the possibility of qualitative changes in the D antigen of some weak D types. To corroborate the safety of the current D-positive transfusion strategy in weak D, a serologic workup of a representative collection of different weak D types was timely.
With the use of numerous weak D samples of known genotype and more than 80 monoclonal anti-D, we investigated immunohematologic differences of 18 weak D types and 5 partial D, including 5 newly characterizedRHD alleles. The different weak D types presented distinct immunohematologic features. We derived a Rhesus index, showed its use as a rough estimate for an anti-D immunization rate, and analyzed 6 case reports of weak D with anti-D. On the basis of our results, we proposed improved transfusion strategies in weak D patients.
Materials and methods
Blood samples
EDTA- or citrate-anticoagulated blood samples were collected from white blood donors in southwestern Germany or referred to our laboratory. Weak D type 4.1, type 17, and DIM were identified among white blood donors with weak D expression. Seven antigen D–positive samples were sent to the German Rhesus Immunization Registry because of an anti-D. One of these samples, DIII type IV, was classified as DIII subtype, because of a negative crossmatch with DIIIc. The genotype of the DIIIc red cells (kindly provided by Silvano Wendel, Sao Paulo, Brazil) was confirmed using RHD exon-specific PCR-SSP.28 An anti-D was from a different DIIIcproband (kindly provided by Anna Ribera, Barcelona, Spain).
Sequencing of the 10 RHD exons from genomic DNA
DNA was prepared as described previously.28 Nucleotide sequencing was performed using Prism dye terminator cycle-sequencing kit with AmpliTaq FS DNA polymerase and Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Weiterstadt, Germany) with a DNA sequencing unit (ABI 373A and ABI 377, Applied Biosystems). Nucleotide sequencing of genomic DNA stretches representative for all 10 RHD exons was accomplished as described.26 In some samples, modified reactions were used: For exon 1 PCR, re13 was substituted by re04 or re012 (exon 6, rf51 by rf52; exon 7, re617 by re615; exon 10, rr4 by rez2); for exon 3 sequencing re31 and rb20d were substituted by re29. Primer sequences were re012, TCCACTTTCCACCTCCCTGC; re04, AGGTCACATCCATTTATCCCACTG; re29, TCTTCTATTCCCACAGAAAGTAGG; re615, GTAACTCATAGTGTGGTCCGTAG; rez2, CCTTGGTCTGCCAGAATTTTCA, rf52, TGAGAGCTGAGGGTGTCAGA.
Immunohematology
Monoclonal anti-D were provided by the Workshop on Monoclonal Antibodies Against Human Red Blood Cells and Related Antigens.29 Agglutination was tested in a gel matrix test (LISS-Coombs 37°C, DiaMed-ID Micro Typing System; DiaMed, Cressier sur Morat, Switzerland) using the following antibodies: B9A4B2 (number III-1-28), D-89/47 (29), HG/92 (30), D-90/7 (31), D-90/17 (32), D-90/12 (33), HeM-92 (34), 175-2 (35), 17 010C9 (36), NaTH28-3C11 (37), NaTH87-4A5 (38), NaTH53-2A7 (39), AUB-2F7/Fiss (41), CAZ7-4C5 (42), LOR11-2D9 (43), LOR12-E2 (44), LOR17-6C7 (45), LOR17-8D3 (46), LOR28-21D3 (47), LOR28-7E6 (48), LOR29-F7 (49), LORA (50), LORE (51), MAR-1F8 (52), NAU3-2E8 (53), NAU6-1G6 (54), NAU6-4D5 (55), NOI (56), NOU (57), SAL17-4E8 (58), SAL20-12D5 (59), SALSA-12 (60), VOL-3F6 (61), ZIG-189 (62), 822 (68), 819 (69), BTSN4 (71), BTSN6 (72), BTSN10 (73), LHM76/58 (74), LHM76/55 (75), LHM76/59 (76), LHM77/64 (77), LHM59/19 (78), LHM70/45 (79), LHM50/2B (80), LHM169/80 (81), LHM169/81 (82), LHM174/102 (83), LHM50/3.5 (84), LHM59/25 (85), LHM59/20 (86), T3D2F7 (87), C205-29 (88), CLAS1-126 (89), F5S (90), H2D5D2F5 (93), RAB.B15 (94), BIRMA-DG3 (95), BIRMA-D6 (96), BIRMA-D56 (97), P3187 (98), P3F17 (99), P3F20 (100), P3G6 (101), P3AF6 (102), BRAD3 (105), L87.1G7 (108), MS26 (112), MS201 (113), D10 (114), HIRO-1 (115), HIRO-6 (116), HIRO-3 (117), HIRO-4 (118), ID6-H8 (119), HIRO-7 (120), HIRO-8 (121), HIRO-2 (122), MCAD-6 (124), HS114 (134), BS87 (180).
Flow cytometry
Flow cytometry was performed using monoclonal IgG anti-D BS221, BS227, BS228, BS229, BS231, and H41 (Biotest, Dreieich, Germany); P3 × 35, P3 × 241, P3 × 249, P3 × 290, and HM16 (Diagast, Loos, France) and the following Workshop IgG anti-D: number 29, 30, 31, 32, 33, 36, 41, 43, 44, 45, 47, 49, 55, 56, 58, 59, 68, 71, 72, 73, 75, 76, 77, 80, 81, 82, 89, 90, 93, 94, 95, 96, 97, 101, 102, 105, 108, 112, 114, 117, 118, 119, 120, 121, 122, 124 plus BRAD5 (Workshop number 104), and D6D02 (123). The secondary antibody was goat antihuman IgG, Fab-fragment, FITC-conjugated (supplied by Dianova; Jackson Immunoresearch, Hamburg, Germany).
All blood samples were stored on fluid nitrogen. The fluorescence was compared with that of a standard CDe/cde red cell of known antigen density (13 000 RhD antigens per cell).18 The standard cell was measured twice and the geometric mean of both results was used. Measurements with more than 15% difference were repeated. Background fluorescence was determined with RhD-negative samples. Generally, markers were set to count all red cells, even if a fraction of red cells appeared unstained. The weak D type 10 donor displayed about 50% unstained cells in samples from 2 independent donations, for this type the antigen density was derived from the stained red cell population. The number of RhD epitopes detected by a specific monoclonal anti-D was calculated as described previously.18 30
Antigen density
The RhD antigen density (RhD antigens per cell) of a red cell sample was estimated as the median of the epitope densities detected with all antibodies that resulted in epitope densities above a cutoff. This cutoff was defined as 0.1 of the 90 percentile of the epitope densities detected with all monoclonal anti-D used in flow cytometry.
In the experiments involving several samples for weak D type 1 to type 5, antigen densities were estimated as geometric means of the epitope densities detected with the anti-D BS221, BS227, BS228, BS229, BS231, and H41. These estimates varied from the estimate based on the full set of antibodies by a factor of 0.98 (weak D type 2) to 1.23 (weak D type 4) and were therefore considered representative.
Rhesus D similarity index
A Rhesus index was calculated as the ratio of the 10 percentile to the 90 percentile of the epitope densities detected with all anti-D. For 2 samples (weak D type 12 and type 17), no Rhesus index was given because the 90 percentile was less than 200 epitopes per cell.
The Rhesus index was derived as a measure of the variation of the epitopes detected by different antibodies under the assumption of an approximately log-normal distribution. Compared with other measures, like range or SD, it was robust to outliers and minor deviations from the assumed distribution. The Rhesus index may range from 1 in normal D to 0 in partial D that were lacking many D epitopes.
We simulated the possible effect of assay variability on the Rhesus index for normal D: We calculated a standard error of SDctrl = 0.0165 for the logarithms of the epitope densities measured with the second control sample. Similar to all other samples, this control was standardized on the geometric mean of the 2 control samples. Because of the error propagation rule,31the standard error of a sample was expected to be
SDmeas =√([SDctrl × √2]2 + [SDctrl × √2]2/2) = 0.0286
A log-normal distribution with such a SD yields an expected ratio of 10 percentile to 90 percentile of
10z(0.1) × 0.0286/10z(0.9) × 0.0286 = 10−1.2816 × 0.0286/101.2816 × 0.0286 = 0.845
where z(0.1) and z(0.9) denote the 0.1 and 0.9 limits of the standard normal distribution. These calculations indicated that normal D samples yield a Rhesus index of about 0.845.
Analysis of 6 weak D samples carrying anti-D
We differentiated the antibodies in a gel matrix assay (LISS-Coombs 37°C, DiaMed-ID Micro Typing System; DiaMed). In addition, some antibodies were confirmed in a solid phase assay (Capture R; Immucor, Norcross, GA). Because some low-titer auto-anti-D may fail in weak D samples to cause positive direct antiglobulin tests, we performed antibody elutions to determine the presence of red cell–bound antibodies. For all 6 samples, the full-length coding sequence was determined by sequencing all 10 RHD exons from genomic DNA as described previously.
Statistics
For the interpretation of data, the epitope densities were assumed to have an approximate log-normal distribution; however, all inferences were derived using distribution-independent test methods. Different frequencies were compared using the χ2 test for a 2 × 2 contingency table.31 Antigen densities of multiple types were compared using the 2-sided Wilcoxon rank sum test for each comparison31 and the Bonferroni-Holm procedure32 to correct for multiple testing as indicated in “Results.”
Results
Molecular characterization of RHD alleles
The molecular bases of 6 RHD alleles were determined (Table1). Weak D types 4.1, 4.2.1, and 4.2.2 shared both amino acid substitutions, T201R and F223V, that are typical of weak D type 4,26 but each carried 1 additional missense mutation. DIII type IV also displayed multiple dispersed missense mutations. Weak D type 17 and DIM had single missense mutations in the transmembraneous and exofacial protein segments, respectively.
Molecular basis of new RHD alleles described in this study
| Trivial Name . | Allele . | Nucleotide Change . | Effect on Protein Sequence . | Exons Involved . | Predicted Localization in the Cell Membrane . |
|---|---|---|---|---|---|
| Weak D type 4.1* | RHD(W16C, T201R, F223V) | G → C at 48 | Trp to Cys at 16 | 1 | TM |
| C → G at 602, | Thr to Arg at 201 | 4 | IC | ||
| T → G at 667, | Phe to Val at 223 | 5 | TM | ||
| G → A at 819 | no change | 6 | — | ||
| Weak D type 4.2.1 | RHD(T201R, F223V, I342T) | C → G at 602, | Thr to Arg at 201 | 4 | IC |
| T → G at 667, | Phe to Val at 223 | 5 | TM | ||
| G → A at 957 | no change | 6 | — | ||
| T → C at 1025 | Ile to Thr at 342 | 7 | TM | ||
| Weak D type 4.2.2 | RHD(T201R, F223V, I1342T) | C → G at 602, | Thr to Arg at 201 | 4 | IC |
| T → G at 667, | Phe to Val at 223 | 5 | TM | ||
| C → T at 744 | no change | 5 | — | ||
| G → A at 957 | no change | 6 | — | ||
| T → C at 1025 | Ile to Thr at 342 | 7 | TM | ||
| Weak D type 17 | RHD(R114W) | C → T at 340 | Arg to Trp at 114 | 3 | TM |
| DIII type IV | RHD(L62F, A137V, N152T) | G → 186 at T | Leu to Phe at 62 | 2 | TM |
| C → 410 at T | Ala to Val at 137 | 3 | TM | ||
| A → 455 at C | Asn to Thr at 152 | 3 | TM | ||
| DIM | RHD(C285Y) | G → A at 854 | Cys to Tyr at 285 | 6 | EF |
| Trivial Name . | Allele . | Nucleotide Change . | Effect on Protein Sequence . | Exons Involved . | Predicted Localization in the Cell Membrane . |
|---|---|---|---|---|---|
| Weak D type 4.1* | RHD(W16C, T201R, F223V) | G → C at 48 | Trp to Cys at 16 | 1 | TM |
| C → G at 602, | Thr to Arg at 201 | 4 | IC | ||
| T → G at 667, | Phe to Val at 223 | 5 | TM | ||
| G → A at 819 | no change | 6 | — | ||
| Weak D type 4.2.1 | RHD(T201R, F223V, I342T) | C → G at 602, | Thr to Arg at 201 | 4 | IC |
| T → G at 667, | Phe to Val at 223 | 5 | TM | ||
| G → A at 957 | no change | 6 | — | ||
| T → C at 1025 | Ile to Thr at 342 | 7 | TM | ||
| Weak D type 4.2.2 | RHD(T201R, F223V, I1342T) | C → G at 602, | Thr to Arg at 201 | 4 | IC |
| T → G at 667, | Phe to Val at 223 | 5 | TM | ||
| C → T at 744 | no change | 5 | — | ||
| G → A at 957 | no change | 6 | — | ||
| T → C at 1025 | Ile to Thr at 342 | 7 | TM | ||
| Weak D type 17 | RHD(R114W) | C → T at 340 | Arg to Trp at 114 | 3 | TM |
| DIII type IV | RHD(L62F, A137V, N152T) | G → 186 at T | Leu to Phe at 62 | 2 | TM |
| C → 410 at T | Ala to Val at 137 | 3 | TM | ||
| A → 455 at C | Asn to Thr at 152 | 3 | TM | ||
| DIM | RHD(C285Y) | G → A at 854 | Cys to Tyr at 285 | 6 | EF |
IC, intracellular; TM, transmembraneous; EF, exofacial.
Weak D type 4 with RHD(T201R, F223V) is considered prototypical for a group of RHD alleles that carry various additional missense or silent mutations or both. According to the proposed nomenclature, the original weak D type 4 allele is designated weak D type 4.0. The weak D type 4.1 sample was 1 of the 6 samples described previously (see reference 26, Table 3); the other 5 samples represent weak D type 4.0.
Serology of 17 weak D types and 3 partial D
An exhaustive collection of weak D phenotypes and, for comparison, 3 partial D were tested with 82 monoclonal anti-D (Table2). As known for DIIIa and DIIIc,13,16 DIII type IV was agglutinated by all monoclonal anti-D. This finding corroborated the classification as DIII subtype, based on the negative crossmatch with DIIIc (see “Materials and methods”). DIV type III and DIM displayed antibody reactivity patterns as predicted by the known D epitopes. The reaction pattern of DIV type III differed from that reported for DIVb16 by negative results with 4 of 12 anti-D grouped in epitope 15/16, which may represent a serologic split of this epitope. The reaction pattern of DIM was unique and resembled an intermediate of D category VI18 and DFR.16 33
Reactivity patterns of antibodies with new partial and weak D
| Pattern* . | Antibodies† . | Partial D‡ . | Weak D Type . | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-9 . | 1-37 . | IgG . | IgM . | DIII . | DIV . | DIM . | 1 . | 2 . | 3 . | 4.0 . | 4.1 . | 4.2 . | 5 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 15 . | 16 . | 17 . |
| 1 | 1 | 82 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | w | |
| 1 | 2 | 79 | + | − | − | + | + | + | + | + | w | w | + | + | − | + | w | − | w | − | − | − | |
| 83 | + | − | − | w | w | + | + | + | − | w | + | + | − | + | w | − | + | − | − | − | |||
| 2 | 3 | 44 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |
| 51 | + | − | − | + | + | + | + | + | + | + | + | + | w | + | − | − | + | − | w | − | |||
| 2 | 4 | 48 | + | − | − | − | − | w | + | + | − | − | − | w | − | + | − | − | − | − | − | − | |
| 3 | 5 | 75 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | w | |
| 43, 49 | + | − | +w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |||
| 4 | 6 | 45 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |
| 5 | 7 | 116 | w | + | − | w | w | + | + | + | − | w | + | + | − | + | w | − | w | − | − | − | |
| 42 | + | + | − | w | w | + | + | + | + | − | w | w | − | + | − | − | w | − | − | − | |||
| 5 | 10 | 88, 89 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + | w | + | w | |
| 41 | + | + | − | + | w | + | + | + | + | w | w | + | − | + | − | − | + | − | w | − | |||
| 52 | + | w | − | − | − | w | w | + | w | − | − | w | − | w | − | − | − | − | − | − | |||
| 5 | 11 | 69 | + | + | − | − | − | w | w | w | − | − | + | − | − | + | − | − | − | − | − | − | |
| 6/7 | 12 | 102 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |
| 46 | + | + | w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | w | − | |||
| 113 | + | + | − | w | w | + | + | + | + | w | + | + | − | + | w | − | w | − | w | − | |||
| 35 | + | + | − | w | w | + | + | + | + | − | + | w | − | + | − | − | w | − | − | − | |||
| 6/7 | 13 | 29, 36, 47, 90, 93, 105 | +w | + | +w | + | +w | + | + | + | + | + | + | + | + | + | + | +w | + | +w | + | w | |
| 6/7 | 15/16 | 71, 81, 97 | + | + | +w | + | + | + | + | + | +w | + | + | + | + | + | + | w | + | + | + | w | |
| 32, 58, 108, 114 | + | + | w | + | + | + | + | +w | + | + | + | + | + | + | + | w | + | +w | + | − | |||
| 56 | + | + | w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |||
| 31, 95 | +w | − | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | w | |||
| 59 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |||
| 80 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | w | − | |||
| 6/7 | 17 | 134 | + | w | − | w | w | w | w | − | − | w | + | + | w | + | w | − | w | − | − | − | |
| 115 | w | + | − | w | w | w | w | w | − | w | + | + | − | + | w | − | w | − | − | − | |||
| 99 | + | + | − | w | w | + | + | + | w | w | + | + | − | + | w | − | − | − | − | − | |||
| 98 | w | + | − | w | w | + | + | + | − | w | + | + | − | + | w | − | − | − | − | − | |||
| 28, 34 | + | + | − | w | − | + | + | + | + | − | + | +w | − | + | − | − | w | − | − | − | |||
| 37 | + | + | − | w | w | + | + | + | + | − | + | w | − | + | − | − | w | − | − | − | |||
| 39 | + | w | − | − | − | + | + | + | − | − | − | + | − | + | − | − | − | − | − | − | |||
| 6/7 | 18 | 30 | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | w | + | − | + | − | |
| 119 | + | + | − | + | + | + | + | + | + | w | + | + | w | + | w | − | + | − | − | − | |||
| 84, 85 | + | + | − | w | w | + | + | + | w | w | + | + | − | + | w | − | w | − | − | − | |||
| 86, 87 | w | w | − | w | w | +w | w | w | − | w | +w | w | − | + | w | − | − | − | − | − | |||
| 100 | w | w | − | w | w | w | w | − | − | w | + | + | − | + | w | − | − | − | − | − | |||
| 6/7 | 20/21 | 33 | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | w | + | w | + | − | |
| 94 | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | w | − | |||
| 122 | + | + | w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | + | − | |||
| 61 | + | − | − | w | w | + | w | w | − | − | − | + | − | w | − | − | w | − | − | − | |||
| 8 | 22 | 74, 78 | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | +w | − | |
| 9 | 23 | 112 | + | − | w | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | w | |
| 101, 118 | +w | − | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + | +w | + | − | |||
| 120, 121 | + | − | + | + | + | + | + | +w | + | + | + | + | + | + | + | − | + | +w | + | − | |||
| 96 | + | − | w | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | − | |||
| 77 | + | − | w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |||
| N/A | 31 | 54, 57 | +w | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| N/A | 32 | 62 | + | + | − | w | w | + | w | w | − | − | − | w | − | + | − | − | w | − | − | − | |
| N/A | 33 | 38 | + | w | − | − | − | + | + | + | − | − | − | + | − | w | − | − | − | − | − | − | |
| N/A | 34 | 50 | + | − | − | w | + | + | + | w | − | − | − | w | − | w | − | − | w | − | − | − | |
| N/A | 35 | 60 | + | − | − | + | + | + | + | + | + | w | + | + | w | + | − | − | + | − | w | − | |
| N/A | 36 | 72, 76 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | w | |
| 55 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |||
| 53 | + | − | − | + | + | + | + | + | + | + | w | + | − | + | w | − | + | − | + | − | |||
| N/A | 37 | 68, 73, 117, 124, 180 | + | + | +w | + | + | + | + | + | + | + | + | + | + | + | + | w | + | +w | + | +w | |
| Pattern* . | Antibodies† . | Partial D‡ . | Weak D Type . | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-9 . | 1-37 . | IgG . | IgM . | DIII . | DIV . | DIM . | 1 . | 2 . | 3 . | 4.0 . | 4.1 . | 4.2 . | 5 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 15 . | 16 . | 17 . |
| 1 | 1 | 82 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | w | |
| 1 | 2 | 79 | + | − | − | + | + | + | + | + | w | w | + | + | − | + | w | − | w | − | − | − | |
| 83 | + | − | − | w | w | + | + | + | − | w | + | + | − | + | w | − | + | − | − | − | |||
| 2 | 3 | 44 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |
| 51 | + | − | − | + | + | + | + | + | + | + | + | + | w | + | − | − | + | − | w | − | |||
| 2 | 4 | 48 | + | − | − | − | − | w | + | + | − | − | − | w | − | + | − | − | − | − | − | − | |
| 3 | 5 | 75 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | w | |
| 43, 49 | + | − | +w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |||
| 4 | 6 | 45 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |
| 5 | 7 | 116 | w | + | − | w | w | + | + | + | − | w | + | + | − | + | w | − | w | − | − | − | |
| 42 | + | + | − | w | w | + | + | + | + | − | w | w | − | + | − | − | w | − | − | − | |||
| 5 | 10 | 88, 89 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + | w | + | w | |
| 41 | + | + | − | + | w | + | + | + | + | w | w | + | − | + | − | − | + | − | w | − | |||
| 52 | + | w | − | − | − | w | w | + | w | − | − | w | − | w | − | − | − | − | − | − | |||
| 5 | 11 | 69 | + | + | − | − | − | w | w | w | − | − | + | − | − | + | − | − | − | − | − | − | |
| 6/7 | 12 | 102 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |
| 46 | + | + | w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | w | − | |||
| 113 | + | + | − | w | w | + | + | + | + | w | + | + | − | + | w | − | w | − | w | − | |||
| 35 | + | + | − | w | w | + | + | + | + | − | + | w | − | + | − | − | w | − | − | − | |||
| 6/7 | 13 | 29, 36, 47, 90, 93, 105 | +w | + | +w | + | +w | + | + | + | + | + | + | + | + | + | + | +w | + | +w | + | w | |
| 6/7 | 15/16 | 71, 81, 97 | + | + | +w | + | + | + | + | + | +w | + | + | + | + | + | + | w | + | + | + | w | |
| 32, 58, 108, 114 | + | + | w | + | + | + | + | +w | + | + | + | + | + | + | + | w | + | +w | + | − | |||
| 56 | + | + | w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |||
| 31, 95 | +w | − | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | w | |||
| 59 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |||
| 80 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | w | − | |||
| 6/7 | 17 | 134 | + | w | − | w | w | w | w | − | − | w | + | + | w | + | w | − | w | − | − | − | |
| 115 | w | + | − | w | w | w | w | w | − | w | + | + | − | + | w | − | w | − | − | − | |||
| 99 | + | + | − | w | w | + | + | + | w | w | + | + | − | + | w | − | − | − | − | − | |||
| 98 | w | + | − | w | w | + | + | + | − | w | + | + | − | + | w | − | − | − | − | − | |||
| 28, 34 | + | + | − | w | − | + | + | + | + | − | + | +w | − | + | − | − | w | − | − | − | |||
| 37 | + | + | − | w | w | + | + | + | + | − | + | w | − | + | − | − | w | − | − | − | |||
| 39 | + | w | − | − | − | + | + | + | − | − | − | + | − | + | − | − | − | − | − | − | |||
| 6/7 | 18 | 30 | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | w | + | − | + | − | |
| 119 | + | + | − | + | + | + | + | + | + | w | + | + | w | + | w | − | + | − | − | − | |||
| 84, 85 | + | + | − | w | w | + | + | + | w | w | + | + | − | + | w | − | w | − | − | − | |||
| 86, 87 | w | w | − | w | w | +w | w | w | − | w | +w | w | − | + | w | − | − | − | − | − | |||
| 100 | w | w | − | w | w | w | w | − | − | w | + | + | − | + | w | − | − | − | − | − | |||
| 6/7 | 20/21 | 33 | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | w | + | w | + | − | |
| 94 | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | w | − | |||
| 122 | + | + | w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | + | − | |||
| 61 | + | − | − | w | w | + | w | w | − | − | − | + | − | w | − | − | w | − | − | − | |||
| 8 | 22 | 74, 78 | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | +w | − | |
| 9 | 23 | 112 | + | − | w | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | w | |
| 101, 118 | +w | − | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + | +w | + | − | |||
| 120, 121 | + | − | + | + | + | + | + | +w | + | + | + | + | + | + | + | − | + | +w | + | − | |||
| 96 | + | − | w | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | − | |||
| 77 | + | − | w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |||
| N/A | 31 | 54, 57 | +w | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| N/A | 32 | 62 | + | + | − | w | w | + | w | w | − | − | − | w | − | + | − | − | w | − | − | − | |
| N/A | 33 | 38 | + | w | − | − | − | + | + | + | − | − | − | + | − | w | − | − | − | − | − | − | |
| N/A | 34 | 50 | + | − | − | w | + | + | + | w | − | − | − | w | − | w | − | − | w | − | − | − | |
| N/A | 35 | 60 | + | − | − | + | + | + | + | + | + | w | + | + | w | + | − | − | + | − | w | − | |
| N/A | 36 | 72, 76 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + | + | w | |
| 55 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w | + | − | |||
| 53 | + | − | − | + | + | + | + | + | + | + | w | + | − | + | w | − | + | − | + | − | |||
| N/A | 37 | 68, 73, 117, 124, 180 | + | + | +w | + | + | + | + | + | + | + | + | + | + | + | + | w | + | +w | + | +w | |
Pattern as described previously by Lomas et al14(1-9) and Scott16 (1-37), who did not differentiate epitopes 15 from 16 and epitopes 20 from 21 (Table116).
Antibody numbers as defined in “Materials and methods” section. Reactivity was tested in a gel matrix test. + indicates a normal positive result; “w,” a weak positive result; +w, either + or w, depending on the antibody; and −, a negative result.
DIII, D category III type IV described in this study; DIV, D category IV type III described previously.26
In contrast to the 3 partial D, the fit of the antibody reaction “patterns” with known D epitopes was not satisfactory in most of the 17 weak D types. There were 189 negative results obtained with 22 IgM antibodies, but only 117 negative results with the 60 IgG antibodies investigated (χ2 = 190;P < .001). Two IgM antibodies (numbers 54 and 57) did not agglutinate any weak D sample. Possible “epitope splits” were conspicuously found with anti-D that were associated with an overall weak reactivity. These observations could easily be explained, if the determination of reaction patterns was confounded by the low and variable antigen densities in all weak D.
RhD antigen density of weak D is type-specific
A large number of samples with the prevalent weak D types were analyzed for their RhD antigen densities (Table3). The antigen density was type-specific. With the exception of weak D type 3 and 4, the antigen densities of all prevalent weak D types differed significantly for each pairwise comparison (P < .01; 2-sided Wilcoxon rank sum test with Bonferroni-Holm correction for multiple testing, n = 10).
RhD antigen densities of the prevalent weak D types
| Weak D Type . | RhD Antigen Densities (Antigens Per Cell)3-150 . | Phenotype . | Samples Tested (n) . | ||
|---|---|---|---|---|---|
| Median . | Mean3-151 . | Range . | |||
| Type 1 | 759 | 795 | 533-1283 | CcDee | 25 |
| Type 2 | 674 | 625 | 446-818 | ccDEe | 24 |
| Type 3 | 1948 | 1926 | 1333-2650 | CcDee | 11 |
| Type 4 | 1872 | 1919 | 1687-2406 | ccDee | 7 |
| Type 5 | 316 | 311 | 218-386 | ccDEe | 6 |
| Weak D Type . | RhD Antigen Densities (Antigens Per Cell)3-150 . | Phenotype . | Samples Tested (n) . | ||
|---|---|---|---|---|---|
| Median . | Mean3-151 . | Range . | |||
| Type 1 | 759 | 795 | 533-1283 | CcDee | 25 |
| Type 2 | 674 | 625 | 446-818 | ccDEe | 24 |
| Type 3 | 1948 | 1926 | 1333-2650 | CcDee | 11 |
| Type 4 | 1872 | 1919 | 1687-2406 | ccDee | 7 |
| Type 5 | 316 | 311 | 218-386 | ccDEe | 6 |
Estimated antigen density based on the geometric mean of the number of epitopes detected with 6 IgG anti-D (BS221, BS227, BS228, BS229, BS231, H41).
Geometric mean of the antigen densities of the different samples.
Suppressive effect of C in trans
The antigen densities of 2 CCDee weak D type 1 samples (deduced genotype based on the known CDe haplotype association26 of weak D type 1: CDe/Cde), 1 CcDEe weak D type 2 sample (deduced genotype: Cde/cDE), and 1 CcDee type 4 sample (deduced genotype: Cde/cDe) were determined (Table 4). These samples had considerably lower antigen densities than the controls with cde in trans. For comparison, a weak D type 3 sample with cdE in trans expressed an antigen density similar to its controls.
Effect of the Cde and cdE haplotypes in trans position
| Weak D Type . | Haplotype In Trans . | RhD Antigen Densities (Antigens Per Cell)4-150 . | ||
|---|---|---|---|---|
| Sample . | Reference4-151 . | % of Reference . | ||
| Type 1 | Cde | 235 | 759 | 31 |
| Type 1 | Cde | 369 | 759 | 49 |
| Type 2 | Cde | 112 | 674 | 17 |
| Type 4 | Cde | 360 | 1872 | 19 |
| Type 3 | cdE | 2206 | 1948 | 113 |
| Weak D Type . | Haplotype In Trans . | RhD Antigen Densities (Antigens Per Cell)4-150 . | ||
|---|---|---|---|---|
| Sample . | Reference4-151 . | % of Reference . | ||
| Type 1 | Cde | 235 | 759 | 31 |
| Type 1 | Cde | 369 | 759 | 49 |
| Type 2 | Cde | 112 | 674 | 17 |
| Type 4 | Cde | 360 | 1872 | 19 |
| Type 3 | cdE | 2206 | 1948 | 113 |
Epitope density profiles
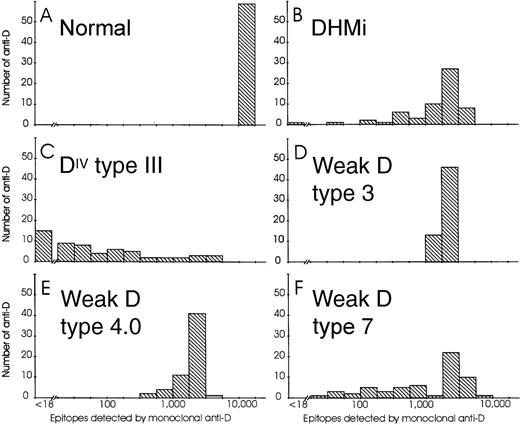
For antigen density determination, epitope density profiles were established with 59 IgG monoclonal anti-D as described previously.18,30 The regular D antigens of a CcDee control showed a single narrow peak (Figure 1, panel A).18 Two partial D, DHMi34 (panel B) and DIV type III26 (panel C), had considerably to extremely broadened peaks. Three representative weak D types (panels D to F) showed peaks ranging from single and narrow (weak D type 3), like that of the regular D antigen, to broadened (weak D type 4.0 and type 7), that was reminiscent of partial D.
Epitope density profiles of selected normal D, partial D, and weak D samples.
On the abscissa, ranges of epitope densities (sites/cell) as detected by various anti-D are given using a logarithmic scale. On the ordinate, the number of anti-D representing the particular ranges of sites/cell are shown. The following phenotypes are depicted: Panel A, normal CcDee sample; Panel B, DHMi; Panel C, DIV type III; Panel D, weak D type 3; Panel E, weak D type 4.0; Panel F, weak D type 7.
Epitope density profiles of selected normal D, partial D, and weak D samples.
On the abscissa, ranges of epitope densities (sites/cell) as detected by various anti-D are given using a logarithmic scale. On the ordinate, the number of anti-D representing the particular ranges of sites/cell are shown. The following phenotypes are depicted: Panel A, normal CcDee sample; Panel B, DHMi; Panel C, DIV type III; Panel D, weak D type 3; Panel E, weak D type 4.0; Panel F, weak D type 7.
Rh antigen densities of rare weak D types
The epitope density profiles as shown in Figure 1 were determined for a larger number of control, partial D, and weak D phenotypes. These data were used to calculate the antigen density representing the quantitative expression of the D antigen. The antigen densities of controls and known partial D (Table 5) were consistent with previous reports.30,35 36 Of the 2 partial D defined in this study, DIII type IV had an enhanced antigen density compared with its appropriate control (ccDee). The second partial D had a very much diminished antigen density and, hence, was dubbed DIM. Its antigen density was lower than DVI type I and represented one of the lowest antigen densities ever reported for partial D.
Antigen density and Rhesus D similarity index
| RhD Phenotype . | Antigen Density . | Rhesus D Similarity Index . | Percentiles of RhD Epitopes Detected . | Antibodies . | |||
|---|---|---|---|---|---|---|---|
| 10% . | 50% . | 90% . | Tested . | >Cutoff5-150 . | |||
| Controls | |||||||
| CcDee | 13 283 | 0.90 | 12 534 | 13 238 | 13 986 | 59 | 59 |
| CcDEe | 24 509 | 0.84 | 21 982 | 24 509 | 26 322 | 59 | 59 |
| CCDee | 22 778 | 0.84 | 20 124 | 22 778 | 24 079 | 59 | 59 |
| ccDEe | 19 710 | 0.87 | 18 120 | 19 710 | 20 803 | 59 | 59 |
| ccDee | 23 240 | 0.88 | 21 884 | 23 240 | 24 984 | 59 | 59 |
| Partial D types | |||||||
| DIIIc | 26 889 | 0.69 | 23 654 | 26 889 | 34 085 | 59 | 59 |
| DIII type IV | 33 255 | 0.45 | 17 200 | 33 255 | 38 515 | 59 | 59 |
| DIV type III5-151 | 607 | 0 | 0 | 46 | 1 552 | 59 | 17 |
| DVI type I | 1 050 | 0 | 0 | 0 | 1 279 | 595-152 | 14 |
| DVI type II | 2 886 | 0 | 0 | 0 | 3 303 | 595-152 | 18 |
| DVI type III | 14 502 | 0 | 0 | 0 | 17 674 | 595-152 | 17 |
| DVII | 8 398 | 0.48 | 4 788 | 8 394 | 9 936 | 59 | 58 |
| DHMi | 2 292 | 0.12 | 413 | 2 200 | 3 465 | 59 | 54 |
| DIM | 192 | 0 | 0 | 0 | 245 | 59 | 20 |
| Weak D types | |||||||
| Type 1 | 1 285 | 0.57 | 823 | 1 285 | 1 454 | 59 | 59 |
| Type 2 | 489 | 0.68 | 378 | 489 | 555 | 59 | 59 |
| Type 3 | 1 932 | 0.77 | 1 597 | 1 932 | 2 081 | 59 | 59 |
| Type 4.0 | 2 288 | 0.36 | 977 | 2 288 | 2 707 | 59 | 59 |
| Type 4.1 | 3 811 | 0.48 | 2 140 | 3 811 | 4 453 | 59 | 59 |
| Type 4.2 | 1 650 | 0.21 | 456 | 1 650 | 2 121 | 58 | 58 |
| Type 5 | 296 | 0.39 | 137 | 295 | 355 | 59 | 57 |
| Type 6 | 1 053 | 0.79 | 857 | 1 053 | 1 088 | 5 | 5 |
| Type 7 | 2 409 | 0.03 | 105 | 1 950 | 3 450 | 59 | 44 |
| Type 8 | 972 | 0.75 | 824 | 972 | 1 105 | 59 | 59 |
| Type 9 | 248 | 0.43 | 135 | 248 | 312 | 59 | 59 |
| Type 105-153 | 1 186 | 0.34 | 867 | 1 186 | 2 579 | 58 | 58 |
| Type 11 | 183 | 0.53 | 127 | 183 | 238 | 59 | 59 |
| Type 12 | 96 | nc5-154 | 0 | 76 | 153 | 41 | 30 |
| Type 13 | 946 | 0.33 | 375 | 946 | 1 124 | 59 | 59 |
| Type 15 | 297 | 0.21 | 92 | 297 | 435 | 59 | 58 |
| Type 16 | 235 | 0.44 | 151 | 234 | 346 | 55 | 54 |
| Type 17 | 66 | nc | 40 | 63 | 176 | 59 | 58 |
| RhD Phenotype . | Antigen Density . | Rhesus D Similarity Index . | Percentiles of RhD Epitopes Detected . | Antibodies . | |||
|---|---|---|---|---|---|---|---|
| 10% . | 50% . | 90% . | Tested . | >Cutoff5-150 . | |||
| Controls | |||||||
| CcDee | 13 283 | 0.90 | 12 534 | 13 238 | 13 986 | 59 | 59 |
| CcDEe | 24 509 | 0.84 | 21 982 | 24 509 | 26 322 | 59 | 59 |
| CCDee | 22 778 | 0.84 | 20 124 | 22 778 | 24 079 | 59 | 59 |
| ccDEe | 19 710 | 0.87 | 18 120 | 19 710 | 20 803 | 59 | 59 |
| ccDee | 23 240 | 0.88 | 21 884 | 23 240 | 24 984 | 59 | 59 |
| Partial D types | |||||||
| DIIIc | 26 889 | 0.69 | 23 654 | 26 889 | 34 085 | 59 | 59 |
| DIII type IV | 33 255 | 0.45 | 17 200 | 33 255 | 38 515 | 59 | 59 |
| DIV type III5-151 | 607 | 0 | 0 | 46 | 1 552 | 59 | 17 |
| DVI type I | 1 050 | 0 | 0 | 0 | 1 279 | 595-152 | 14 |
| DVI type II | 2 886 | 0 | 0 | 0 | 3 303 | 595-152 | 18 |
| DVI type III | 14 502 | 0 | 0 | 0 | 17 674 | 595-152 | 17 |
| DVII | 8 398 | 0.48 | 4 788 | 8 394 | 9 936 | 59 | 58 |
| DHMi | 2 292 | 0.12 | 413 | 2 200 | 3 465 | 59 | 54 |
| DIM | 192 | 0 | 0 | 0 | 245 | 59 | 20 |
| Weak D types | |||||||
| Type 1 | 1 285 | 0.57 | 823 | 1 285 | 1 454 | 59 | 59 |
| Type 2 | 489 | 0.68 | 378 | 489 | 555 | 59 | 59 |
| Type 3 | 1 932 | 0.77 | 1 597 | 1 932 | 2 081 | 59 | 59 |
| Type 4.0 | 2 288 | 0.36 | 977 | 2 288 | 2 707 | 59 | 59 |
| Type 4.1 | 3 811 | 0.48 | 2 140 | 3 811 | 4 453 | 59 | 59 |
| Type 4.2 | 1 650 | 0.21 | 456 | 1 650 | 2 121 | 58 | 58 |
| Type 5 | 296 | 0.39 | 137 | 295 | 355 | 59 | 57 |
| Type 6 | 1 053 | 0.79 | 857 | 1 053 | 1 088 | 5 | 5 |
| Type 7 | 2 409 | 0.03 | 105 | 1 950 | 3 450 | 59 | 44 |
| Type 8 | 972 | 0.75 | 824 | 972 | 1 105 | 59 | 59 |
| Type 9 | 248 | 0.43 | 135 | 248 | 312 | 59 | 59 |
| Type 105-153 | 1 186 | 0.34 | 867 | 1 186 | 2 579 | 58 | 58 |
| Type 11 | 183 | 0.53 | 127 | 183 | 238 | 59 | 59 |
| Type 12 | 96 | nc5-154 | 0 | 76 | 153 | 41 | 30 |
| Type 13 | 946 | 0.33 | 375 | 946 | 1 124 | 59 | 59 |
| Type 15 | 297 | 0.21 | 92 | 297 | 435 | 59 | 58 |
| Type 16 | 235 | 0.44 | 151 | 234 | 346 | 55 | 54 |
| Type 17 | 66 | nc | 40 | 63 | 176 | 59 | 58 |
Number of monoclonal anti-D detecting at least 0.1 of the 90% percentile of the epitope density detected by all anti-D.
Single sample of CCDee phenotype; the antigen density may be depressed by the presence of Cde in trans.
Results derived from a reanalysis of published data.18Thirty-nine of the 59 antibodies used in this study were known to be nonreactive to DVI, and their results were assumed to be 0.
Result of the positive subpopulation (see “Materials and methods”); analysis including nonstaining cells resulted in antigen density 157; D similarity index, 0.39.
nc, not calculated, because 90 percentile was less than 200 antigens per cell.
The antigen densities of weak D samples varied between less than 100 in weak D type 12 and about 4000 RhD antigens per cell in weak D type 4.1. The highest antigen densities were observed for weak D type 3 and types 4. Most rare weak D types had low antigen densities, like weak D type 12 and type 17, which expressed the lowest antigen densities of all aberrant D antigens tested. The 4 most frequent weak D types (weak D type 1 to type 4) representing 95% of all weak D26 had antigen densities higher than 400 RhD antigens per cell.
Rhesus D similarity index
A Rhesus D similarity index (Rhesus index) was defined as the ratio of the 10 percentile and 90 percentile of the epitope densities (Table5). The Rhesus index measured a qualitative difference to the D antigen of the standard phenotype CcDee. Ideally, normal D antigens would have an index of 1, grossly aberrant partial D an index of 0. Because of the known assay variability, the Rhesus index for normal D antigens was expected to be approximately 0.845 (see “Materials and methods”). All normal D antigens tested showed Rhesus indices of 0.84 to 0.90 (Table 5). In contrast, all partial D tested had Rhesus indices of less than 0.7. The Rhesus index allowed us to discriminate DIIIsamples, which are agglutinated by all monoclonal anti-D, from normal D. Partial D lacking many D epitopes, like DVI or DIV, had a Rhesus index of 0.
In weak D, the Rhesus indices ranged from 0.78 in weak D type 3 to 0.03 in weak D type 7 (Table 5). Interestingly, the Rhesus indices of 14 of 16 weak D types were equal or lower than the Rhesus index of DIIIc, indicating that their D antigen deviated from normal D at least as much as DIIIc, which is known to allow an anti-D immunization. The 3 most prevalent weak D types (weak D type 1 to type 3) had Rhesus indices greater than the DVII, whose carriers are frequent among white individuals37 and generally transfused D-positive without clinical problems.
Distinct immunohematologic features of weak D types
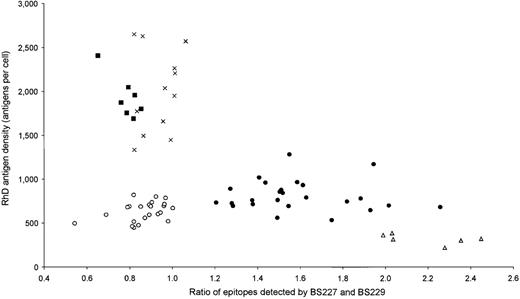
As shown in Table 3, weak D type 1 and type 2 can be distinguished from weak D type 3 and types 4 by their different antigen densities. To further differentiate weak D type 1 from type 2, we determined the ratios of epitopes detected by the 2 IgG monoclonal anti-D BS227 and BS229 (Figure 2), similar to our previous approach with the 3 DVI types.18 All 25 weak D type 1 samples had ratios greater than 1.2, whereas all 24 weak D type 2 samples had ratios equal to or less than 1.0, which allowed a ready discrimination by immunohematologic methods. Likewise, a discrimination of weak D type 3 from type 4 was possible by the ratio of epitopes detected by the 2 IgG monoclonal anti-D BS227 and H41 (data not shown).
Distinct immunohematologic features of the 5 most frequent weak D types.
The RhD antigen density is plotted on the ordinate. On the abscissa, the ratio of RhD epitopes detected by the 2 monoclonal anti-Ds, BS227 and BS229, is shown. Data of 74 weak D samples are shown. •: weak D type 1, n = 25; ○: weak D type 2, n = 24 (only 23 different positions are discernible because 2 samples overlapped); ×: weak D type 3, n = 12; ▪: weak D type 4.0, n = 7; Δ: weak D type 5, n = 6.
Distinct immunohematologic features of the 5 most frequent weak D types.
The RhD antigen density is plotted on the ordinate. On the abscissa, the ratio of RhD epitopes detected by the 2 monoclonal anti-Ds, BS227 and BS229, is shown. Data of 74 weak D samples are shown. •: weak D type 1, n = 25; ○: weak D type 2, n = 24 (only 23 different positions are discernible because 2 samples overlapped); ×: weak D type 3, n = 12; ▪: weak D type 4.0, n = 7; Δ: weak D type 5, n = 6.
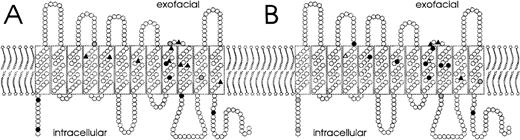
Molecular structure and phenotype
For 18 RHD alleles with single nucleotide substitutions, we correlated the involved amino acid positions with the antigen densities (Figure 3, panel A) and the Rhesus indices (Figure 3, panel B). Many of the weak D types with strongly reduced antigen densities of less than 500 RhD antigens per cell had amino acid exchanges in the transmembraneous part of the RhD protein. Most of these types also displayed moderately reduced Rhesus indices ranging from 0.2 to 0.6. Likewise, many weak D types with intracellular substitutions had antigen densities in the range of 500 to 2000 RhD antigens per cell, and most of them displayed almost normal RhD indices. Interestingly, weak D type 7, for which the substituted amino acid was predicted to be deeply buried in the transmembraneous helix 11, had a very low Rhesus index of 0.03 and a rather high antigen density of 2400 RhD antigens per cell. There was no simple relation of the type of substitution to the antigen density or the Rhesus index. For example, the seemingly conservative M295I substitution in weak D type 11 caused a very low antigen density.
Relationship of involved amino acid position, antigen density, and Rhesus index.
A model for the orientation of the RhD protein in the red-cell membrane is shown.26 The amino acid positions involved and their effect on antigen density (A) and on Rhesus index (B) are indicated for 15 weak D and 3 partial D alleles with single missense mutations. Symbols denote major changes (▴, antigen density less than 500 RhD antigens/cell or Rhesus index less than 0.2, respectively), moderate changes (•, 500-2000 RhD antigens/cell; Rhesus index, 0.2-0.6), and minor changes (○, more than 2000 RhD antigens/cell; Rhesus index more than 0.6). RhD antigen density was too low for the determination of the Rhesus indices in 2 weak D types (Δ, weak D type 12 and type 17).
Relationship of involved amino acid position, antigen density, and Rhesus index.
A model for the orientation of the RhD protein in the red-cell membrane is shown.26 The amino acid positions involved and their effect on antigen density (A) and on Rhesus index (B) are indicated for 15 weak D and 3 partial D alleles with single missense mutations. Symbols denote major changes (▴, antigen density less than 500 RhD antigens/cell or Rhesus index less than 0.2, respectively), moderate changes (•, 500-2000 RhD antigens/cell; Rhesus index, 0.2-0.6), and minor changes (○, more than 2000 RhD antigens/cell; Rhesus index more than 0.6). RhD antigen density was too low for the determination of the Rhesus indices in 2 weak D types (Δ, weak D type 12 and type 17).
Six case reports of weak D with anti-D
The overlap of the Rhesus indices of weak D and partial D with documented anti-D production fostered a search for samples with weak D phenotype and anti-D.38 Six samples were referred to us.39 Four samples could be assigned to the previously described RHD alleles weak D type 1, type 2, and type 15. Two samples carried new alleles that belonged to the group of weak D type 4 alleles and were designated weak D type 4.2 (Table6). The anti-D observed in the frequent weak D type 1 and type 2 were shown to represent auto-anti-D by antibody elution. In contrast, weak D type 15 and 1 sample of type 4.2 had allo-anti-D; it should be noted that these 2 types had Rhesus indices lower than 0.3, which was lower than all other weak D types (Table 3) with the single exception of weak D type 7.
Samples with weak D phenotype and anti-D
| Case . | Sex . | Phenotype . | Anti-D (Titer) . | Other Antibodies . | Eluate6-150 . | RHD Allele . |
|---|---|---|---|---|---|---|
| RIR-1 | female | ccDEe | 1:2 | none | positive | weak D type 2 |
| RIR-5 | female | ccDEE | 1:1 | none | negative | weak D type 15 |
| RIR-6 | male | CcDee | 1:1 | anti-E | positive | weak D type 1 |
| RIR-11 | female | ccDEe | 1:4 | anti-C | positive | weak D type 2 |
| RIR-14 | female | ccDee | 1:128 | none | NA6-151 | weak D type 4.2.26-152 |
| RIR-15 | male | ccDee | 1:1 | none | positive | weak D type 4.2.16-152 |
| Case . | Sex . | Phenotype . | Anti-D (Titer) . | Other Antibodies . | Eluate6-150 . | RHD Allele . |
|---|---|---|---|---|---|---|
| RIR-1 | female | ccDEe | 1:2 | none | positive | weak D type 2 |
| RIR-5 | female | ccDEE | 1:1 | none | negative | weak D type 15 |
| RIR-6 | male | CcDee | 1:1 | anti-E | positive | weak D type 1 |
| RIR-11 | female | ccDEe | 1:4 | anti-C | positive | weak D type 2 |
| RIR-14 | female | ccDee | 1:128 | none | NA6-151 | weak D type 4.2.26-152 |
| RIR-15 | male | ccDee | 1:1 | none | positive | weak D type 4.2.16-152 |
Red cell eluate tested for the presence of anti-D.
NA, no red cells available for elution.
Discussion
The 17 analyzed weak D types expressed distinct phenotypes. Each weak D type was associated with a certain range of RhD antigen densities. In contrast to previous conjectures, weak D types possessed slightly to considerably altered D antigens. The extent of qualitative changes observed in weak D overlapped with that observed in partial D. We provided experimental evidence that the risk of allo-anti-D immunization in the frequent weak D types was, however, low. The data allowed us to formulate a rational framework for a transfusion strategy in weak D patients.
For many years phenotypic differences among weak D samples have been noted and comprised quantitative changes, like antigen density variation,15,20,35,40-48 and qualitative changes, like variable epitope presentation.20,25,47 Apparently, these differences were too subtle to base a satisfying classification of weak D on serologic criteria alone. Antibody dissociation constants in weak D were reported to be indistinguishable from normal D.21 A qualitative classification failed in the majority of weak D types (Table 2), often because of confounding effects like low antigen density17 and slight antigenic differences (Table4). Antigen densities of different weak D types overlapped (Table 3), which may have previously been mistaken to represent a continuum of D antigen densities in weak D.42,43 49
Altered binding characteristics of monoclonal anti-D were previously reported for several partial D,18,30,35,50 even if the respective epitopes were known to be present. Prompted by these observations, we proposed a Rhesus index as a complementary tool for discerning qualitative changes of the D antigen. The technical features of the Rhesus index were designed to minimize confounding by low antigen density. The Rhesus index achieved a discrimination of DIII from normal D, that was previously beyond the scope of any monoclonal antibody–based technique. The Rhesus index gauged the loss or deviation of the target epitopes compared with normal antigen D for a large number of IgG monoclonal anti-D. Clinical observations confirmed that this parameter represented a rough estimate of the anti-D immunization rate: DVI, of known high immunization risk, had a Rhesus index of 0. In contrast, the Rhesus index of DVII was 0.48 and indicated a much less altered antigen D than in DVI. This finding correlated well with the infrequent anti-D immunization in DVII, despite generally D-positive transfusion in this most prevalent partial D in white individuals.37 51
The concurrent quantification of antigen density and antigen variability allowed us to put the previously separate concepts of partial D and weak D into context. Normal D samples had normal antigen densities and normal Rhesus indices. Aberrant RhD phenotypes with about normal antigen density were detectable by virtue of their qualitative changes only, even if the changes were discrete, and have been dubbed partial D.52 We will not be surprised if many such aberrant RhD were not yet recognized by serologic methods. In contrast, aberrant RhD with reduced antigen density could be detected by their low antigen density or by their qualitative changes. Because of the difficulties in discerning qualitative abnormalities in samples with weak D antigen expression, only those presenting prominent qualitative changes were previously prone to be identified as separate entities and considered partial D. Most other such samples were lumped together and loosely marked “weak D.” Hence, in the group of aberrant RhD with less than 5000 antigens per cell, it is not surprising to find a continuum of qualitative changes varying from an almost normal D antigen, like in weak D type 3, to an extremely altered D antigen, like in DVI type I.
We evaluated epidemiologic data to corroborate the use of the Rhesus index for predicting an anti-D immunization risk in weak D. In the Rhesus Immunization Registry, RhD-positive samples with suspected anti-D production were collected and systematically analyzed at the molecular level.39 Two allo-anti-D were shown in weak D type 4.2 and type 15 for which the Rhesus index indicated considerably altered D antigens. Three other weak D samples proved to be auto-anti-D in weak D type 1 and type 2, which have an almost normal Rhesus index. Both weak D types represent 88% of all weak D, whereas weak D type 4.2 and type 15 comprise less than 2% in white individuals.26The inverse relationship of population frequency and allo-anti-D observation was intriguing and suggested that the Rhesus index may be representative of the immunization risk.
In many countries,25,53 transfusion recipients are currently typed and transfused D-positive, if their red cells are agglutinated by 2 IgM monoclonal anti-D that do not react with DVI. This policy ensures D-negative transfusions in DVI patients that are at risk of anti-D immunization.5,18 25 There is no defined antigen density threshold for D-positive transfusions in weak D patients. Rather, the transfusion strategy in weak D patients is not controlled and depends on the accidental sensitivity of the preferred typing reagents and methods.
We propose an RhD typing and transfusion strategy based on scientifically deduced criteria: An improved strategy should take account of the population frequencies of weak D types26 and the qualitative changes of their antigen D in conjunction with the observed anti-D immunization events. On the basis of our data, we conclude that an optimized antigen density threshold in white individuals would provide for D-positive transfusions in patients with such weak D types only that have an antigen density similar or higher than weak D type 2. This antigen density threshold of about 400 RhD antigens per cell is well within the detection limits of currently available IgM monoclonal anti-D typing reagents and methods, like tubes and gels without requiring an antiglobulin test. Then, weak D type 2 red cells would be preferred for quality control of anti-D sera and typing methods.
Applying the proposed threshold would provide for a D-positive transfusion in 97%26 of all white weak D patients, the vast majority of whom carry weak D type 1, type 2, and type 3. For these 3 most prevalent weak D types, D-positive transfusion can be considered safe because these types have a low predicted immunization risk, are frequent, and no immunization events have been documented yet. In contrast, even a moderate anti-D immunization risk may have been missed in the rarer weak D types, like weak D type 9, and Rhesus indices implicated a potential of anti-D immunization in several of these types, like weak D type 15.
Lowering the threshold below 400 RhD antigens per cell would be feasible, for example, by applying the antiglobulin test, and marginally increase the number of weak D patients transfused D-positive. However, this increase would affect patients carrying a multitude of rare weak D types for which an anti-D immunization risk was likely or could not be excluded. We propose to consider Rh-negative transfusion of individuals carrying those weak D types of lower frequency and lower antigen density representing less than 3% of all weak D in white individuals.26
There were rare weak D types, like type 7 and type 4.2, with antigen densities well above the proposed threshold, which were likely or known to be at risk for anti-D immunization. D-negative transfusions in such weak D types could currently only be achieved at the expense of D-negative transfusions in all weak D patients. Such a strategy may still be rational in populations with a high frequency of these weak D types. Further advances may be brought forth by improved typing reagents with a low affinity for weak D type 4.2 and type 7 and a high affinity for weak D type 1, type 2, and type 3. Phage display technology might allow the specific selection and cloning of those antibodies54 by differential panning with suitable weak D types. Once established, such improved typing strategies with novel reagents would enhance the transfusion safety without incurring additional typing costs.
For donor typing, all potentially immunogenic RHD-positive samples should be recognized as RhD-positive. The safety of weak D red cell unit transfusion to D-negative patients remains equivocal.20,55 Hence, a typing method with a high sensitivity,53 56 like the antiglobulin test in combination with oligoclonal anti-D and gel, may be recommended.
The phenotypic analysis of more than 20 aberrant RhD with single amino acid substitutions revealed that in general, transmembranous mutations hindered membrane integration and antigen expression most severely. Intracellular and extracellular mutations were less impeding in this regard. Amino acid substitutions on or close to the red cell surface had usually the most pronounced qualitative effects. The notion26 that the substitution of the RHD-specific amino acids in the transmembranous parts of RhD coded by exons 4 and 5 with their RHCE-specific counterparts lowers antigen density, whereas the N152T substitution enhances D expression, was further supported by the phenotypes of weak D type 4.1, type 4.2, and DIII type IV. The reduction of the antigen density by a Cde haplotype in trans, known as Ceppellini effect,57 was also effective in weak D. The seeming paradoxical lower antigen density of most ccDEe weak D samples compared with CcDee weak D samples35,46 simply indicated that, like in the 3 DVI types,18 the major influence of the molecular type overrided the modulating effect of C in trans.
Current epitope models attributed no58,59 or a few60 D epitopes to RhD exofacial loop 5. In contrast, most, but not all,61 older studies with polyclonal62-64 and monoclonal65 anti-D predicted that the conservation of the only exofacial cysteine at position 285 in loop 5 was very critical for anti-D immunoreactivity. A site-directed mutagenesis study66 showed that a D with a C285A substitution retained most D epitopes. In contrast, DIM carrying a C285Y substitution lacked many D epitopes currently mapped to loop 3, like epD2, epD17, epD18, and epD22, and to loop 4, like epD1, epD2, epD3, and epD20/21. This indicated that these epitopes are sensitive for major structural changes of the D protein. In contrast, the possibly linear epitopes of loop 6,67 like epD5, epD6, and epD23, were retained in DIM.
We characterized 4 RHD alleles, DIII type IV, weak D type 4.1, type 4.2.1, and type 4.2.2, with amino acid changes in 2 or more exons. Four similar RHD alleles, DIIIa,68 DIVa,69ARRO-1,27 and weak D type 4.0,26 had been known previously. ARRO-1,27 now called DAR, may be identical to weak D type 4.2. It is interesting to note that 7 of these 8RHD alleles, whose multiple residue substitutions cannot be explained by 1 simple gene conversion event, certainly arose in the cDe haplotype. All 8 haplotypes may represent branches of the RHevolutionary tree separate from the branches leading to the prevalentRH haplotypes in white individuals. The L62F and A137V substitutions in DIII type IV were likely remnants of very old RHD alleles because identical amino acids at these positions are present in extant RH alleles of the great apes.70 71
Note added in proof.
After the revised version of our manuscript was submitted, Hemker et al72 published the full coding sequence of the partial D DAR. It shares the 3 missense mutations found in weak D type 4.2.1 and type 4.2.2 but lacks their silent mutations. Hence, in “weak D parlance,” DAR might be dubbed weak D type 4.2.0. But we acknowledge the priority of the DAR nomenclature.
Acknowledgments
We thank Hans-Hermann Sonneborn and Manfred Ernst, Biotest AG, Dreieich, Germany, for generously supplying us with their monoclonal anti-D. We are greatly indebted to all contributors of the Workshop on Monoclonal Antibodies Against Human Red Blood Cells and Related Antigens, who provided most other monoclonal anti-D. We thank Silvano Wendel, Sao Paulo, Brazil, and Anna Ribera, Barcelona, Spain, for red cells and antibodies of the rare DIIIcphenotype and Christoph Gassner and Diether Schönitzer, Innsbruck, Austria, for the weak D type 13 sample. We thank Anita Hacker, Marianne Lotsch, Katharina Schmid, Sabine Zahn, and Olga Zarupski for expert technical assistance.
Supported by the DRK-Blutspendedienst Baden-Württemberg, Stuttgart, Germany, and by the University of Ulm (Forschungsförderungsprojekt P 422 and P 531), Ulm, Germany.
T.H.M. is now at Franz-Volhard-Klinik, Max-Delbrück-Centrum für Molekulare Medizin, Berlin-Buch, Germany.
Reprints:Willy A. Flegel, Abteilung Transfusionsmedizin, Universitätsklinikum Ulm, and DRK-Blutspendedienst Baden-Württemberg, Institut Ulm, Helmholtzstrasse 10, D-89081 Ulm, Germany; e-mail: waf@ucsd.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal