Abstract
The band 1q21 is among the most common sites affected by chromosomal translocations in lymphoid, myeloid, epithelial, and sarcomatous lesions. In non-Hodgkin's lymphoma (NHL), translocations and duplications affecting this chromosomal site are frequently, but not exclusively, seen in association with primary abnormalities such as the t(14;18)(q32;q21) and t(8;14)(q24;q32) translocations, suggesting a role for 1q21 rearrangements in tumor progression. We report here the characterization and cloning of breakpoints in a case of extranodal ascitic B-cell lymphoma with a t(1;14)(q21;q32) translocation. The breakpoints on the der(1) and der(14) chromosomes were mapped by fluorescence in situ hybridization and Southern blot analysis and cloned using an IGHG (Cγ) probe. The translocation linked theIGHG4 switch (Sγ4) sequences of the productively rearranged allele to chromosome 1 sequences downstream of MUC1, leaving the MUC1 transcriptional unit intact. MUC1 was markedly overexpressed in the tumor at the mRNA and protein levels relative to lymphoma cell lines lacking a 1q21 rearrangement. Presumably,MUC1 transcription is aberrantly regulated by the IGHA(C) 3′ enhancer element retained on the same chromosome. Screening of a panel of B-cell lymphomas by Southern blot analysis identified a subset with a 3′ MUC1 breakpoint and another with low-level amplification of MUC1. MUC-1 mucin has previously been shown to be frequently overexpressed in human epithelial cancers and to be associated with tumor progression and poor clinical outcome. Thus, MUC1 activation by chromosomal translocation, rearrangement, and amplification, identified here for the first time in NHL, is consistent with its suggested role in tumorigenesis.
The chromosomal band 1q21 is the third most frequent site of rearrangement in non-Hodgkin's lymphoma (NHL).1-3Translocations and duplications affecting this chromosomal site are usually detected along with other translocations, such as t(14;18)(q32;q21) and t(8;14)(q24;q32), that are considered to be primary genetic alterations in NHL. These data have suggested that 1q21 abnormalities may be associated with tumor progression.1-3Rearrangement breaks affecting band 1q21 are also common in leukemia, sarcoma, and epithelial tumors.4 Thus, identification of the genes involved in 1q21 rearrangements is of importance to the understanding of tumorigenesis in multiple lineages. Recently, a candidate gene, BCL9, was cloned from a case of acute lymphoblastic leukemia (ALL) with a t(1;14)(q21;q32) translocation.5 However, BCL9 rearrangements were rare in NHL,5 suggesting that other genes at this band may be involved in 1q21 rearrangements in lymphomas. We report here the cloning of a t(1;14)(q21;q32) translocation breakpoint in a B-cell lymphoma. The translocation linked sequences 2.4 kbp 3′ of theMUC1 gene on chromosome 1 to the IGHG4 switch (Sγ4) region on chromosome 14. MUC1 was markedly overexpressed in the tumor with respect to lymphoma cell lines that did not contain a 1q21 rearrangement. This observation is consistent with aberrant activation of MUC1 expression by an IGHenhancer, a mechanism of gene activation well established in other 14q32-associated translocations in B-cell leukemias and lymphomas.6 Screening of a large panel of B-cell lymphomas by Southern blot analysis using probes derived from the MUC1genomic locus identified a subset with recurrent DNA rearrangements clustering 3′ of the MUC1 gene and another subset with multiple MUC1 gene copies. MUC-1 mucin has previously been shown to be frequently overexpressed in human epithelial cancers and associated with tumor progression and poor clinical outcome.7-10MUC1 activation by chromosomal translocation, a novel phenomenon in lymphomas, is thus consistent with its previously recognized role in tumorigenesis.
Materials and methods
Tumor specimens and cell lines
Case 2385 was from a patient with an extranodal ascitic lymphoma at presentation that exhibited the G-banded karyotype: 47-50, XY,+X, t(1; 14)(q21;q32),add(6)(q21),+7,+10,der(10)t(?;10)(?;p15)t(?;2)(?;p11.2), add(11)(q23),+13,add(15)(p13),der(17)t(?;17)(?;q25)t(?;8)(?;q13),add (18)(q21)[cp20]. The tumor cells were positive for CD19, CD20, and CD21 with κ clonal excess and μ-chain expression, confirming B-lineage derivation. Other B-cell NHL specimens used in this study were derived from the ongoing prospective ascertainment and cytogenetic analysis of NHL cases at the Memorial Sloan-Kettering Cancer Center.2 Two groups were analyzed for MUC1 rearrangement and copy number. One group that exhibited a 1q21 rearrangement by G-banding karyotype analysis comprised 72 B-cell lymphomas of all histologic types (small lymphocytic, follicular, diffuse small noncleaved, diffuse large cell, and immunoblastic). In the second group of 106 biopsies, 27 cases were cytogenetic failures, 7 cases yielded normal metaphases, and 72 cases were clonally abnormal but did not exhibit a 1q21 aberration. In this group, 12 had follicular histologies and 94 were diffuse large-cell lymphomas. OSI-Ly 3, a B-cell lymphoma cell line, and OSI-Ly 12, a T-cell lymphoma cell line11 (gift of Mark Minden) that did not contain 1q21 rearrangements, were maintained as suspension cultures.
DNA probes
The probes used for fluorescence in situ hybridization (FISH), Southern blotting, and genomic library screening comprised the following: a 5.5-kbp BamHI–HindIII fragment containingIGHJ (JH), a 1.3-kbp EcoRI fragment containing the first 2 exons of IGHM (Cμ), a 7-kbpHindIII–BamHI fragment containing IGHG1(Cγ), and a 13-kbp HindIII fragment containing IGHA1(Cα). The PAC clones 288D9 and 35A12 containing, respectively, theBCL9 and MUC1 loci were obtained from Genome Systems (St Louis, MO), and the YAC clone 876B11, mapping 8 cM telomeric to BCL9, was obtained from Research Genetics (Huntsville, AL).
Fluorescence in situ hybridization
DNA probes were labeled with biotin-14-dATP (Gibco BRL, Rockville, MD) and hybridized to metaphase spreads from normal human lymphocytes and tumor cells as previously described.12 Hybridization signals and the corresponding chromosomal bands were visualized with fluorescein isothiocyanate-conjugated avidin (Oncor, Gaithersburg, MD) after staining with 4′,6-diamidino-2-phenylindole dihydrochloride.
Southern and Northern blot analyses
Genomic DNA was extracted and subjected to restriction enzyme digestion, gel electrophoresis, and Southern blot analysis as previously described, using normal placental DNA as control.13 Mapping of subcloned genomic fragments was performed by Southern blot analysis of monochromosomal cell hybrid DNA (Coriell Cell Repositories, Camden, NJ). DNA copy number was estimated relative to a control probe (D2S48) as previously described.14 Total RNA was extracted from the ascitic fluid or cultured lymphoma cell lines, electrophoresed, and hybridized as previously described.15 Northern filters were initially hybridized with a probe derived from the tandem repeat segment ofMUC1 cDNA,16 followed by THBS3 (generated by polymerase chain reaction [PCR]), and then with GAPDH to control for loading. Multiple-tissue Northern filters were obtained from Clontech (Palo Alto, CA).
Molecular cloning of the translocation breakpoint
A genomic library of the tumor DNA was constructed in λGEM-11 phage vector (Promega, Madison, WI) by partial Sau3A digestion,17 and was screened using the probes detailed above. Fragments of phage clones were subcloned in pBluescript.
Polymerase chain reaction
PCR was performed in a 50-μL reaction volume using AmpliTaq DNA–polymerase (Perkin-Elmer, Norwalk, CT) under the following conditions: 94°C (1 minute) followed by 30 cycles of 94°C (30 seconds), 60°C (30 seconds), 72°C (2 minutes), and a final extension at 72°C (10 minutes). The following primers were used to isolate PACs from the Genome Systems PAC library: 5′-TTTGTGGACTTGGGTATCAATGA-3′ and 5′-AAAAACAGAAAA GAATCCTGGGAGAAG-3′ for BCL9; 5′-CCCACCAATTTCTCGGACACTTCTCA-3′ and 5′-GTCCAGTTCAGGATCCCC GCTATCTC-3′ for germline chromosome 1 region containing MUC1. The primers 5′-ACCTGGGACCTGAGCTGTGATTTCCT-3′ and 5′-GACACCCACCTTCCCAACATCCTTTC-3′, derived from sequences on chromosomes 14 and 1, respectively, were used for cloning the breakpoint on der(1). The isolated PAC 35A12 and YAC 876B11 were confirmed to be nonchimeric by FISH analysis of normal human metaphases.
Flow cytometric analysis
Expression of MUC-1 mucin on live ascitic fluid cells and cultured cell lines was assayed by FACS as described.18 Cells were either incubated with normal mouse serum or with a mouse monoclonal antihuman MUC-1 mucin antibody (HMFG-2)19 as a primary antibody. FITC-conjugated rabbit antimouse IgG (H+L) was used as a secondary antibody for detection.
Results
Mapping of translocation breakpoints by FISH
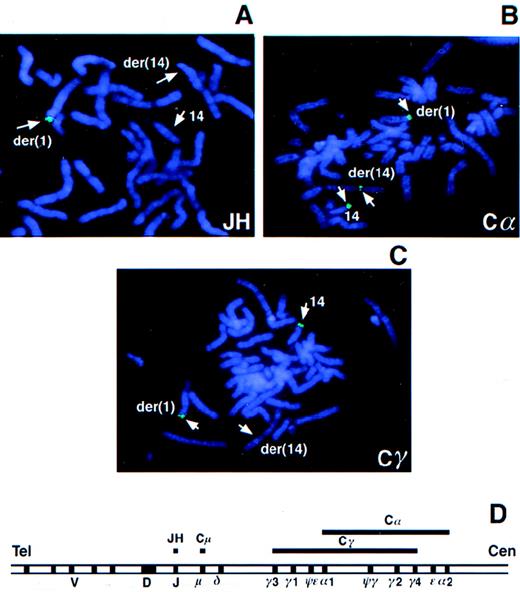
To determine whether BCL9 was affected by the t(1;14)(q21;q32) translocation in tumor 2385, PAC 288D9 was used as a probe in FISH analysis of tumor metaphases. Hybridization signals were observed on the normal chromosome 1 and the der(1) chromosome, but not on the der(14) chromosome, indicating that the breakpoint was distal to BCL9 (data not shown). In contrast, YAC 876B11, which mapped distal to BCL9, hybridized to normal chromosome 1 and the der(14) chromosome, but not to the der(1) chromosome, narrowing the breakpoint region to approximately 8 cM telomeric to BCL9 (data not shown). The JH probe hybridized only to the der(1) chromosome without a signal either on the der(14) or on the apparently normal chromosome 14, suggesting a deletion of this region in the latter (Figure1A). The Cα probe hybridized to the der(1) and der(14) chromosomes and to the apparently normal chromosome 14 (Figure 1B). The Cα probe hybridized to both copies of Cα in each homolog; therefore, this result indicated that the breakpoint in the der(14) chromosome occurred in an approximately 150-kbp region between Cα1 and Cα2, with Cα1 translocated to der(1) and Cα2 retained on the der(14) chromosome. The Cγ probe was detected on the apparently normal chromosome 14 and the der(1) chromosome, but not on the der(14) chromosome (Figure 1C). Absence of Cγ signal on the der(14) chromosome suggested that the breakpoint was within or downstream of Cγ4 (Figure 1D). These results indicated the translocation breakpoint to be in the region containing Cγ4, Cε, and Cα2.
FISH analysis of the t(1;14) translocation in tumor 2385.
Metaphase chromosome spreads from tumor 2385 were hybridized with theIGH gene probes JH (A), Cα (B), and Cγ (C). The IGHlocus map is shown (D). Lines above the map indicate the target regions for hybridization by the JH, Cμ, Cα, and Cγ probes.
FISH analysis of the t(1;14) translocation in tumor 2385.
Metaphase chromosome spreads from tumor 2385 were hybridized with theIGH gene probes JH (A), Cα (B), and Cγ (C). The IGHlocus map is shown (D). Lines above the map indicate the target regions for hybridization by the JH, Cμ, Cα, and Cγ probes.
Mapping the translocation breakpoints by Southern blot analysis
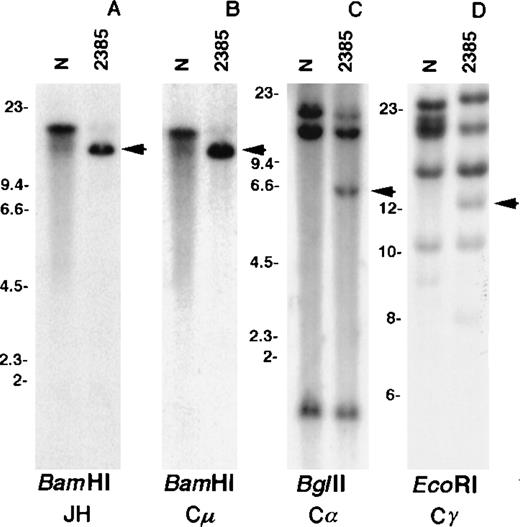
Southern blot analysis using the JH probe revealed a single rearranged band in the BamHI digest of tumor DNA (Figure2A). A band of the same size was detected by hybridization with the Cμ probe as expected for a productive rearrangement in a μ-chain producing tumor (Figure 2B). Consistent with the FISH data, a germline BamHI fragment containing JH and Cμ sequences was not detected, indicating a deletion of this region. Because the only copy of JH mapped to the der(1) chromosome, it was concluded that the translocation affected the productive IGHallele with a breakpoint centromeric to JH. No rearrangement was detected in Southern blot analysis using the Cε probe (data not shown), whereas the Cα (Figure 2C) and Cγ (Figure 2D) probes detected rearranged bands; these 2 rearrangements were selected for molecular cloning.
Southern blot analysis of tumor 2385 DNA.
DNA extracted from normal placenta (N) and tumor fluid (2385) were digested with the respective enzymes. The blots were hybridized with the IGH gene probes JH (A), Cμ (B), Cα (C), and Cγ (D). Arrows indicate rearranged bands in the tumor DNA. Size markers are given in kbp.
Southern blot analysis of tumor 2385 DNA.
DNA extracted from normal placenta (N) and tumor fluid (2385) were digested with the respective enzymes. The blots were hybridized with the IGH gene probes JH (A), Cμ (B), Cα (C), and Cγ (D). Arrows indicate rearranged bands in the tumor DNA. Size markers are given in kbp.
Cloning of the translocation breakpoints
A genomic library of the tumor DNA partially digested withSau3A was constructed and screened with the Cα and Cγ probes. Positive clones were analyzed by Southern blotting, FISH, restriction enzyme mapping, and partial sequencing. A Cα-positive phage clone containing a 6-kbp Cα-positive BglII fragment corresponding to the BglII rearranged band in the tumor DNA (Figure 2C) was isolated. These clones contained an intrachromosomal breakpoint linking IGHA1 switch (Sα1) sequences to theIGHD region downstream of the D3-3 segment (data not shown). The ensuing deletion included the JH and Cμ regions and, together with FISH data described above, established a nonproductive IGHallele on the karyotypically normal chromosome 14.
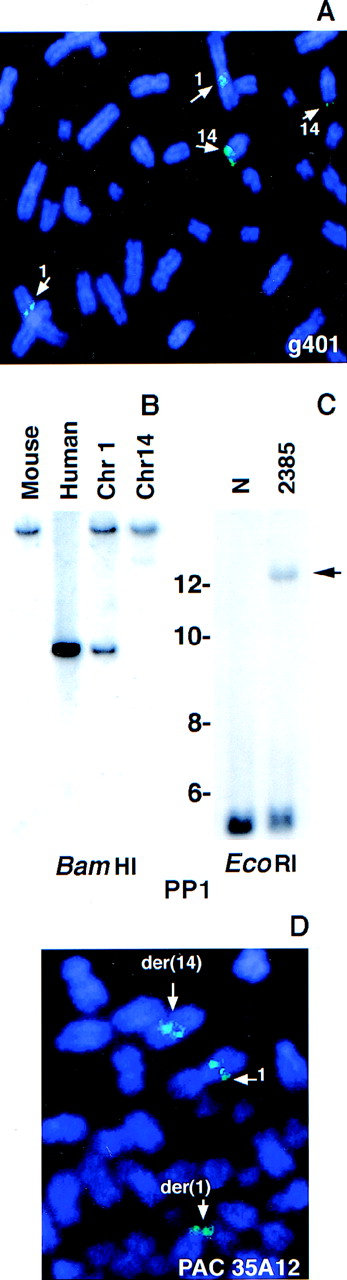
FISH of the Cγ-positive phage clone g401 to normal human metaphases revealed signals on both chromosomes 1 and 14 (Figure3A), confirming that this clone contained sequences from both chromosomes. A repeat-free, non-IGH, 1.1-kbp PstI–PstI fragment (PP1) was isolated from clone g401 and used as a probe in Southern blot analysis. Hybridization of this probe to a somatic cell hybrid DNA panel established that it was derived from chromosome 1 (Figure 3B). When hybridized to anEcoRI digest of tumor 2385 DNA, it recognized the same rearranged band as the Cγ probe (Figure 2D), confirming the clonal nature of the interchromosomal breakpoint in clone g401 (Figure 3C). PAC 35A12, which was obtained by screening a PAC library using g401-derived chromosome 1 sequences, hybridized to the normal chromosome 1 as well as to the der(1) and der(14) chromosomes in the tumor (Figure 3D).
Analysis of the breakpoint-containing clone g401.
Normal metaphase spreads were hybridized with the phage clone g401 (A). A repeat-free fragment derived from g401 (PP1) was hybridized to a somatic cell hybrid panel (B) and to tumor DNA (C). Arrow indicates the rearranged band in tumor DNA. Size markers are given in kbp. Tumor metaphase spreads were hybridized with a chromosome 1 PAC (PAC 35A12) that spanned the breakpoint (D).
Analysis of the breakpoint-containing clone g401.
Normal metaphase spreads were hybridized with the phage clone g401 (A). A repeat-free fragment derived from g401 (PP1) was hybridized to a somatic cell hybrid panel (B) and to tumor DNA (C). Arrow indicates the rearranged band in tumor DNA. Size markers are given in kbp. Tumor metaphase spreads were hybridized with a chromosome 1 PAC (PAC 35A12) that spanned the breakpoint (D).
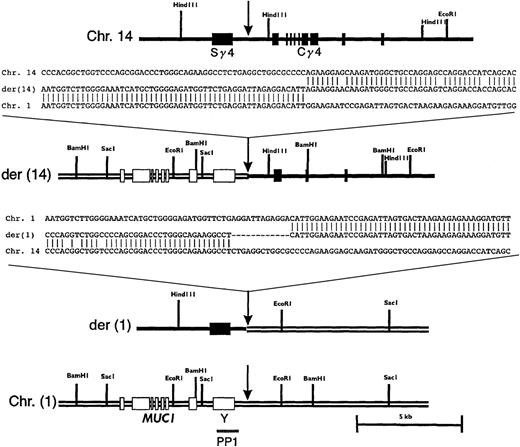
Restriction mapping and partial sequencing of the breakpoint region showed that the translocation occurred between Sγ4 and Cγ4 (Figure4). Sequence analysis revealed a 27-bp deletion in Cγ immediately 3′ of the breakpoint. An additional 1.5-kbp deletion, including exons 1-2 of Cγ, and a 51-bp duplication 5′ of exon 3 were also noted (data not shown). Partial deletion of the target for the Cγ probe is consistent with the lack of signal on der(14) with this probe in the FISH experiment described above. Restriction mapping and sequence analysis of subcloned PAC fragments showed that sequences from the 5′ part of clone g401 were colinear to the corresponding germline region of chromosome 1 (Figure4).
Restriction maps and sequences of the breakpoint regions on normal and derivative chromosomes.
The respective regions of germline chromosomes 1 and 14 are shown, with sites of breakpoint indicated by arrows. The location of PP1 is indicated. Sequences shown are chromosome 14, a germline IGHG4heavy chain switch region (GenBank accession number 56,796); chromosome 1; PAC 35A12; der(14); phage clone g401; and der(1), a PCR product amplified as detailed in “Materials and methods.”
Restriction maps and sequences of the breakpoint regions on normal and derivative chromosomes.
The respective regions of germline chromosomes 1 and 14 are shown, with sites of breakpoint indicated by arrows. The location of PP1 is indicated. Sequences shown are chromosome 14, a germline IGHG4heavy chain switch region (GenBank accession number 56,796); chromosome 1; PAC 35A12; der(14); phage clone g401; and der(1), a PCR product amplified as detailed in “Materials and methods.”
To clone the reciprocal breakpoint on the der(1) chromosome by PCR, a primer pair was designed such that a forward primer from chromosome 14 (an Sγ4 sequence) and a reverse primer based on the germline chromosome 1 sequence proximal to the breakpoint identified on the der(14) chromosome would amplify a DNA fragment from the tumor DNA. As expected, the PCR reaction did not yield a band with placental DNA, but a 930-bp band was amplified from the tumor DNA (data not shown). Sequence analysis of the PCR product revealed sequences colinear with Sγ4 and germline chromosome 1, as predicted for the reciprocal breakpoint region on the der(1) chromosome. A 12-bp deletion of Sγ4 sequence and a duplication of a CATT tetramer originating from chromosome 1 were found in the immediate vicinity of the breakpoint (Figure 4).
Identification of transcriptional units in the breakpoint region
Because IGH gene-associated translocations result in aberrant activation of genes immediately adjacent to the breakpoint, we sought to identify transcriptional units in the breakpoint region. Repeat-free restriction fragments derived from the breakpoint region on chromosome 1 were hybridized to human multiple tissue Northern blots. Northern hybridization analysis using the PP1 probe revealed a 2.7-kb transcript expressed predominantly in human brain (data not shown). A number of human and mouse cDNA clones homologous to the PP1 sequence were identified by searching EST databases. A more detailed sequence analysis showed that the PP1 fragment contained the last exon of the human homolog of the putative mouse gene Y. The partial sequences of both human and mouse homologs contained a continuing open-reading frame with no homology to any known protein. The exon contained in the PP1 fragment measured 1050-bp and did not account for the entire 2.7-kb transcript observed in the Northern hybridization analysis. The 5′ end of gene Y containing the ATG codon is presumably retained on the der(1) chromosome and remains to be cloned and characterized. Gene Y was not considered a candidate for an activated gene because its open-reading frame was disrupted by the t(1;14)(q21;q32) translocation in the tumor.
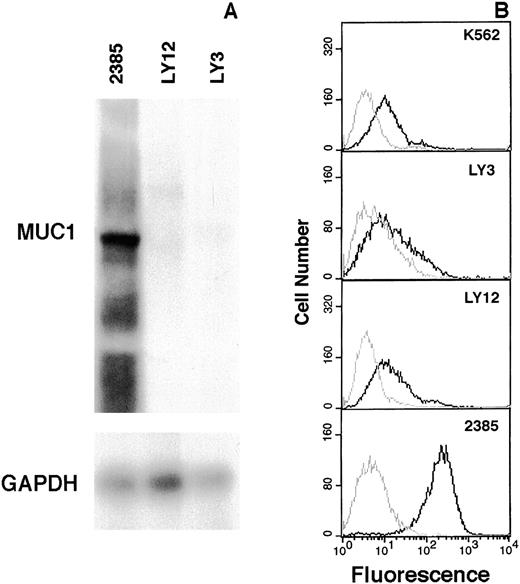
Immediately centromeric to gene Y, 2 genes, MUC1 andTHBS3,20 were identified. In tumor 2385, theTHBS3 gene, which encodes thrombospondin 3, a glycoprotein that mediates cell-to-cell and cell-to-matrix interactions, showed an mRNA expression level similar to that of the lymphoma cell lines (data not shown). The MUC1 gene encodes a high molecular weight glycoprotein containing multiple copies of a tandemly repeated 20-amino acid mucin-like domain.21,22 Allelic differences in the number of repeat units have been observed that account for differences in the size of MUC1 mRNA species (4kb-7kb).21 In tumor 2385, marked expression of a single mRNA species was observed in comparison to both lymphoma cell lines (Figure 5A). Flow cytometric analysis using a MUC-1 mucin-specific antibody revealed marked expression in tumor 2385, no expression in K562 cells as previously reported,23 and low levels in the lymphoma cell lines (Figure 5B). The 3′ end of MUC1 was located 2.4 kbp telomeric to the der(14) chromosomal breakpoint. MUC1, with a telomeric-to-centromeric direction of transcription, remained intact in the translocated allele. The presence of an immunoglobulin 3′ Cα enhancer sequence, presumably retained on the same chromosome, suggests a mechanism for MUC1 gene deregulation. Since MUC-1 mucin has previously been implicated in human cancer and found to be overexpressed in tumor 2385, it was considered to be the target gene deregulated and activated by the t(1;14)(q21;q32) translocation in this case.
Elevated expression of MUC-1 mucin in tumor 2385.
Total RNA extracted from tumor 2385 and lymphoma cell lines OCI-Ly 3 and OCI-Ly 12 were hybridized with an MUC1 tandem repeat probe and subsequently with GAPDH to control for loading (A). Expression of MUC-1 mucin in K562, OCI-Ly 3, OCI-Ly 12, and tumor 2385 cells by flow cytometric analysis (B). Live cells were incubated either with normal mouse serum (gray line) or a MUC-1 mucin antibody (HMFG-2) (black line) and quantitated by FACS analysis. MUC-1 mucin staining is plotted as a histogram.
Elevated expression of MUC-1 mucin in tumor 2385.
Total RNA extracted from tumor 2385 and lymphoma cell lines OCI-Ly 3 and OCI-Ly 12 were hybridized with an MUC1 tandem repeat probe and subsequently with GAPDH to control for loading (A). Expression of MUC-1 mucin in K562, OCI-Ly 3, OCI-Ly 12, and tumor 2385 cells by flow cytometric analysis (B). Live cells were incubated either with normal mouse serum (gray line) or a MUC-1 mucin antibody (HMFG-2) (black line) and quantitated by FACS analysis. MUC-1 mucin staining is plotted as a histogram.
Translocation breakpoint cluster near MUC1 in B-cell lymphoma
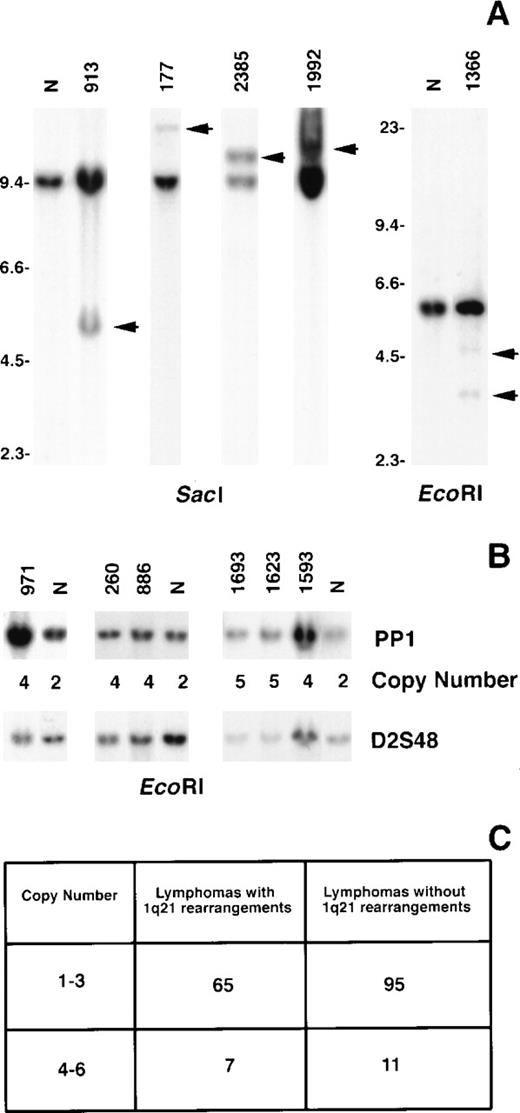
The incidence of MUC1 rearrangements was ascertained by Southern blot analysis of 2 groups of B-cell lymphoma using the PP1 fragment as the probe. One group comprised 72 biopsies that included all histologic types (small lymphocytic, follicular, diffuse small noncleaved, diffuse large cell, and immunoblastic), each of which exhibited a rearrangement affecting band 1q21 by G-banding analysis. In this group, 4 tumors (6%) exhibited MUC1 DNA rearrangements (Figure 6A). These comprised 1 each of diffuse small cleaved (177), follicular mixed (1992), immunoblastic (1366), and extranodal ascitic (2385) lesions. Case 1992 was a relapsed tumor, whereas the remaining were ascertained at presentation. The second group comprised 106 biopsies. Of these, 12 had follicular histologies and 94 were diffuse large-cell lymphomas that were either cytogenetic failures (27 cases), yielded only normal metaphases (7 cases), or were clonally abnormal but without a 1q21 aberration (72 cases). In this group, 1 diffuse mixed lymphoma that exhibited only a normal karyotype (913) showed a MUC1 rearrangement (Figure 6A).
Rearrangement and increased copy number of MUC1in B-cell NHL.
Tumor DNA was digested with the respective enzymes and hybridized with the PP1 probe. Rearranged bands in the tumor DNA compared to normal placenta (N) are indicated by arrows (A). Size markers are given in kbp. The calculated copy numbers of MUC1 are listed relative to normal placenta (N) using the hybridization signal from the probe D2S48 to control for loading (B).14 The numbers of tumors in each study group with 1 to 3 (normal) and 4 to 6 (increased) copy numbers ofMUC1 are shown (C).
Rearrangement and increased copy number of MUC1in B-cell NHL.
Tumor DNA was digested with the respective enzymes and hybridized with the PP1 probe. Rearranged bands in the tumor DNA compared to normal placenta (N) are indicated by arrows (A). Size markers are given in kbp. The calculated copy numbers of MUC1 are listed relative to normal placenta (N) using the hybridization signal from the probe D2S48 to control for loading (B).14 The numbers of tumors in each study group with 1 to 3 (normal) and 4 to 6 (increased) copy numbers ofMUC1 are shown (C).
Increased copy number of MUC1 in B-cell lymphoma
We also assayed for copy number changes of MUC1 in the 2 groups of B-cell lymphoma biopsies evaluated for rearrangements. Seven (10%) in the first group and 11 (10%) in the second group showed 4 to 6 copies of MUC1 (Figures 6B, 6C). We have previously found that this level of copy number increase represents either nondisjunctional gain of the chromosomal region or a low level of amplification of the gene.14
Discussion
The third most common site of chromosomal rearrangement in B-cell lymphoma after bands 14q32 and 18q21 is band 1q21. This site exhibits a promiscuous pattern of rearrangement involving multiple other chromosomal sites.1,2 Translocations, duplications, and amplifications affecting 1q21 are usually, but not exclusively, seen in lymphomas in addition to primary abnormalities such as t(14;18)(q32;q21) and t(8;14)(q24;q32), suggesting that 1q21-associated abnormalities may play a role in tumor progression.1-3Rearrangements involving this chromosomal site have also been noted in other tumor systems; thus, the gene(s) involved in 1q21 rearrangements may play an important role in tumor biology per se. Previous studies have identified BCL9 as a gene possibly deregulated in 1q21-associated translocations. However, only 1 lymphoma with a DNA rearrangement involving this gene was identified,5indicating the presence of additional gene(s) that may be more relevant in lymphoma translocations. We report here the identification ofMUC1 as a gene recurrently involved in 1q21-associated rearrangements in B-cell lymphomas.
In tumor 2385, the t(1;14)(q21;q32) translocation resulted in linkage of the Sγ4 region to the region 3′ of the MUC1 gene, 8 cM distal to BCL9. Interestingly, the IGH allele involved in the translocation also was the productively rearranged allele, an event not previously reported. The karyotypically normal chromosome 14 in this tumor did in fact have a deletion that resulted in a nonproductive IGH allele. Sequence analysis of the cloned der(14) chromosomal breakpoint revealed that it was located within a poorly characterized gene Y. Based on the transcript size and direction of transcription of gene Y, it was predicted that the translocation would result in disruption of this gene. Hence, geneY was discounted as a candidate for the gene activated in the translocation. The transcriptional unit of the immediately adjacentMUC1 gene was found to be intact in the rearranged allele and thus became a candidate for the involved gene. Indeed, the steady state levels of MUC1 mRNA and protein in this tumor were elevated compared to lymphoma cell lines that did not exhibit a 1q21 rearrangement. Presumably, this elevated MUC1 expression resulted from aberrant activation by the 3′ Cα enhancer sequence retained on the der(14) chromosome.
We show that the MUC1 region is rearranged in 6% of tumors with a documented 1q21 cytogenetic aberration. Thus, although recurring, this molecular breakpoint does not account for all the 1q21-associated cytogenetic breakpoints. We also found increased copy numbers of the MUC1 region in 10% of B-cell lymphomas of all histologies. The amplification itself was of a relatively low level (4 to 6 copies), and its effect on MUC1 expression remains to be determined. Of significance, however, is the observation that up to 16% of B-cell lymphomas show a molecular perturbation of theMUC1 region that can potentially lead to its deregulated overexpression. The clinical significance of these perturbations remains to be established.
The MUC-1 mucin protein can be expressed as a transmembrane or secreted protein as a result of alternate splicing. Short MUC-1 mucin isoforms, MUC-1/Y and MUC-1/Z, devoid of tandem repeats, have also been described.16,24 MUC-1 mucin has previously been implicated in human carcinogenesis. It is frequently overexpressed in both epithelial7-10 and nonepithelial tumors,16,23,25 and is associated with cell invasiveness and poor outcome. An important role for MUC1 in tumorigenesis has been demonstrated in Muc-1 null mice, which show a delayed progression of primary tumors.26 Therefore, a role forMUC1 in the progression of NHL is consistent with 1q21 as a site of secondary chromosomal abnormalities in these tumors. Although the precise role of MUC-1 mucin protein in tumorigenesis is not well understood, a few putative functions have been proposed that may confer cells overexpressing MUC-1 mucin a selective advantage. MUC-1 mucin has been shown to inhibit cell adhesion mediated by integrin and E-cadherin that may facilitate metastatic expansion of tumors.27 High serum levels of the secreted form of MUC-1 mucin, frequently found in patients with epithelial tumors, can suppress T-cell proliferative responses blocking T cells in an “anergic” state.28The MUC-1/Y isoform has a cytokine receptor-like domain and has been suggested to play a role in signal transduction.29
The mechanisms shown to date that are responsible for MUC-1 mucin overexpression in epithelial tumors include gene amplification30 or activation by transcription factors.31 We now show deregulation of MUC1expression by chromosomal translocation. Translocations affecting band 1q21 are frequent in leukemias, lymphomas, sarcomas, and epithelial tumors,1-4 which may indicate a previously unrecognized mechanism for MUC1 deregulation in these tumor types. Aberrant, high expression of MUC-1 mucin on the surface of cancer cells makes it an attractive target gene for immunotherapy of epithelial tumors.32 33 The possibility that its expression may also be frequently deregulated in NHL makes it a target for immunotherapeutic approaches for the treatment of lymphomas.
Supported by National Institutes of Health–National Cancer Institute grants CA34775 and CA66999 (R.S.K.C.) and CA52477 (K.O.L.).
Reprints:R. S. K. Chaganti, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10021; e-mail:chagantr@mskcc.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal