Abstract
The GATA-1 transcription factor is capable of suppressing the myeloid gene expression program when ectopically expressed in myeloid cells. We examined the ability of GATA-1 to repress the expression and function of the PU.1 transcription factor, a central regulator of myeloid differentiation. We found that GATA-1 is capable of suppressing the myeloid phenotype without interfering with PU.1 gene expression, but instead was capable of inhibiting the activity of the PU.1 protein in a dose-dependent manner. This inhibition was independent of the ability of GATA-1 to bind DNA, suggesting that it is mediated by protein-protein interaction. We examined the ability of PU.1 to interact with GATA-1 and found a direct interaction between the PU.1 ETS domain and the C-terminal finger region of GATA-1. Replacing the PU.1 ETS domain with the GAL4 DNA-binding domain removed the ability of GATA-1 to inhibit PU.1 activity, indicating that the PU.1 DNA-binding domain, rather than the transactivation domain, is the target for GATA-1–mediated repression. We therefore propose that GATA-1 represses myeloid gene expression, at least in part, through its ability to directly interact with the PU.1 ETS domain and thereby interfere with PU.1 function.
Blood formation, or hematopoiesis, is the regulated development of at least 8 distinct cellular lineages from a common precursor, the hematopoietic stem cell. This process involves fundamental changes in gene expression, resulting in each cell type expressing a characteristic complement of genes necessary for its function. This is achieved through the action of transcriptional regulators with both general and restricted expression patterns in the hematopoietic system. A particular cell type will express a subset of transcription factors characteristic for the lineage, and these will positively regulate genes specifically expressed in this lineage. Thus, myeloid cells express PU.1 and CCAAT/enhancer binding proteins (C/EBPs), which together with the more widely expressed factors AML1, Ets-1, and c-Myb, activate myeloid-specific promoters, such as the murine neutrophil elastase, granulocyte colony-stimulating factor (G-CSF), macrophage (M)-CSF, and GM-CSF promoters.1 In eosinophils, which express GATA-1 and C/EBP, the EOS47 promoter is activated by these factors in collaboration with c-Myb and Ets-1/Fli-1.2 Erythroid- and thrombocyte-specific genes are regulated by GATA factors, erythroid Kruppel-like factor, and Maf family members, which are abundantly expressed in these cells.3 4
However, it is becoming clear that another level of regulation is superimposed on this pattern of combinatorial activation, which involves repression of genes specific for one lineage by transcription factors promoting other lineages. This is illustrated by the repression of erythroid-specific genes, as well as erythroid differentiation, by the MafB protein, which in the hematopoietic system is expressed at high levels only in myeloid cell types. This is due to the ability of MafB to interact with and repress the activity of Ets-1 on erythroid-specific promoters, such as that of the transferrin receptor gene.5 An even more striking example is the incompatibility of the myeloid phenotype with GATA-1 expression. Ectopic GATA-1 expression in chicken myeloid cell lines, which do not express this factor, leads to transdifferentiation into eosinophils (at intermediate GATA-1 levels) or retrodifferentiation into multipotent progenitors (at high GATA-1 levels).6 Similar reprogramming of the 416B murine myeloid cell line by GATA-1 has been reported.7These results strongly indicate that down-regulation of GATA-1, which is expressed at high levels in multipotent myeloid-erythroid precursor cells,6 8 is a prerequisite for the proper execution of the myeloid gene expression program. However, the molecular mechanism behind the ability of GATA-1 to suppress myeloid gene expression has not been established.
The antagonism between transcription factors driving competing differentiation programs may provide a developmental switch in the choice between two lineages, because up-regulation of one program automatically leads to repression of the other, thereby rendering lineage commitment irreversible and preventing ectopic expression of lineage-specific genes. However, these mechanisms may also be important in understanding the differentiation blocks that contribute to many types of leukemia. A correlation between expression of GATA-1 and a bad prognosis has been found in myeloid leukemias,9 suggesting that the inability to extinguish GATA-1 expression may be part of the malignant phenotype in some types of immature acute myeloid leukemia. Conversely, in the spleen focus-forming virus mouse erythroleukemia model, about 95% of the malignant clones have ectopically activated the PU.1 locus through retroviral insertion.10 This leads to a block of differentiation at the proerythroblast stage, a block similar to that observed in differentiating erythroid GATA-1- cells,11,12 suggesting a mutually antagonistic relationship between the 2 factors. This was further substantiated by the finding that PU.1 could directly inhibit the activity of GATA-1 in erythroid cells through direct protein-protein interaction.13 Also consistent with this idea, GATA-1 expression is downregulated during PU.1-mediated myeloid lineage commitment of multipotent progenitors,14 and GATA-1–mediated reprogramming of chicken myeloid cells leads to extinction of endogenous PU.1 gene expression.2
We previously observed down-regulation of myeloid-specific gene expression after expression of GATA-1 in chicken myeloid cells.6 The most crucial factor known to be required for myeloid differentiation in vivo is PU.1.15,16 Furthermore, expression of PU.1 in multipotent progenitor cells is sufficient to mediate their commitment to the myeloid lineage,14suggesting a role for PU.1 in establishing and maintaining the myeloid phenotype. We therefore addressed the role of PU.1 in repression of myeloid gene expression by GATA-1. We found that a decrease in PU.1 gene expression is not a prerequisite for down-regulation of myeloid markers upon activation of a GATA-1–estrogen receptor (GER) fusion in chicken myeloblasts. Rather, the activity of the PU.1 protein was directly inhibited by GATA-1 in a dosage-dependent manner. This inhibition did not require the ability of GATA-1 to bind DNA, indicating that it takes place through protein-protein interaction. We found that the highly conserved C-terminal zinc finger of GATA-1 was sufficient to mediate direct interaction between GATA-1 and the PU.1 ETS domain, which is the DNA-binding domain of the factor. Replacing the PU.1 ETS domain with the yeast GAL4 DNA-binding domain rendered PU.1-mediated transactivation insensitive to GATA-1 repression. We therefore propose that negative regulation of PU.1 activity by GATA-1 takes place through direct interaction of the C-finger of GATA-1 with the PU.1 ETS domain.
Materials and methods
Generation of DNA constructs
GATA-1 expression constructs were based on the cytomegalovirus (CMV) expression vector pSPCMV (H.K., unpublished data). The chicken GATA-1 coding sequence was excised from pNEO-GATA-1 expression vector17 with ClaI and EcoRI and inserted into pBluescript SK- (Stratagene, La Jolla, CA). From this vector, a BamHI-XbaI fragment was excised and inserted into pSPCMV to generate pSPCMV-GATA-1. Deletion mutants were obtained by in-frame deletion of an XmnI fragment encoding amino acids 227 to 299 (D3′227) or by using polymerase chain reaction (PCR) to generate an NcoI site at GATA-1 amino acid 71 (a methionine) and deleting the NcoI fragment encoding amino acids 1-70 (D5′70). For the double-deletion mutant (D5′,3′), the 2 deletions were combined. Point mutation of the zinc fingers was done using overlapping PCR. For the N-finger mutant (cysteines 131 and 134 to serines), the outer primers were 5′-GCC ACC CCC ATG GAG CC and 5′-TTT GCG GTT TCG GGG TTT GG and the mutagenesis primers 5′-GCC CGG AGG CGT TGC CCA G and 5′-CTG GGC AAC GCC TCC GGG C. For the C-finger mutant (cysteines 164 and 167 to serines), the same outer primers were used, and the mutagenesis primers were 5′-CTG GCT GTT GCT GGA CAC TGT and 5′-ACA GTG TCC AGC AAC AGC CAG. In both cases, the product was used to replace the internalNcoI-HincII fragment of the GATA-1 coding sequence.
GST-GATA-1 fusion protein expression vectors were generated by PCR amplification using Pfu polymerase (Stratagene) and the following primers:
N-finger (amino acids 91 to 150): NF5: 5′-GTG GAA TTC TCC TCC GGG CCC CTA CTG and NF3′: 5′-GCG CTC GAG TCA GGG GCG GAT GAG CGG GCG.
C-finger (amino acids 143-206): CF5: 5′-GTG GAA TTC CAG AAC CGC CCG CTC ATC and CF3: 5′-GCG CTC GAG TCA GTC TTT GCG CAT CGT GAG.
N-terminus+N-finger (amino acids 1-150): G15: 5′-GTG GAA TTC ATG GAG TTC GTG GCG CTG GGG and NF3.
N-finger+C-finger: NF5 and CF3.
C-finger+C-terminus (amino acids 143-304): CF5 and G13: GCG CTC GAG TCA AAT CTG CGG GCT CAG CCC.
Templates were XbaI-linearized pSPCMV-GATA-1 or point-mutated versions thereof, as appropriate. PCR products were cloned into pGEX-4T-1 (Amersham Pharmacia, Uppsala, Sweden) using EcoRI andXhoI.
The pNEO-GATA-1-ER251 expression vector has been previously described.6 The pNEO-GATA-1282 expression vector was derived from pNEO-GATA-1-ER251 by replacing theBamHI-EcoRI fragment encoding the human estrogen receptor ligand binding domain with that derived from pHE14.18 The GATA-1-ER251 was used initially, but the GATA-1-ER282 was subsequently found to be more efficient in induction of eosinophil gene expression. The 2 forms are equally efficient in suppression of myeloid gene expression (H.K., unpublished data).
The human PU.1 expression vector pCMV-MTPU.1 and derivatives pGDPU1-170 as well as the pPU3-TK-LUC reporter have been described.14 The pPUDC198 plasmid was generated by PCR with Pfu polymerase using EcoRI-linearized pCMV-MTPU.1 as template and the following primers: PU.1 5′: 5′-GGA GGG CGC GCC GAT GTT ACA GGC GTG CAA AAT G; PUDC198 3′: 5′-GCG CTT AAT TAA TCA GGT GCC CTT GTC CTT GTC CAC. The product was digested with AscI and PacI and used to replace the PU.1 sequence in pCMV-MTPU.1. The pCMV-MTPU1-170GD plasmid, encoding the PU.1-GAL4 hybrid activator, was constructed by PCR amplifying the PU.1 coding sequence (using pCMV-MTPU.1 as template) with the PU.1 5′ primer described above and the PU170 3′ primer: 5′-GAG GAA TTC GAT CTT CTT CTT GCT GCC. The GAL4 DNA-binding domain was amplified (using pGD-PU1-170 as template) with 5′-GAG GAA TTC ATG AGC TAC TCT TCT TCT and 5′-GCG CTT AAT TAA TCA CGA TAC AGT CAA CTG TCT. The PU.1 fragment was digested withAscI and EcoRI, the GAL4 fragment with EcoRI and PacI, and these 2 used to replace theAscI-PacI fragment of pCMV-MTPU.1, yielding pCMV-MTPU1-170GD. The pGATA-1P-LUC reporter plasmid was generated by PCR amplification of the chicken GATA-1 promoter19 using the following primers: GATA-P 5′-GAG CTC GAG CAT GGC TGT CCC TGA TAA GCA and GATA-P 3′-GAG CGA AGC TTG GGG GGG GGC CTG AAA GAA. The resulting product was cloned into the pGL2-basic vector (Promega, Madison, WI) using XhoI and HindIII.
Cell lines and tissue culture
HD50M and HD57M myeloblasts8 and GATA-1-ER–expressing derivatives thereof were grown in blastoderm medium composed of DMEM supplemented with 10% fetal calf serum, 2.5% chicken serum, 0.15% NaHCO3, 56 μg/mL conalbumin, 80 μM 2-mercaptoethanol, 0.9 mg/mL insulin, and the standard complement of antibiotics at 37°C in 5% carbon dioxide. Medium was supplemented with approximately 10 units/mL of recombinant chicken myelomonocytic growth factor.20 Stable GATA-1-ER251–expressing HD50M and GATA-1-ER282–expressing HD57M myeloblast cell lines were generated by electroporation and G418 selection as described.6 For induction, media were supplemented with 1 μM of β-estradiol from a 10-mM stock dissolved in ethanol.
Indirect immunofluorescence and flow cytometry
Western blotting
Western blotting was performed on total cell lysates of transfected cells transferred to Immobilon polyvinylidine difluoride membranes (Millipore, Bedford, MA) using anti-GATA–1 antiserum (ref. 6; kindly provided by Dr T. Evans and used at a 1:2000 dilution) and the 9E10 monoclonal antibody (a gift from Dr P. Orban; used at a 1:2000 dilution). Secondary antibodies were horseradish peroxidase–coupled antimouse immunoglobulin and antirabbit immunoglobulin (Amersham Pharmacia, Uppsala, Sweden). Blots were developed using enhanced luminescence (ECL) (Amersham Pharmacia, Uppsala, Sweden).
RNA extraction and Northern blotting
Total cellular RNA was prepared according to Chomczynski and Sacchi.23 After electrophoresis through a 1.2% formaldehyde-agarose gel, RNA was transferred to Duralose (Stratagene, La Jolla, CA) by capillary blotting and hybridized to chicken PU.1 (a kind gift from Dr Jaques Ghysdael), MHC class II γ chain,6 β-actin,24 and GAPDH25complementary DNAs 32P-labeled by random priming.
Transfection and reporter gene assays
Transient transfection into Q2bn fibroblasts was performed using calcium phosphate coprecipitation and luciferase and β-galactosidase activities assayed as previously described using pRSV-βgal as an internal control plasmid.26
In vitro translation
In vitro translation and 35S-labeling was performed using the TNT T7 Quick system (Promega, Madison, WI) and35S-ProMix (Amersham Pharmacia, Uppsala, Sweden) according to the instructions provided by the manufacturer.
GST fusion protein preparation and GST pulldown analysis
GST (from pGEx4T-1) and GST-GATA-1 fusion proteins were expressed in XL-1 BLUE (Stratagene, La Jolla, CA) containing the pRI952 plasmid (kindly provided by SmithKline Beecham, King of Prussia, PA) essentially as described.2 Briefly, overnight cultures were diluted 1:10 into LB medium containing 100 μg/mL of ampicillin and 50 μg/mL of chloramphenicol. After shaking at 30°C for 1 hour, isopropyl thiogalactose was added to a final concentration of 0.4 mM and induction allowed to proceed for 2 hours, after which fusion proteins were extracted and batch-purified on glutathione sepharose (Pharmacia). After washing, beads were resuspended in NETN buffer (10 mM Tris, pH 7.5; 100 mM NaCl; 1mM EDTA; 0.5% NP40). Pulldown reactions were carried by preincubating 75-μL bead suspension in NETN (corresponding to about 500-ng fusion protein) with 50 μL of 10% bovine serum albumin and 375 μL of pulldown buffer (20 mM Tris, pH 7.5; 250 mM NaCl; 0.5% NP40; 1mM EDTA; 0.1 mM ZnCl2; 1 mM DTT) for 30 minutes with gentle shaking at room temperature before adding 3 to 5 μL of 35S-labeled in vitro–translated protein. Incubation was continued for 1 hour, beads washed 3 times in NETN, boiled in sodium dodecyl sulfate (SDS) sample buffer, and eluted proteins run on 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gels. After drying the gels, radiolabeled proteins were detected on a Fuji BAS2500 Phosphorimager.
Results
GATA-1 down-regulates myeloid-specific antigens without repressing PU.1 gene expression
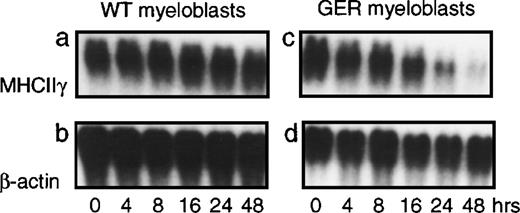
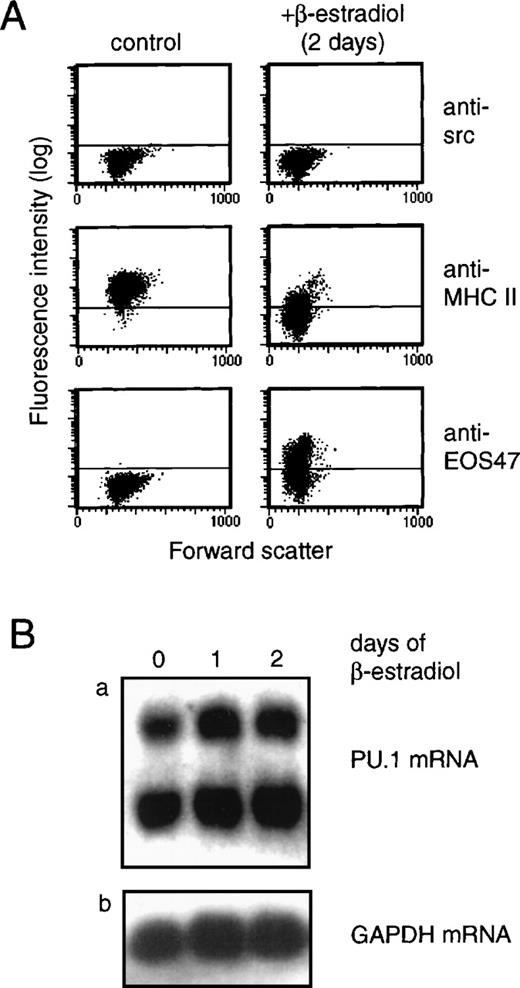
The ability of GATA-1 to directly suppress myeloid gene expression was analyzed by activation of a conditional GATA-1 allele, in this case a fusion between the chicken GATA-1 protein and the hormone-binding domain of the human estrogen receptor (GER). If this fusion is introduced into chicken myeloblasts, no effect is observed in the absence of estrogen agonists. However, upon addition of β-estradiol, transdifferentiation toward the eosinophil lineage—involving a rapid down-regulation of myeloid gene expression as well as up-regulation of the eosinophil-specific EOS47 antigen—is observed. An example of this is shown in Figure 1, where GER-expressing and control myeloblasts were exposed to β-estradiol followed by extraction of total RNA at the indicated time points. Expression of the γ chain of the MHC class II antigen (MHCIIγ), which in the nonlymphoid compartment is expressed specifically in myeloid cell types,21 was assayed by Northern blotting. This showed down-regulation of MHCIIγ expression in GER-expressing, but not control, myeloblasts. To determine if down-regulation of PU.1 gene expression was a requirement for the observed repression, we analyzed PU.1 mRNA expression upon induction of the GER fusion in chicken myeloblasts (Figure 2). Here, activation of the GER chimera led to up-regulation of the eosinophil-specific EOS47 surface antigen, concomitant with the down-regulation of myeloid-specific markers, such as the MHC class II (Figure 2A) and MYL51/2 (not shown) antigens, as demonstrated by indirect immunoflourescence using specific monoclonal antibodies. After 2 days, greater than 90% loss of MHCII expression was observed, consistent with the down-regulation of the mRNA observed in Figure 1. However, during this process, PU.1 messenger RNA (mRNA) levels were maintained (as measured relative to the β-actin internal control; Figure 2B), demonstrating that repression of myeloid genes does not require extinction of PU.1 gene expression.
GATA-1 down-regulates the myeloid phenotype without repressing PU.1 gene expression.
HD50M myeloblasts containing a GATA-1–estrogen receptor (GER) fusion (right panels) as well as control myeloblasts (left panels) were treated with 1 μM of β-estradiol or left untreated, as indicated. Total RNA was extracted from cells at the indicated time points, and 10-μg aliquots were subjected to Northern blot analysis. The blot was probed sequentially with probes for chicken MHC class II γ chain and β-actin.
GATA-1 down-regulates the myeloid phenotype without repressing PU.1 gene expression.
HD50M myeloblasts containing a GATA-1–estrogen receptor (GER) fusion (right panels) as well as control myeloblasts (left panels) were treated with 1 μM of β-estradiol or left untreated, as indicated. Total RNA was extracted from cells at the indicated time points, and 10-μg aliquots were subjected to Northern blot analysis. The blot was probed sequentially with probes for chicken MHC class II γ chain and β-actin.
PU.1 gene expression is not affected during GER-mediated down-regulation of myeloid markers.
(A) Expression of myeloid-specific (MHCII) and eosinophil-specific (EOS47) antigens was monitored after β-estradiol stimulation of GER-expressing myeloblasts by indirect immunofluorescence staining and flow cytometry. The time point shown represents 2 days of induction, but significant changes were observed after 18 hours (not shown). (B) PU.1 gene expression was analyzed in the same cells as in (A) by Northern blotting as described in Figure 1. RNA was isolated after 1 and 2 days of induction and compared to the level in uninduced cells (0 days of induction) (panel a). Two species of PU.1 mRNA are found in avian cells.2 To normalize RNA levels, the blot was probed for GAPDH as an internal standard (panel b).
PU.1 gene expression is not affected during GER-mediated down-regulation of myeloid markers.
(A) Expression of myeloid-specific (MHCII) and eosinophil-specific (EOS47) antigens was monitored after β-estradiol stimulation of GER-expressing myeloblasts by indirect immunofluorescence staining and flow cytometry. The time point shown represents 2 days of induction, but significant changes were observed after 18 hours (not shown). (B) PU.1 gene expression was analyzed in the same cells as in (A) by Northern blotting as described in Figure 1. RNA was isolated after 1 and 2 days of induction and compared to the level in uninduced cells (0 days of induction) (panel a). Two species of PU.1 mRNA are found in avian cells.2 To normalize RNA levels, the blot was probed for GAPDH as an internal standard (panel b).
GATA-1 represses PU.1 activity
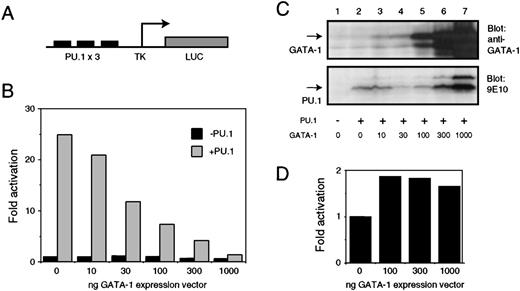
To test if the activity of the PU.1 protein was affected by the presence of GATA-1, we used a PU.1-responsive reporter construct containing 3 PU.1 binding sites upstream of the herpes simplex virus minimal promoter (pPU3-TK-LUC; Figure 3A). When cotransfected with a PU.1 expression vector (pCMV-MTPU.1) into Q2bn fibroblasts, activation of this reporter construct is observed; this activation decreased in a dose-dependent manner when increasing amounts of GATA-1 expression vector (pSPCMV-GATA-1) was included in the transfection (Figure 3B). No repression of basal promoter activity by GATA-1 was observed in the absence of PU.1. The amount of GATA-1 expressed was monitored by Western blotting with an anti–GATA-1 antibody (Figure 3C, upper panel). The lack of PU.1 transactivation was not due to an absence of PU.1 protein expression. Western blotting of extracts from transfected cells (using the 9E10 monoclonal antibody, which recognizes the Myc tag on the PU.1 protein) showed PU.1 levels in cells expressing GATA-1 equivalent to, or higher than, those found in the absence of GATA-1 (Figure 3C, lower panel). That high levels of GATA-1 are not by themselves repressive was tested by observing the response of the chicken GATA-1 promoter (which is itself activated by GATA-119) to increasing levels of GATA-1 protein. The promoter is activated (about 2-fold) even by low levels of cotransfected GATA-1, and no significant decrease in this activation is observed when GATA-1 levels are increased (Figure 3D).
Inhibition of PU.1 activity by GATA-1.
(A) Structure of the PU.1 responsive pPU3-TK-LUC luciferase reporter. (B) pPU3-TK-LUC (1 μg) was cotransfected into Q2bn fibroblasts in the presence (solid bars) or absence (hatched bars) of 0.25 μg of CMV–based PU.1 expression vector (pCMV-MTPU.1) or empty pcDNAI expression vector as indicated. Increasing amounts of GATA-1 expression vector (pSPCMV-GATA-1) were titrated into the experiment. Empty pSPCMV expression vector was added to a total of 1 μg. The luciferase activity was measured and is shown normalized to the β-galactosidase activity obtained from the pRSV-βgal internal control plasmid (0.25 μg); the values are expressed relative to that obtained with pPU3-TK-LUC in the presence of empty expression vectors only (lane 1). The data shown are the average of 2 determinations from a representative experiment. (C) Equal amounts of total cell lysate (corresponding to about 1 × 105 cells) from cells transfected as in (A) were electrophoresed on a 12% SDS-PAGE gel and subjected to Western analysis with anti–GATA-1 antiserum (panel a) or 9E10 monoclonal antibody (detecting the MycTag on the PU.1 protein; panel b). Arrows indicate the positions of the full-length GATA-1 and PU.1 proteins. (D) pGATA-1P-LUC (1 μg) was cotransfected with the indicated amounts of pSPCMV-GATA-1 expression vector and pRSV-βgal internal control plasmid (0.25 μg) and fold activation calculated as described in (B).
Inhibition of PU.1 activity by GATA-1.
(A) Structure of the PU.1 responsive pPU3-TK-LUC luciferase reporter. (B) pPU3-TK-LUC (1 μg) was cotransfected into Q2bn fibroblasts in the presence (solid bars) or absence (hatched bars) of 0.25 μg of CMV–based PU.1 expression vector (pCMV-MTPU.1) or empty pcDNAI expression vector as indicated. Increasing amounts of GATA-1 expression vector (pSPCMV-GATA-1) were titrated into the experiment. Empty pSPCMV expression vector was added to a total of 1 μg. The luciferase activity was measured and is shown normalized to the β-galactosidase activity obtained from the pRSV-βgal internal control plasmid (0.25 μg); the values are expressed relative to that obtained with pPU3-TK-LUC in the presence of empty expression vectors only (lane 1). The data shown are the average of 2 determinations from a representative experiment. (C) Equal amounts of total cell lysate (corresponding to about 1 × 105 cells) from cells transfected as in (A) were electrophoresed on a 12% SDS-PAGE gel and subjected to Western analysis with anti–GATA-1 antiserum (panel a) or 9E10 monoclonal antibody (detecting the MycTag on the PU.1 protein; panel b). Arrows indicate the positions of the full-length GATA-1 and PU.1 proteins. (D) pGATA-1P-LUC (1 μg) was cotransfected with the indicated amounts of pSPCMV-GATA-1 expression vector and pRSV-βgal internal control plasmid (0.25 μg) and fold activation calculated as described in (B).
GATA-1 DNA binding is not required for repression of PU.1 function
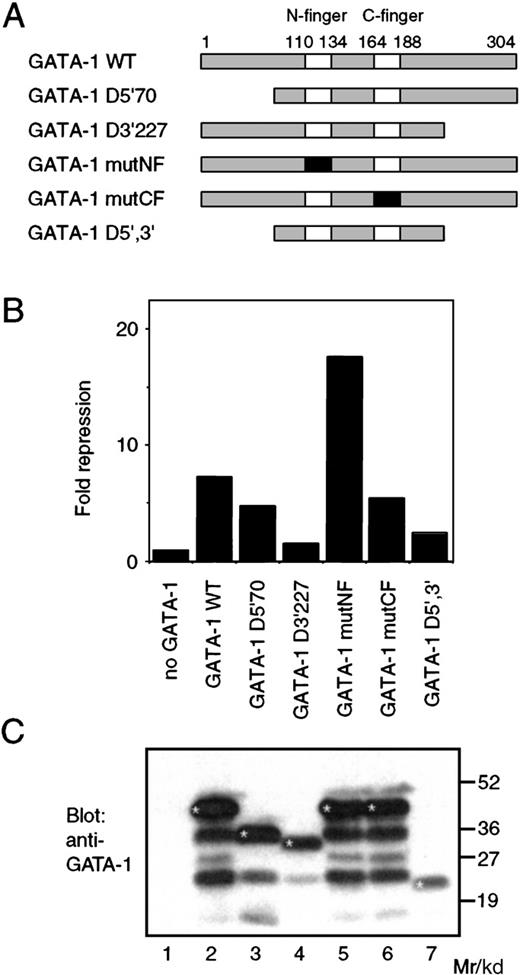
To address the mechanism involved, we analyzed the ability of various mutants of GATA-1 to repress PU.1 activity. The GATA-1 protein consists of a zinc finger domain containing 2 Cys4-type zinc fingers (designated the N-terminal (N-) finger and C-terminal (C-) finger, respectively). This domain is highly conserved between GATA family members, whereas the flanking regions are more variable.27 The C-finger is necessary and sufficient for binding to the WGATAR consensus sequence. The N-finger mediates interaction with the FOG (friend of GATA-1) family of GATA cofactors and stabilizes GATA-1 DNA binding.28-30 We introduced point mutations and deletions into the GATA-1 protein, as outlined in Figure4A. Deletions were introduced that remove the N-terminal 70 amino acids (D5′/70) and C-terminal 77 amino acids (D3′/227). The zinc fingers were mutated by altering 2 of the zinc coordinating cysteines to alanine (generating mutNF and mutCF, respectively), disrupting the finger secondary structures but leaving the primary protein sequence largely undisturbed. All of the GATA-1 mutants, except for the C-finger mutant (mutCF), were capable of binding to the WGATAR consensus sequence (data not shown), as expected from previously published results.31 We used cotransfection experiments in the Q2bn cell line to analyze the GATA-1 mutants for their ability to repress PU.1-mediated activation (Figure 4B). Interestingly, mutation of the C-finger does not reduce repression significantly, and mutation of the N-finger actually increased repression. GATA-1 molecules in which the C-terminal domain (amino acids 227-304) had been deleted showed a severe reduction in their ability to mediate repression. These results showed that no correlation exists between the ability of GATA-1 to bind DNA and its ability to repress PU.1 function. The lack of repression by both the C-terminal deletion mutant and the D5′,3′ mutant may be a function of protein instability, because Western blot analysis of extracts from transfected cells showed that these GATA-1 mutants were expressed at somewhat lower levels than the full-length GATA-1 proteins (Figure 4C). This analysis, and in particular the lack of requirement for GATA-1 DNA binding for inhibition of PU.1, strongly suggests that the mechanism of repression involves protein-protein interaction rather than, for example, activation by GATA-1 of a gene encoding an antagonist of PU.1 or direct binding of GATA-1 to the target promoter. We therefore analyzed the ability of PU.1 to interact with GATA-1 in vitro.
Structural requirement for GATA-1 mediated PU.1 repression.
(A) The structure of the chicken GATA-1 protein and mutant proteins used in this study. The 2 zinc fingers are shown as boxes; numbers indicate amino acids. Mutated zinc fingers are shown in black. (B) The pPU3-TK-LUC reporter (100 ng), pCMV-MTPU.1 expression vector (0.25 μg), and pRSV-βgal (100 ng) internal control plasmids were cotransfected into Q2bn cells with 1 μg of pSPCMV-GATA-1 (WT) or the corresponding expression vectors for the mutant GATA-1 proteins as in Figure 3. The fold repression relative to the value obtained by cotransfection of pCMV-MTPU.1 in the absence of GATA-1 was calculated after normalization of the luciferase values to the β-galactosidase activity. The data shown are the average of 2 determinations from a representative experiment. (C) The expression of the wild-type and mutant GATA-1 proteins in the Q2bn cells transfected in (B) was analyzed by Western blotting as described in Figure 3B. Bands corresponding to the undegraded GATA-1 proteins have been marked with an asterisk (*).
Structural requirement for GATA-1 mediated PU.1 repression.
(A) The structure of the chicken GATA-1 protein and mutant proteins used in this study. The 2 zinc fingers are shown as boxes; numbers indicate amino acids. Mutated zinc fingers are shown in black. (B) The pPU3-TK-LUC reporter (100 ng), pCMV-MTPU.1 expression vector (0.25 μg), and pRSV-βgal (100 ng) internal control plasmids were cotransfected into Q2bn cells with 1 μg of pSPCMV-GATA-1 (WT) or the corresponding expression vectors for the mutant GATA-1 proteins as in Figure 3. The fold repression relative to the value obtained by cotransfection of pCMV-MTPU.1 in the absence of GATA-1 was calculated after normalization of the luciferase values to the β-galactosidase activity. The data shown are the average of 2 determinations from a representative experiment. (C) The expression of the wild-type and mutant GATA-1 proteins in the Q2bn cells transfected in (B) was analyzed by Western blotting as described in Figure 3B. Bands corresponding to the undegraded GATA-1 proteins have been marked with an asterisk (*).
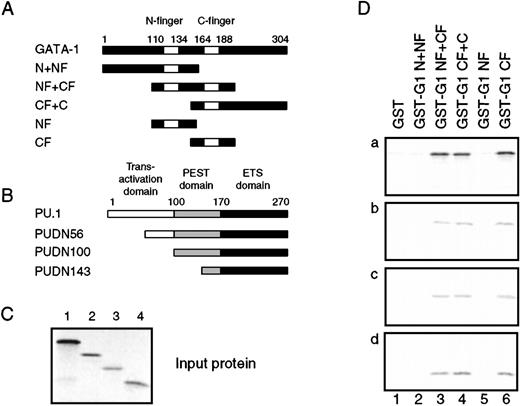
The PU.1 ETS domain and the GATA-1 C-finger region interact in vitro
Overlapping fragments of GATA-1 were fused to glutathione-S-transferase (GST), as outlined in Figure5A, and expressed in Escherichia coli. The N+NF construct includes the N-terminus and the N-finger of GATA-1, the NF+CF constructs the entire finger domain, and the CF+C the C-finger and C-terminal part of GATA-1. Also, the 2 fingers were fused to GST by themselves (NF and CF, respectively). The resulting fusion proteins were purified and used in GST pulldown experiments with in vitro–translated 35S-labeled PU.1 (Figure 5C, lane 1). We found that all GATA-1 fragments containing the C-finger were capable of interacting with PU.1 (Figure 5D, panel a). When deletion mutants of PU.1 (Figure 5B and 5C) were analyzed, we found that neither the N-terminal transactivation domain nor the PEST domain were required for interaction with the GATA-1 C-finger (Figure 5B, panels b-d) while deletion of the C–terminal 71 amino acids of PU.1, corresponding to most of the ETS domain (PUDC198), abolished GATA-1 binding (Figure6C, lower panel). This showed that the PU.1 ETS domain was necessary and sufficient for interaction with the GATA-1 C-finger region. Finally, the observation that mutation of the C-finger, while destroying DNA binding, did not abolish the ability of GATA-1 to repress PU.1 activity, suggested that the integrity of the C-finger was not required for PU.1 binding. We therefore tested the ability of PU.1, as well as PU.1 N-terminal deletion mutants (Figure7A), to interact with the wild-type GATA-1 finger domain (NF+CF in Figure 4A) and with finger domains where the 2 zinc fingers had been individually mutated. Mutation of the C-finger had no detectable effect on binding to either full-length PU.1 or to the ETS domain alone (Figure 7E) compared with that observed with the wild-type finger of mutant N-finger regions (Figure 7C and 7D). This demonstrated that the secondary structure of the C-finger region is not essential for the interaction and is consistent with the lack of requirement for an intact C-finger for the ability of GATA-1 to repress PU.1.
Interaction between PU.1 and GATA-1 in vitro.
(A) The indicated fragments of the GATA-1 protein were fused to the C-terminus of GST, expressed in E. coli, and purified on glutathione-sepharose beads. (B) Full-length and 3 N-terminal deletion mutants of PU.1 (the name indicates the first amino acid still present) were translated in vitro in the presence of 35S-labeled methionine and cysteine. The positions of the transactivation domain, PEST domain, and DNA-binding, or ETS, domain are shown above the coding sequence. The translation products are shown in (C), as detected by phosphorimaging after electrophoresis on a 12% SDS-PAGE gel. (D) The35S-labeled PU.1 proteins were subjected to GST pulldown assays using GST alone or fused to the GATA-1 fragments from (A). GST fusion protein, 500 ng, was used in each assay, along with 4 μL of in vitro–translated protein. After electrophoresis of the35S-labeled bound proteins, they were detected by phosphorimaging as in (C). About 10% to 20% of the input protein was retained for all PU.1 derivatives.
Interaction between PU.1 and GATA-1 in vitro.
(A) The indicated fragments of the GATA-1 protein were fused to the C-terminus of GST, expressed in E. coli, and purified on glutathione-sepharose beads. (B) Full-length and 3 N-terminal deletion mutants of PU.1 (the name indicates the first amino acid still present) were translated in vitro in the presence of 35S-labeled methionine and cysteine. The positions of the transactivation domain, PEST domain, and DNA-binding, or ETS, domain are shown above the coding sequence. The translation products are shown in (C), as detected by phosphorimaging after electrophoresis on a 12% SDS-PAGE gel. (D) The35S-labeled PU.1 proteins were subjected to GST pulldown assays using GST alone or fused to the GATA-1 fragments from (A). GST fusion protein, 500 ng, was used in each assay, along with 4 μL of in vitro–translated protein. After electrophoresis of the35S-labeled bound proteins, they were detected by phosphorimaging as in (C). About 10% to 20% of the input protein was retained for all PU.1 derivatives.
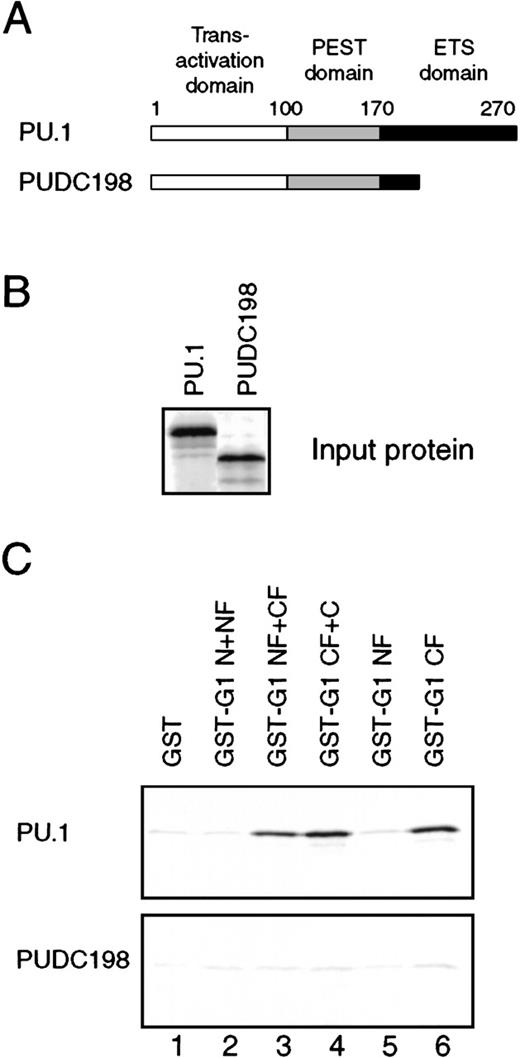
The ETS domain of PU.1 is required for interaction with GATA-1.
(A) The structure of the PUDC198 protein, in which most of the ETS domain has been deleted. (B) Wild-type PU.1 and PUDC198 were35S-labeled by in vitro translation, and 4-μL aliquots were separated on a 12% SDS-PAGE gel and detected by phosphorimaging. (C) Four-microliter 35S-labeled PU.1 (upper panel) and PUDC198 (lower panel) proteins were subjected to GST pulldown analysis and bound proteins detected as in Figure 5D.
The ETS domain of PU.1 is required for interaction with GATA-1.
(A) The structure of the PUDC198 protein, in which most of the ETS domain has been deleted. (B) Wild-type PU.1 and PUDC198 were35S-labeled by in vitro translation, and 4-μL aliquots were separated on a 12% SDS-PAGE gel and detected by phosphorimaging. (C) Four-microliter 35S-labeled PU.1 (upper panel) and PUDC198 (lower panel) proteins were subjected to GST pulldown analysis and bound proteins detected as in Figure 5D.
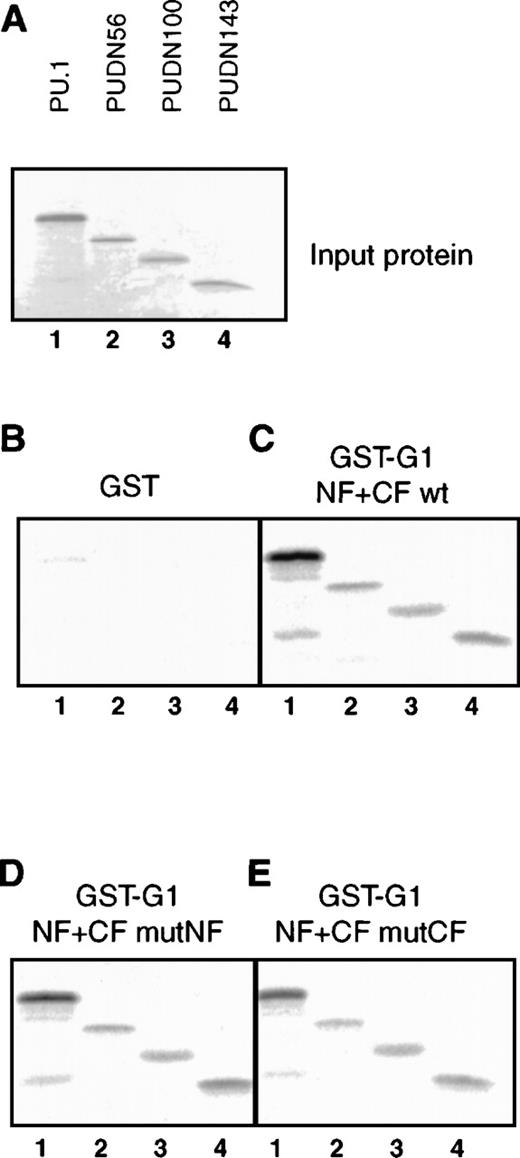
The GATA-1C–finger secondary structure is not required for interaction with PU.1.
(A) In vitro–translated 35S-labeled PU.1 and N-terminal deletion mutant input proteins (3 μL). Lane 1: PU.1; lane 2: PUDN56; lane 3: PUDN100; lane 4: PUDN143. (B) Pulldown of35S-labeled PU.1 proteins (5 μL) with GST-GATA-1 fusions. Lanes 1-4 as in (A). (C) Pulldown as in (B) with GST fusion containing the wild-type GATA-1 finger region (GST-G1 NF+CF wt). (D) Pulldown as in (B) with GST fusion of GATA-1 finger region carrying point mutations in the N-finger (GST-G1 NF+CF mutNF). (E) Pulldown as in (B) with GST fusion of GATA-1 finger region carrying point mutations in the C-finger (GST-G1 NF+CF mutCF).
The GATA-1C–finger secondary structure is not required for interaction with PU.1.
(A) In vitro–translated 35S-labeled PU.1 and N-terminal deletion mutant input proteins (3 μL). Lane 1: PU.1; lane 2: PUDN56; lane 3: PUDN100; lane 4: PUDN143. (B) Pulldown of35S-labeled PU.1 proteins (5 μL) with GST-GATA-1 fusions. Lanes 1-4 as in (A). (C) Pulldown as in (B) with GST fusion containing the wild-type GATA-1 finger region (GST-G1 NF+CF wt). (D) Pulldown as in (B) with GST fusion of GATA-1 finger region carrying point mutations in the N-finger (GST-G1 NF+CF mutNF). (E) Pulldown as in (B) with GST fusion of GATA-1 finger region carrying point mutations in the C-finger (GST-G1 NF+CF mutCF).
The PU.1 ETS domain is required for GATA-1–mediated repression
The above results pointed to the interaction between the GATA-1 C-finger and the PU.1 ETS domain as being instrumental to the observed repression. To confirm this, we used hybrid proteins in which the PU.1 ETS domain had been replaced with the DNA-binding domain from the yeast GAL4 protein by fusing the transactivation domain either C- or N-terminally to the GAL4 DNA-binding domain, yielding GAL4-PU.1 and PU.1-GAL4, respectively (Figure8A). The ability of GATA-1 to repress the activity of the GAL4-PU.1 fusion proteins (Figure 8C and 8D) was compared to the repression of wild-type PU.1 (Figure 8B). This showed that, whereas PU.1 itself was repressed even at moderate GATA-1 levels, the GAL4-PU.1 hybrid activators were only marginally affected by GATA-1. Similar results were obtained when only the transactivation domain (PU.1 amino acids 1-99) or the core transactivation domain (amino acids 1-55) were fused to GAL4 (data not shown). These results further support a role for the direct interaction between the ETS domain and GATA-1 in the observed repression of PU.1 activity.
The PU.1 ETS domain is required for repression of PU.1 by GATA-1.
(A) Fusion of PU.1 transactivation domain and PEST domain to, either C- or N-terminally, the GAL4 amino acids 1-147 generates GAL4-PU.1 and PU.1-GAL4 hybrid activators, respectively. (B) Repression of PU.1 by GATA-1: 0.25 μg of pCMV-MTPU.1 or empty pcDNAI expression vector was cotransfected with 100 ng of pPU3-TK-LUC, 100 of ng pRSVβgal, and the indicated amount of pSPCMV-GATA-1 expression vector. The total amount of DNA was kept constant by addition of empty pSPCMV vector. Results are expressed as luciferase activity normalized to β-galactosidase, setting the level obtained with empty expression vector to 1. Each column represents the average of 2 determinations. (C) Parallel experiment to (B), where pPU3-TK-LUC was replaced with the GAL4-responsive reporter pG5B-LUC (100 ng) and pCMV-MTPU.1 with pGD-PU 1-170 expressing GAL4-PU.1 (0.25 μg). (D) Similar experiment as in (C), except that the GAL4-PU.1 activator was replaced by PU.1-GAL4 encoded by pCMV-MTPU1-170GD.
The PU.1 ETS domain is required for repression of PU.1 by GATA-1.
(A) Fusion of PU.1 transactivation domain and PEST domain to, either C- or N-terminally, the GAL4 amino acids 1-147 generates GAL4-PU.1 and PU.1-GAL4 hybrid activators, respectively. (B) Repression of PU.1 by GATA-1: 0.25 μg of pCMV-MTPU.1 or empty pcDNAI expression vector was cotransfected with 100 ng of pPU3-TK-LUC, 100 of ng pRSVβgal, and the indicated amount of pSPCMV-GATA-1 expression vector. The total amount of DNA was kept constant by addition of empty pSPCMV vector. Results are expressed as luciferase activity normalized to β-galactosidase, setting the level obtained with empty expression vector to 1. Each column represents the average of 2 determinations. (C) Parallel experiment to (B), where pPU3-TK-LUC was replaced with the GAL4-responsive reporter pG5B-LUC (100 ng) and pCMV-MTPU.1 with pGD-PU 1-170 expressing GAL4-PU.1 (0.25 μg). (D) Similar experiment as in (C), except that the GAL4-PU.1 activator was replaced by PU.1-GAL4 encoded by pCMV-MTPU1-170GD.
Discussion
GATA-1 is a repressor of PU.1 function
We have in this report found that the GATA-1 transcription factor is capable of functionally interfering with the PU.1 protein and have provided evidence that this interference is mediated through interaction between the PU.1 ETS domain and the GATA-1 C-finger region. This finding provides a molecular basis for the observed ability of the GATA-1 protein to inhibit the expression of myeloid-specific genes and, thereby, for the necessity for down-regulation of GATA-1 expression during normal myeloid (ie, neutrophil granulocyte and macrophage) development. Our results on PU.1–GATA-1 interactions are consistent with those obtained by Rekthman et al13 when analyzing the ability of PU.1 to inhibit GATA-1 activity. These authors also show that interaction between the 2 factors is mediated by their DNA-binding domains. It is interesting that expression of the C-finger domain of GATA-1 or -2 was sufficient to induce limited megakaryocytic differentiation of the myeloid 416B cell line,32 suggesting that the C-finger is capable of mediating some aspects of GATA-1 function by itself, possibly through interaction with PU.1. Finally, it is worth noting that nothing in this study excludes the possibility that other GATA-1 targets are involved in the observed extinction of myeloid gene expression. C/EBPα and C/EBPβ, which are required for proper neutrophil and macrophage differentiation, respectively,33,34 are still expressed in eosinophils and are active in the presence of GATA-1 on the eosinophil-specific EOS47 promoter.2 Other C/EBP regulated genes, like mim-1, are expressed in both myeloid and eosinophilic cells.6 These particular factors therefore do not seem likely targets, but others may yet be identified.
We found that replacing the PU.1 ETS domain with the GAL4 DNA-binding domain rendered the resulting GAL4-PU.1 hybrid protein virtually insensitive to GATA-1 inhibition. This, together with the lack of requirement for GATA-1 DNA binding for repression, provides substantial evidence that the observed direct protein-protein interaction between the GATA-1 C-finger and the ETS domain of PU.1 is instrumental to repression. Recently, results similar to those presented here were published by Zhang et al35 using different cell lines and PU.1-responsive promoters. These authors found that GATA-1 (and -2) repress PU.1 function in a manner dependent on an interaction between the GATA C-finger and the PU.1 ETS domain. In addition, the C-finger was found to interact with the PU.1 transactivation domain, but because no repression by GATA factors was observed when the PU.1 transactivation domain was fused to the GAL4 DNA-binding domain, it was concluded that the latter interaction is not sufficient for repression to take place. The results presented by Zhang et al are thus fully consistent with those presented here and, together with our results, seem to rule out that the observed repression is due to promoter or cell-type specific effects.
We consistently found that mutation of the GATA-1 N-finger increased the repression of PU.1 activity 2- to 3-fold. Although the N-finger does not appear to play a direct role in the interaction between PU.1 and GATA-1, it is required for interaction of GATA-1 with FOG29 and is implicated in the formation of GATA-1 dimers through N-finger–C-finger interactions.36 A possible explanation for this observation, therefore, is that lowering the ability of GATA-1 to dimerize increases the level of GATA-1 monomers capable of other protein-protein interactions, such as that with PU.1.
ETS domains as protein-protein interaction domains
The ability of GATA-1 to interact with PU.1 is the latest in a number of protein-protein interactions shown to be mediated by the PU.1 ETS domain in particular and ETS domains in general. PU.1 has previously, along with other ETS proteins, been found to interact with Jun family members37,38 and proteins of the C/EBP family.2,39 Other basic region–leucine zipper (BR-LZ)-containing factors, such as USF and MafB, seem to bind Ets-1 but not PU.1.5 40 The GATA-1–PU.1 interaction, however, seems to be the first between a zinc finger domain and an ETS domain. Whether this represents a general type of interaction, as in the case of BR-LZ-ETS interactions, will require work to determine the specificities of interaction.
Negative regulatory interactions in hematopoietic lineage commitment and leukemia
The PU.1-GATA–1 interaction is another example of a negative regulatory interaction between hematopoietic transcription factors. Previous examples include the inhibition of Ets-1 on erythroid promoters by MafB5 and the inhibition of the ability of GATA-1 transactivation by FOG-mediated recruitment of the CtBP repressor.30 Although the precise functional relevance of these inhibitory interactions is yet to be established, they all seem to be involved in maintaining the correct expression boundaries for lineage-specific genes, as exemplified by the repression of erythroid-specific promoters by the myeloid-specific MafB protein. It is, however, possible that these negative interactions also play an important role in lineage commitment. It is believed that lineage commitment of multipotent cells is preceded by a phase during which the genes characteristic of the various possible final gene expression programs are in an accessible chromatin configuration and are expressed at low levels.41 This phase can be visualized as a competition between the various programs where eventually one program prevails and is activated but the others are shut down. The transcriptional shutdown of heterologous gene expression programs is a hallmark of a truly committed cell, and mechanisms capable of mediating such repression are likely to be of importance during the commitment process. One of the initial choices of a multipotent myeloid/erythroid precursor is between GATA-1– (and GATA–2) expressing lineages (in particular, erythroid and thrombocytic cells) and PU.1–expressing lineages (granulocytes and macrophages), and the PU.1–GATA-1 (or -2) antagonism may therefore play a role in this decision. The recent results of Skoultchi et al13 showing inhibition of GATA-1 activity by PU.1 further supports this idea. It should be noted that commitment to the myeloid lineage can take place in the absence of PU.116 but appears to be significantly reduced. Indeed, C/EBPβ has also been shown to be able to mediate myeloid lineage choice in multipotent cells, albeit less efficiently than PU.1.42
Additionally, the GATA-1–PU.1 antagonism may provide a molecular explanation for the observation that expression of GATA-1 correlates with a bad prognosis and immature phenotype in acute myeloid leukemia.9 Because PU.1 activity is required for the proper development of macrophages and neutrophils, inhibition of PU.1 by GATA-1 may help institute the differentiation block characteristic of leukemic cells; the efficiency of the block would then depend on the level of GATA-1 expression. While GATA-1 may obviously promote a malignant phenotype in other ways, or its expression be an effect rather than a cause thereof, it may be worthwhile to consider the PU.1–GATA-1 interaction as a target for therapeutic intervention. It is also interesting to note that a similar mechanism could operate in the lymphoid compartment, where PU.1 and GATA-3 are exclusively expressed in and required for the development of B cells and T cells, respectively.15,16 43 The possibility thus exists that these factors may play a role in the B- versus T-lymphoid decision, a hypothesis currently under investigation.
Acknowledgment
The authors are grateful to G. Döderlein for expert technical assistance.
Supported by the Deutsche Forschungsgemeinschaft, the Danish Medical Research Council, and the Novo Nordisk Foundation.
Reprints:Claus Nerlov, Laboratory of Gene Therapy Research, Copenhagen University Hospital, RH9322, Juliane Mariesvej 20, 2100 Copenhagen Ø, Denmark; e-mail: nerlovv@rh.dk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal