Abstract
Heterodimeric cytokine receptors generally consist of a major cytokine-binding subunit and a signaling subunit. The latter can transduce signals by more than 1 cytokine, as exemplified by the granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-2 (IL-2), and IL-6 receptor systems. However, often the signaling subunits in isolation are unable to bind cytokines, a fact that has made it more difficult to obtain structural definition of their ligand-binding sites. This report details the crystal structure of the ligand-binding domain of the GM-CSF/IL-3/IL-5 receptor β-chain (βc) signaling subunit in complex with the Fab fragment of the antagonistic monoclonal antibody, BION-1. This is the first single antagonist of all 3 known eosinophil-producing cytokines, and it is therefore capable of regulating eosinophil-related diseases such as asthma. The structure reveals a fibronectin type III domain, and the antagonist-binding site involves major contributions from the loop between the B and C strands and overlaps the cytokine-binding site. Furthermore, tyrosine421 (Tyr421), a key residue involved in receptor activation, lies in the neighboring loop between the F and G strands, although it is not immediately adjacent to the cytokine-binding residues in the B-C loop. Interestingly, functional experiments using receptors mutated across these loops demonstrate that they are cooperatively involved in full receptor activation. The experiments, however, reveal subtle differences between the B-C loop and Tyr421, which is suggestive of distinct functional roles. The elucidation of the structure of the ligand-binding domain of βc also suggests how different cytokines recognize a single receptor subunit, which may have implications for homologous receptor systems.
Cytokine receptors exist largely as homodimers or heterodimers.1-3 Homodimeric receptors, of which the human growth hormone receptor (GH) is the prototype, bind to a single cytokine that bridges 2 identical subunits and causes receptor activation.2 In contrast, heterodimeric receptors comprise 2 or 3 subunits that subserve distinct and specialized functions: a major ligand-binding subunit (the α subunit) and a signaling subunit (the β or γ subunit). Importantly, a signaling subunit is able to recognize several cytokines complexed to the appropriate α-chain and to transduce their signals. This is exemplified by the common β-chain (βc) of the granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3), and IL-5 receptors; the common IL-2 receptor γ-chain (shared by the IL-2, IL-4, IL-7, IL-9, and IL-15 receptors); and gp130 (shared by the receptors for IL-6, IL-11, Leukemia Inhibitory Factor (LIF), ciliary neurotrophic factor, oncostatin M, and cardiotrophin).
The GM-CSF, IL-3, and IL-5 receptors are the only receptors known to transduce signals leading to eosinophil production and, significantly, the corresponding cytokines can be found concomitantly at elevated levels in lungs affected by allergic inflammation. The simultaneous elevation of the GM-CSF, IL-3, and IL-5 receptors may increase eosinophil numbers, contribute to the overall degree of eosinophil activation, cause the different phases of eosinophil infiltration, and determine a localized versus a generalized eosinophil-mediated inflammation.4-6 This may be particularly important in the pathology of certain diseases, such as asthma, where the eosinophil plays a major effector role. Thus, an antagonist directed against βc would simultaneously inhibit the function of all 3 eosinophilopoietic cytokines and may prove a useful therapeutic agent.
The extracellular part of βc comprises 2 pairs of a conserved cytokine receptor module (CRM),3 a membrane-spanning region, and a cytoplasmic domain. Each CRM comprises 2 domains of a fibronectin type III structure with features especially conserved among cytokine receptors.3 Although βc does not bind cytokines by itself, its coexpression with the α-chains enhances the affinity of cytokine binding, a process termed affinity conversion. Extensive mutational analysis has localized the affinity-converting (cytokine-binding) region to residues in the fourth extracellular domain (D4βc) and has shown that this domain and in particular the residues Tyr365, His367, Ile368, and Tyr421 within it are crucial for receptor activation.7-9
A major problem in seeking structural data of the binding site of a communal subunit complexed to cytokines is that, unlike homodimeric receptors or isolated α-chains of heterodimeric receptors that can bind directly to cytokines, communal subunits often cannot bind to cytokines by themselves. We therefore used an antagonistic monoclonal antibody (mAb), BION-1, which we have shown to reciprocally inhibit cytokine binding to βc10, to form a complex for crystallographic studies. BION-1, which was raised against D4βc, has been shown to inhibit the high-affinity binding of GM-CSF, IL-3, and IL-5 to human eosinophils and their production and functional activation in vitro.10 Within βc, residues Glu366, Arg418, and Met363 or Arg364 were found to be required for binding BION-1.10
BION-1 thus represents the first common antagonist of the GM-CSF, IL-3, and IL-5 receptors, and it is a unique tool with which to explore the cytokine-binding site in the common βc. Here we report the crystal structure of the activation domain of the GM-CSF/IL-3/IL-5 receptor signaling subunit bound to the mAb antagonist, BION-1. The structure provides a molecular basis for understanding ligand recognition and receptor assembly. Furthermore, the structure of the complex provides leads for the design of novel therapeutics against allergic diseases.
Materials and methods
Crystallization and data collection
D4βc (residues 338-438 with an additional amino-terminal Met) was expressed using the pEC611 vector inEscherichia coli and purified by reverse-phase high-performance liquid chromatography (HPLC). The expressed protein was insoluble, but it could be recovered from the bacteria by dissolution in 6 mol/L guanidine hydrochloride and 50 mmol/L sodium acetate buffer (pH 4.0). After HPLC the protein was dialyzed exhaustively against 5 mmol/L 2-[N-morpholino]ethanesulfonic acid (MES) buffer (pH 6.0). The BION-1 mAb was raised against D4βc,10 and Fab fragments were generated and purified by standard methods. The complex was produced by mixing BION-1 Fab and D4βc to give a 1:1 (mol/mol) complex, which was purified on a gel filtration column (Superdex 75; Amersham Pharmacia, Little Chalfont, England).
Crystals of the complex were grown by the hanging-drop vapor diffusion method at 22°C. We mixed 2-μL droplets of protein solution (protein concentration of 5-7 mg/mL) with 1.5 μL of the reservoir solution. The solution was then equilibrated against a 1-mL reservoir consisting of 100 mmol/L citrate buffer (pH 5.5) containing 12% (wt/vol) polyethylene glycol 4000. The crystals reached maximum size of approximately 0.6 mm × 0.2 mm × 0.2 mm over 10 days. The crystals belonged to space group P41 212 and had the following cell dimensions: a andb were 7.76 nm, and c was 29.49 nm. The crystals were micromanipulated, washed several times in reservoir buffer, and dissolved in sodium dodecyl sulfate–N-tris[hydroxymethyl]methylglycine (SDS-Tricine) sample buffer. Polyacrylamide gel electrophoresis (PAGE) was performed to confirm that the crystals were of the intact complex. The crystals proved to be sensitive to radiation, and hence, cryocooling was essential. However, they were fragile, and an array of commonly used cryoprotectants caused disordering of the crystals. A flash-freezing protocol was finally established. This involved soaking the crystals in 5% (vol/vol) increments of 2-methyl-2,4-pentanediol for 2 minutes to a final concentration of 15% (vol/vol).
A native data set was initially collected in-house on an imaging plate area detector (MARResearch, marUSA, Evanston, IL) with CuKα x-rays generated by a rotating anode generator (Rigaku RU-200, Molecular Structure Corp., The Woodlands, TX). A better native data set was subsequently collected from a single crystal frozen to –273°C using synchrotron radiation (BioCARS beamline, 14-BM-C; Advanced Photon Source, Chicago, IL). The diffraction data were processed and analyzed using DENZO and SCALEPACK11 and programs in the CCP4 suite12 (Table1).
Crystallographic analysis
| Data collection | |||
| Temperature of collection (°C) Resolution limit (nm) Observations Unique reflections Completeness (%) | 173 0.28 58 732 21 211 88.1 | Multiplicity I/ςI No. of data > 2ςI (%) *Rmerge (%) | 2.8 11.4 66.2 9.8 |
| Refinement statistics | |||
| Resolution range (nm) †Rfactor (%) †Rfree (%) | ∞ − 0.28 22.8 28.8 | SD from ideality Bond lengths (nm) Bond angles (°) Impropers (°) Dihedrals (°) | 0.0010 1.55 0.95 27.1 |
| Atoms in model | |||
| Protein (nonhydrogen) Water Carbohydrate | 4146 124 14 | ||
| Data collection | |||
| Temperature of collection (°C) Resolution limit (nm) Observations Unique reflections Completeness (%) | 173 0.28 58 732 21 211 88.1 | Multiplicity I/ςI No. of data > 2ςI (%) *Rmerge (%) | 2.8 11.4 66.2 9.8 |
| Refinement statistics | |||
| Resolution range (nm) †Rfactor (%) †Rfree (%) | ∞ − 0.28 22.8 28.8 | SD from ideality Bond lengths (nm) Bond angles (°) Impropers (°) Dihedrals (°) | 0.0010 1.55 0.95 27.1 |
| Atoms in model | |||
| Protein (nonhydrogen) Water Carbohydrate | 4146 124 14 | ||
| Residues in most favored regions13 of Ramachandran plot (%) | 80 |
| Residues in additionally allowed regions13of Ramachandran plot (%) | 19 |
| Residues in most favored regions13 of Ramachandran plot (%) | 80 |
| Residues in additionally allowed regions13of Ramachandran plot (%) | 19 |
Rmerge indicates ΣhklΣi‖Ii − 〈I 〉‖/‖〈I 〉‖, where Ii is the intensity for theith measurement of an equivalent reflection, with indices h, k, and l.
Rfactor indicates 100(Σ‖‖Fo‖ − ‖Fc‖‖/Σ‖Fo‖) using all data except 6%, which were used for theRfree calculation.
Structure determination
The crystal structure was solved by molecular replacement using AmoRe14 and the in-house native data set. Nonredundant Fab fragments were downloaded from the protein databank (PDB) and systematically tested as molecular replacement search probes. The second search probe tested, a mouse Fab fragment with PDB identifier 1YEC,15 proved successful. The 10th peak in the rotation function (peak height of 3.3 ς) produced the highest peak in the translation function (with a correlation coefficient of 27.9 and anRfactor of 54.1% compared with the next highest peak, which had a correlation coefficient of 17.3 and anRfactor of 57.5%). The statistics indicated that P41 212 was the correct enantiomorphic space group. Rigid body refinement of the initial solution lead to a model with a correlation coefficient of 28.7 and anRfactor of 49.9% (resolution range, 1.0-0.45 nm). Further refinement, in which the Fab domains were treated as separate rigid bodies, resulted in further improvement of the statistics (anRfactor of 46.1% and a drop ofRfree from 50.9% to 43.8%). Maps calculated from this solution yielded readily interpretable density for D4βc.
The model of the complex was then built with the help of skeletonized maps using the program O16 and refined using the maximum likelihood target in the program package CNS.17The refinement was completed with the synchrotron native data set (Table 1). In the final stages a bulk solvent correction and restrained individual isotropic B-factors were applied. The quality of the final map was very good, with no breaks in the main-chain connectivity, and the real space fit16 of residues into the map never fell below 0.7. The final model comprises residues 338-438 for D4βc; all residues for the Fab fragment; 124 solvent molecules; and 1 carbohydrate unit, an N-acetylglucosamine unit off the BION-1 residue Asn26L.
The choice of solvent molecules was conservative. The molecules were only accepted if they appeared as peaks, with a signal of more than 3 times the SD error in difference maps; reappeared in subsequent 2Fo-Fc maps; took part in at least 1 hydrogen-bonding interaction; and had temperature factors of less than 0.8 nm2. The stereochemical quality of the final model is good (Table 1), and other stereochemical parameters, such as side-chain chi angle values, peptide bond planarity, alpha-carbon tetrahedral distortions, and nonbonded interactions, are all significantly better than the ranges allowed according to the program PROCHECK.13 The correctness of the tracing is supported by residue omit maps in which 10% of the model was deleted, a round of simulated annealing was performed to reduce bias, and the resultant map was examined in the region of omission. The tracing is also supported by 3D-1D scores that never fall below 0.2, which indicates that there are no residues in chemically unreasonable environments.18
Functional studies
Human Embryonal Kidney 293T (HEK293T) cells were grown in RPMI-1640 (GIBCO Laboratories, Glen Waverly, Vic., Australia) containing 10% fetal calf serum (FCS) and were cotransfected with expression constructs encoding IL-3 receptor α-chain, βc, and Janus kinase (JAK-2) using a calcium-phosphate precipitation procedure. Briefly, 1.4 × 106 cells were plated onto 6-cm plastic tissue culture dishes the day before transfection and left to adhere overnight. Four hours after a medium change, 10 μg DNA was added in the form of a calcium phosphate precipitate, which was left on the cells for a further 4 hours. The expression constructs used per transfection were 6 μg wild-type or mutant βccomplementary DNA (cDNA) cloned into pcDNA1 (Invitrogen, Groningen, The Netherlands), 3 μg IL-3 receptor α-chain cDNA in pCDM8, and 1 μg JAK-2 in pRcCMV (Invitrogen). The cells were then released from the plates, replated in flasks, and incubated for a further 40 hours before use in an activation assay. On the day of the functional experiment, the cells were released and washed in cold phosphate-buffered saline (PBS) and subsequently stimulated with IL-3 at the concentration specified for 5 minutes on ice. Lysates were prepared, precleared and immunoprecipitation was carried out with an anti-βc mAb, 8E4, essentially as described previously.19 After extensive washing, immunoprecipitates were separated on a 7.5% SDS-PAGE gel under reducing conditions, transferred to nitrocellulose, and immunoblotted with antiphosphotyrosine antibody, PY20 (Transduction Laboratories, Lexington, KY). The blot was developed by enhanced chemiluminescence (ECL) (Amersham, Pharmacia) and then stripped and reprobed with anti-βc antibody 1C1 to control the amount of βc present.
Epitope mapping
African green monkey COS cells were maintained in RPMI with 10% FCS and transfected with 10 μg of wild-type or mutant βcexpression construct by electroporation essentially as described previously.8 The cells were washed in PBS and lysed as detailed elsewhere19 48 hours after transfection. Lysates were run on a 7.5% SDS-PAGE under reducing conditions, and immunoblotting was carried out after electro-blot transfer using either the antagonistic mAb, BION-1, or anti-βc mAb, 1C1, at 1 μg/mL. This was followed by the addition of antimouse Ig coupled to horseradish peroxidase (Pierce, Rockford, IL). Blots were developed by ECL (Amersham) following manufacturer's instructions.
Results
Crystal structure of the GM-CSF/IL-3/IL-5 receptor βcactivation domain
We expressed D4βc in E coli and purified it to homogeneity by reverse-phase HPLC. BION-1 mAb was digested with ficin to generate Fab fragments that were purified by chromatography on protein A sepharose. Titration of D4βc and the BION-1 Fab produced a stoichiometric 1:1 complex that subsequently formed crystals. These crystals diffracted well enough to allow a full crystallographic structure determination to proceed.
We determined the structure of the BION-1/D4βc complex to a resolution limit of 0.28 nm. The structure showed that the D4βc molecule has a compact globular shape with overall dimensions of 4.5 nm × 2.5 nm × 2.0 nm (Figure1). The amino-terminus and carboxy-terminus represent the sites of attachment for the remainder of the extracellular region and the membrane-spanning domain, respectively. The molecule adopts the topology of a fibronectin type III module with 2 antiparallel β-sheets (42% sheet) packing against each other via a multitude of hydrophobic interactions including 2 clusters of aromatic residues Trp434, Tyr354, and Tyr376; Trp358, Phe372, and His370). Sheet A consists of 3 beta-strands: strand A is comprised of residues 344-350, B of residues 353-359, and E of residues 396-398. Sheet B consists of 4 strands: strand C is comprised of residues 369-378, D of residues 389-392, F of residues 406-417, and G of residues 432-436). The longest strand, F, almost spans the entire length of the molecule.
Structure of D4βc.
(A) Structure of the Fab receptor D4βc complex shown in ribbon representation. The mAb light chain is shown in cyan blue, the heavy chain in blue, and the receptor in yellow. The major structural features of D4βc are labeled, and the locations of key residues are denoted by stick representation. These pictures were produced using the Molscript20 and Raster3D21programs. (B) Structure as for (A) but reoriented 90° about the vertical axis. (C) Surface representation of the receptor using the program GRASP.22 The green surface indicates the location of hydrophobic-aromatic patch H1. The molecule is tilted approximately 20° counterclockwise relative to (A). (D) View of hydrophobic-aromatic patch H2 prepared as for (C). The molecule is tilted approximately 20° clockwise and rotated approximately 60° clockwise from above, about a vertical axis relative to (B).
Structure of D4βc.
(A) Structure of the Fab receptor D4βc complex shown in ribbon representation. The mAb light chain is shown in cyan blue, the heavy chain in blue, and the receptor in yellow. The major structural features of D4βc are labeled, and the locations of key residues are denoted by stick representation. These pictures were produced using the Molscript20 and Raster3D21programs. (B) Structure as for (A) but reoriented 90° about the vertical axis. (C) Surface representation of the receptor using the program GRASP.22 The green surface indicates the location of hydrophobic-aromatic patch H1. The molecule is tilted approximately 20° counterclockwise relative to (A). (D) View of hydrophobic-aromatic patch H2 prepared as for (C). The molecule is tilted approximately 20° clockwise and rotated approximately 60° clockwise from above, about a vertical axis relative to (B).
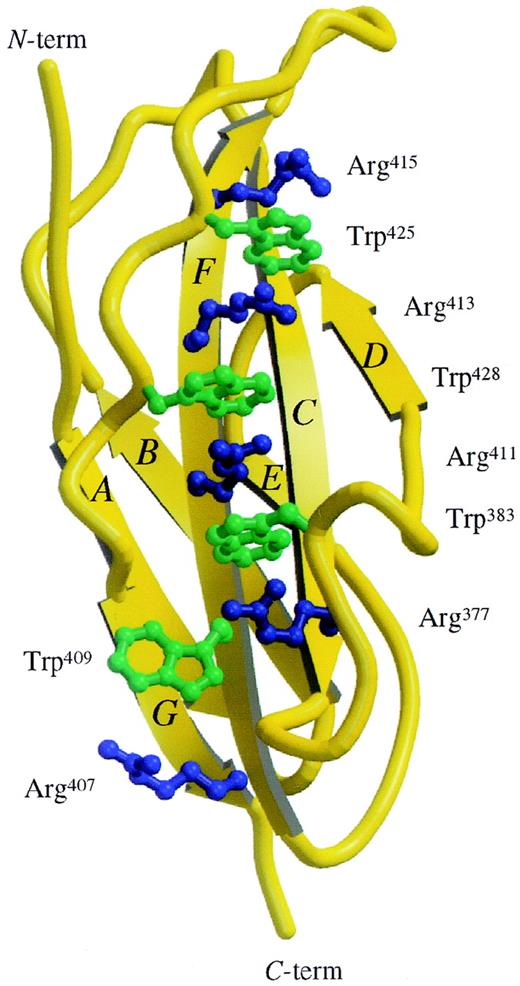
The amino acid sequence motif WSXWS (tryptophan-serine-any residue-tryptophan-serine) is a characteristic feature of many cytokine receptors. WSXWS is located between F and G strands and adopts a double β-bulge structure (Figure 2) with the tryptophan side chains interdigitated between the arginine side chains from the adjacent F strand. In D4βc, this ladder of alternating basic and aromatic residues is extended and consists of the following side chain: Arg415-Trp425-Arg413-Trp428-Arg411-Trp383-Arg377-Trp409-Arg407. There is a “sidestep” in the ladder at Arg377-Trp409. This 9-rung ladder, measuring 2.9 nm long with rungs of about 0.5 nm wide, represents the only significant electropositive patch on the surface of the molecule.
View of the Trp/Arg “ladder.”
Structure of the D4βc shown in ribbon representation with the side chains of the Trp/Arg stack shown as ball and stick. The molecular graphics were produced using the Molscript20 and Raster3D21 programs.
There are 2 large hydrophobic patches on the surface of D4βc: H1 and H2. The first, H1, forms part of a lip at the end of a pronounced groove on the surface of the molecule (Figure1C). This patch is made up of residues Ile338, Met340, Ala341, Pro342, Met361, Tyr365, the aliphatic moiety of Lys362, Ile368, and Tyr421. The groove is located at the N-terminal end of the molecule, where one wall is formed by the B-C loop and part of the F-G loop, and the other wall is formed by the N-terminus (residues 338-342) (Figure 1C). The second hydrophobic patch, H2, located on the opposite face to the first, is a dense strip of hydrophobic residues located at one edge of the β-sandwich defined by the D and E strands. It measures 2.7 nm × 0.6 nm (Figure 1D).
Of the interstrand loops, only the B-C and F-G loops protrude significantly from the body of the protein; both have been implicated in cytokine binding.3 7-9 The B-C loop adopts significant regular structure with residues 365-368, forming a type I β-turn (Figure 1A). Significant features of the B-C loop of D4βcinclude residues Tyr365 and His367, both of which project out into the solution (Figure 1A, B). The F-G loop adopts a type I′ β-turn at its tip, and the most significant features in this region are Arg418 and Tyr421, both of which project away from the body of D4βc(Figure 1A, B).
Functional roles of the B-C loop and Tyr421
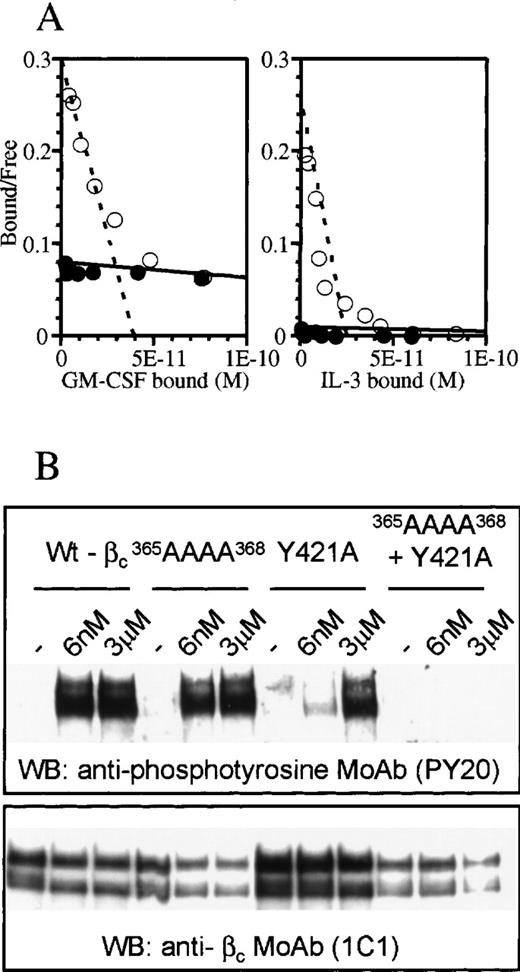
Although the structure of D4βc revealed that Tyr421 is in close proximity to the 3 residues in the B-C loop involved in cytokine binding (Tyr365, His367, and Ile368), the side chain is oriented away from these, possibly reflecting different functional roles. Previous experiments9 suggested that high-affinity binding of IL-3 was sensitive to mutation of Tyr421 but not to replacement of individual residues in the B-C loop.8 We examined whether a multiple mutation in the B-C loop of the residues implicated in binding GM-CSF and IL-5 would affect IL-3 high-affinity binding. The results showed that alanine substitution of residues 365-368 in the B-C loop abrogated high-affinity binding of both GM-CSF and IL-3 (Figure 3A). We next examined phosphorylation of cytoplasmic tyrosine residues, as this is a very sensitive measure of recruitment of βc to a ligand/α-chain complex. Some analogs of βc are unable to affinity-convert due to the affinity of the α-β complex for cytokine being less than or equal to that of the α-chain alone. These same analogs may nevertheless exhibit differences in tyrosine phosphorylation (see “Discussion”). We examined the effects of mutating the B-C loop or Tyr421, either separately or in combination, on the ability of βc to undergo tyrosine phosphorylation in response to IL-3. We found that substitution of Tyr421 had a pronounced effect, with high levels of tyrosine phosphorylation of βc being achieved only at 3 μmol/L IL-3, a concentration about 500-fold higher than that required by the native receptor (6 nmol/L). In contrast, mutation of the B-C loop alone did not impair IL-3–induced phosphorylation of βc at the high concentration used. Nevertheless, a role for the B-C loop in βc activation was demonstrated by a combined mutant of the B-C loop and Tyr421 (Figure 3B), which abrogated IL-3–induced tyrosine phosphorylation of βc.
Differential effects of mutating the B-C loop and/or Tyr421 of the F-G loop in receptor activation.
(A) Scatchard plot transformation of binding isotherms for 125I-GM-CSF and 125I-IL-3 to cells transfected with wild-type βc (○) or365AAAA368 mutant βc (•). (B) Western blot of wild-type and mutant βc after stimulation with various concentrations of IL-3. The blot was probed for phosphotyrosine (upper panel) and βc (lower panel). The double bands in each lane of the gels represent glycosylation variants of βc.23
Differential effects of mutating the B-C loop and/or Tyr421 of the F-G loop in receptor activation.
(A) Scatchard plot transformation of binding isotherms for 125I-GM-CSF and 125I-IL-3 to cells transfected with wild-type βc (○) or365AAAA368 mutant βc (•). (B) Western blot of wild-type and mutant βc after stimulation with various concentrations of IL-3. The blot was probed for phosphotyrosine (upper panel) and βc (lower panel). The double bands in each lane of the gels represent glycosylation variants of βc.23
Antagonist interactions with the βc activation domain
A detailed analysis of the structure of the BION-1/D4βc complex confirmed and extended the observations that BION-1 appeared to form extensive and intimate interactions with the receptor activation domain (Figure 1A, B and Figure 4). The total surface area buried on complex formation is 15 nm2, which is in the range reported for other antibody-protein antigen complexes.24 In total, there are 2 salt bridges (Lys362/AspL94 and Glu366/LysH35), 8 potential hydrogen bonds, and 124 van der Waals (vdw) interactions (Table2). The B-C loop of D4βc is nestled in the shallow antigen-binding groove between the VH and VL domains, whereas the F-G loop forms a more peripheral interaction with complementarity determining region one of the light chain (CDR L1) of BION-1 (Figure 1A, B and Figure 4). The contact surface comprises 14 residues from BION-1, with 9 residues from VH and 5 residues from VL. The majority of contacts are roughly shared between 4 of the CDRs: CDR L1 (1 hydrogen bond and 29 vdw contacts); CDR L3 (1 salt bridge, 3 hydrogen bonds, and 28 vdw contacts); CDR H1 (1 salt bridge, 3 hydrogen bonds, and 36 vdw contacts); and CDR H3 (1 hydrogen bond and 23 vdw contacts). In addition, CDR H2 provides 8 vdw contacts, but CDR L2 makes no contacts with the receptor domain.
Interactions between D4βc and BION-1 or cytokines
| Residue identity . | Residue type . | Vdw contact . | Buried area polar interactions (nm2) . | Required for binding BION-1 . | Required for affinity conversion . | |
|---|---|---|---|---|---|---|
| D4βc | ||||||
| βc 361 | Met | No | 0 | No | No | |
| βc 362 | Lys | Yes | 0.45 | Nζ → L94:Oδ | No | No |
| βc 363 | Met | Yes | 1.21 | Sδ → H57:N | ? | No |
| βc 364 | Arg | Yes | 0.33 | Nη → H33:Oη | ? | No |
| O → H33:Oη | ||||||
| βc 365 | Tyr | Yes | 0.87 | Oη → L94:Oδ | No | Yes |
| βc 366 | Glu | Yes | 1.65 | Oε → H35:Nζ | Yes | No |
| Oε → H33:N | ||||||
| βc 367 | His | Yes | 0.99 | Nε → L91:O | No | Yes |
| βc 368 | Ile | No | 0.16 | No | Yes | |
| βc 369 | Asp | No | 0 | No | No | |
| βc 370 | His | No | 0 | No | No | |
| βc 395 | His | No | 0.26 | ND | ND | |
| βc 416 | Thr | Yes | 0.29 | Oγ → L28:Oη | ND | ND |
| βc 418 | Arg | Yes | 1.01 | Nη → H97:O | Yes | No |
| βc 419 | Thr | No | 0.15 | No | No | |
| βc 420 | Gly | No | 0 | No | No | |
| βc 421 | Tyr | Yes | 0.45 | No | Yes | |
| βc 422 | Asn | No | 0 | No | No | |
| BION-1 light chain | ||||||
| L 28 | Tyr | Yes | 1.62 | Oη → βc416:Oγ | ||
| L 29 | Gly | No | 0.21 | |||
| L 30 | Asp | No | 0.18 | |||
| L 32 | Phe | Yes | 0.39 | |||
| L 91 | Asn | Yes | 0.17 | O → βc367:Nε | ||
| L 92 | Asn | No | 0.14 | |||
| L 93 | Glu | No | 0.13 | |||
| L 94 | Asp | Yes | 0.48 | Oδ → βc362:Nζ | ||
| Oδ → βc365:Oη | ||||||
| L 96 | Trp | Yes | 0.31 | |||
| BION-1 heavy chain | ||||||
| H 32 | Tyr | Yes | 0.60 | |||
| H 33 | Tyr | Yes | 1.00 | Oη → βc364:Oη | ||
| Oη → βc364:O | ||||||
| N → βc366:Oε | ||||||
| H 35 | Lys | No | 0 | Nζ → βc366:Oε | ||
| H 51A | Asn | Yes | 0.21 | |||
| H 53 | Asn | No | 0.37 | |||
| H 55 | Gly | Yes | 0.09 | |||
| H 57 | Thr | Yes | 0.18 | Oγ → βc363:Sδ | ||
| H 58 | Leu | No | 0.52 | |||
| H 96 | Asp | Yes | 0.06 | |||
| H 96A | Gly | Yes | 0.51 | |||
| H 97 | Ile | Yes | 0.13 | O → βc418:Nη | ||
| H 100A | Gly | Yes | 0.16 | |||
| Residue identity . | Residue type . | Vdw contact . | Buried area polar interactions (nm2) . | Required for binding BION-1 . | Required for affinity conversion . | |
|---|---|---|---|---|---|---|
| D4βc | ||||||
| βc 361 | Met | No | 0 | No | No | |
| βc 362 | Lys | Yes | 0.45 | Nζ → L94:Oδ | No | No |
| βc 363 | Met | Yes | 1.21 | Sδ → H57:N | ? | No |
| βc 364 | Arg | Yes | 0.33 | Nη → H33:Oη | ? | No |
| O → H33:Oη | ||||||
| βc 365 | Tyr | Yes | 0.87 | Oη → L94:Oδ | No | Yes |
| βc 366 | Glu | Yes | 1.65 | Oε → H35:Nζ | Yes | No |
| Oε → H33:N | ||||||
| βc 367 | His | Yes | 0.99 | Nε → L91:O | No | Yes |
| βc 368 | Ile | No | 0.16 | No | Yes | |
| βc 369 | Asp | No | 0 | No | No | |
| βc 370 | His | No | 0 | No | No | |
| βc 395 | His | No | 0.26 | ND | ND | |
| βc 416 | Thr | Yes | 0.29 | Oγ → L28:Oη | ND | ND |
| βc 418 | Arg | Yes | 1.01 | Nη → H97:O | Yes | No |
| βc 419 | Thr | No | 0.15 | No | No | |
| βc 420 | Gly | No | 0 | No | No | |
| βc 421 | Tyr | Yes | 0.45 | No | Yes | |
| βc 422 | Asn | No | 0 | No | No | |
| BION-1 light chain | ||||||
| L 28 | Tyr | Yes | 1.62 | Oη → βc416:Oγ | ||
| L 29 | Gly | No | 0.21 | |||
| L 30 | Asp | No | 0.18 | |||
| L 32 | Phe | Yes | 0.39 | |||
| L 91 | Asn | Yes | 0.17 | O → βc367:Nε | ||
| L 92 | Asn | No | 0.14 | |||
| L 93 | Glu | No | 0.13 | |||
| L 94 | Asp | Yes | 0.48 | Oδ → βc362:Nζ | ||
| Oδ → βc365:Oη | ||||||
| L 96 | Trp | Yes | 0.31 | |||
| BION-1 heavy chain | ||||||
| H 32 | Tyr | Yes | 0.60 | |||
| H 33 | Tyr | Yes | 1.00 | Oη → βc364:Oη | ||
| Oη → βc364:O | ||||||
| N → βc366:Oε | ||||||
| H 35 | Lys | No | 0 | Nζ → βc366:Oε | ||
| H 51A | Asn | Yes | 0.21 | |||
| H 53 | Asn | No | 0.37 | |||
| H 55 | Gly | Yes | 0.09 | |||
| H 57 | Thr | Yes | 0.18 | Oγ → βc363:Sδ | ||
| H 58 | Leu | No | 0.52 | |||
| H 96 | Asp | Yes | 0.06 | |||
| H 96A | Gly | Yes | 0.51 | |||
| H 97 | Ile | Yes | 0.13 | O → βc418:Nη | ||
| H 100A | Gly | Yes | 0.16 | |||
vdw contacts are βc with BION-1 or vice versa. The buried area (solvent exposure lost) on formation of the D4βc/BION-1 complex was calculated using dssp.25 The polar interactions include salt bridges and hydrogen bonds and are based on the distance and geometric criteria of Kabsch and Sander.25 The requirement for binding BION-110 and the requirement for affinity conversion8 9 are based on mutations to alanine. ND indicates not determined.
In total, 6 residues from the B-C loop (between residues 362-368) and 3 residues from the F-G loop (between residues 416-422) of D4βc are involved in antibody interactions with those from the B-C loop, accounting for 75% of the total. The B-C loop interacts with CDRs H1, H2, H3, L1, and L3, whereas the F-G loop interacts only with CDRs H3 and L1 (Figure4). There is one small cavity of 0.0099 nm3 in the antibody-antigen interface. The cavity is lined by residues Tyr365, His367, and Ile368 of the receptor and residues Val27, Tyr28, Phe32, and Asn92 of the antibody light chain. Not all of the potential salt bridges and hydrogen bonds identified above are likely to contribute productively to complex formation because substitution analysis has only identified Glu366, Arg418, and Met363 or Arg364 in D4βc as contributing to the epitope for binding BION-110 (Table 2).
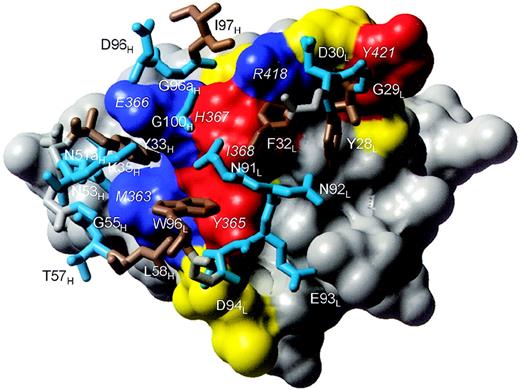
The BION-1/D4βc interface.
D4βc is shown as a surface representation colored according to the functional effect of residue substitution. Blue represents residues whose substitution abrogates binding of BION-1 but does not affect affinity-conversion. Red represents residues whose substitution reduces affinity-conversion but does not affect binding of BION-1. Yellow represents residues whose substitution does not affect binding of BION-1 or cytokines. Gray represents residues that have not been examined by mutation and do not contact BION-1. The identities of key D4βc residues are shown in italics. Residues in BION-1 that contact D4βc are shown in stick representation and colored cyan blue (hydrophilic) or brown (hydrophobic-aromatic). The backbone atoms of residues colored gray show the connectivity of the loops.
The BION-1/D4βc interface.
D4βc is shown as a surface representation colored according to the functional effect of residue substitution. Blue represents residues whose substitution abrogates binding of BION-1 but does not affect affinity-conversion. Red represents residues whose substitution reduces affinity-conversion but does not affect binding of BION-1. Yellow represents residues whose substitution does not affect binding of BION-1 or cytokines. Gray represents residues that have not been examined by mutation and do not contact BION-1. The identities of key D4βc residues are shown in italics. Residues in BION-1 that contact D4βc are shown in stick representation and colored cyan blue (hydrophilic) or brown (hydrophobic-aromatic). The backbone atoms of residues colored gray show the connectivity of the loops.
Discussion
We describe here the structure of the activating domain of the common βc of the GM-CSF/IL-3/IL-5 receptors complexed with the Fab fragment of the antagonistic mAb, BION-1. The structure shows general features typical of the cytokine receptor superfamily as well as unique features that reveal how a single receptor subunit can interact with 3 different cytokines. Functional analyses show the separate but cooperative interplay of the B-C loop and Tyr421 in the F-G loop in receptor activation.
A number of related (class 1) cytokine receptor structures are known: growth hormone receptor (GHR),26 prolactin receptor (PRLR),27 erythropoietin receptor (EPOR),28G-CSF receptor (G-CSFR),29 gp130,30and the IL-4 receptor α-chain (IL-4Rα-chain).31 The pair-wise sequence identities between D4βc and these receptors, after structure-based alignment, range from 12% (G-CSF) to 27% (gp130). There are only 7 residues (Pro343, Trp358, Leu402, Tyr408, Arg413, Gly423, and Ser426) that are strictly conserved across the receptors; all appear to play structural roles. The structural importance of Trp358 is highlighted by the observation that its substitution or the substitution of neighboring Tyr356 by Asn abrogates affinity conversion by βc.32 A structural superposition indicates that D4βc is most closely related to PRLR (0.16 nm studies on 88 Cα atoms, 20% sequence identity) followed by GHR (0.19 nm on 81 Cα atoms, 23% sequence identity). The root mean square deviation of other receptors indicates that (1) the membrane-distal B-C loop of the second domain within a CRM is normally involved in cytokine-binding and (2) the neighboring F-G loop of this domain and the A-B and E-F loops of the first domain also make contributions to cytokine-binding. The B-C loop of D4βc, in particular Tyr365 and His367, has been found to be involved in cytokine binding (Figure 3).7 8 G-CSFR, GHR, PRLR, and IL-4Rα-chain have an aromatic residue in an equivalent position to Tyr365, whereas only the IL-4Rα-chain has an aromatic residue (tyrosine) similar to His367 of βc. The IL-4Rα-chain is also the only receptor of known structure that has an aromatic residue (tyrosine) equivalent to Tyr421 in the F-G loop.
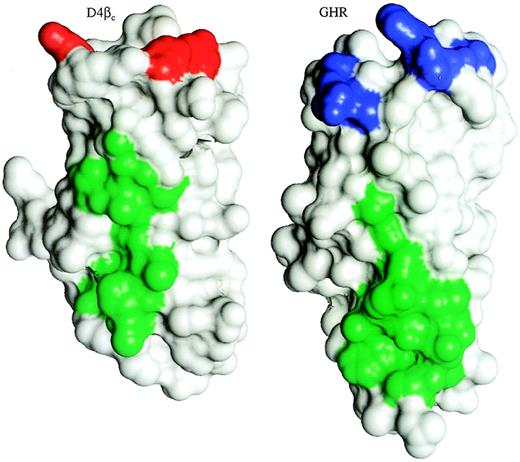
The most salient features of the D4βc crystal structure are the 2 hydrophobic-aromatic patches and the distinct groove, which is formed in part by the B-C and F-G loops and hence located at the putative cytokine-binding site. The hydrophobic-aromatic surface patches, H1 and H2 (Figure 1C, D), have corresponding features in most of the other receptors. With the exception of gp130, all the receptors possess significant hydrophobic-aromatic patches equivalent to the location of H2 (centered about the D-E strand connection), although the degree and extent of hydrophobicity varies greatly. The corresponding H2 patch of GHR (Figure 5) forms part of the surface involved in subunit contacts.26 This is suggestive of a role for the H2 of D4βc in association with α-chains, particularly the GMRα with which it associates spontaneously.23 The equivalent region to H1 is conserved in all but gp130. By analogy with the other receptors, the H1 patch of D4βc might interact with the A-B loop from domain 3 of the intact receptor. The groove is only present in G-CSFR, whereas the N-terminal ends of the equivalent domains of EPOR and gp130 are rather flat, and those of GHR and PRLR are mostly flat, with the exception of a tryptophan residue that protrudes into the solution.
Comparison of D4βc with the membrane-proximal domain of GHR.
D4βc and domain 2 of the subunit of the GHR, which interacts with the helix A/helix C face of GH, were aligned structurally via their core residues and are shown as surface representations using the program InsightII (MSI, San Diego, CA). The hydrophobic-aromatic patch, H2, of D4βc and the location of GHR that interacts with the opposing receptor molecule are indicated by green surfaces. The red surfaces of D4βc indicate the residues required for affinity-conversion, and the blue surfaces of GHR indicate the region known to interact with GH.
Comparison of D4βc with the membrane-proximal domain of GHR.
D4βc and domain 2 of the subunit of the GHR, which interacts with the helix A/helix C face of GH, were aligned structurally via their core residues and are shown as surface representations using the program InsightII (MSI, San Diego, CA). The hydrophobic-aromatic patch, H2, of D4βc and the location of GHR that interacts with the opposing receptor molecule are indicated by green surfaces. The red surfaces of D4βc indicate the residues required for affinity-conversion, and the blue surfaces of GHR indicate the region known to interact with GH.
There are considerable amounts of mutagenesis data available that indicate which regions of the receptor and the cytokine interact with each other. In the cytokines there is an essential glutamate (Glu21 of GM-CSF, Glu22 of IL-3, and Glu13 of IL-5) involved in binding to βc.33-35 The loops of domains 3 and 4 of βc have been the subject of extensive mutagenesis that has led to the following conclusions: (1) Tyr365, His367, and Ile368 of the B-C loop are implicated in cytokine interaction, whereas mutations of other residues in this loop have little or no effect on binding.7,8Substitution of any of these residues by alanine led to a loss of affinity-conversion of GM-CSF and IL-5 binding,8 whereas the Phe365 mutant retained affinity-conversion of GM-CSF binding.7 (2) The major residue in the F-G loop that has been implicated in binding9 is Tyr421. (3) To date, there have been no residues in domain 3 implicated in binding.3
The crystal structure of D4βc provides a molecular explanation of how each cytokine, with less than 15% pair-wise sequence identity, can recognize the signaling subunit. The side-chains of the 3 key residues in the B-C loop that interact with a cytokine, as identified by the mutagenesis studies, are seen to converge closely at their tips (Figure 1A). Thus Tyr365, His367, and Ile368 may play a pivotal role by promoting an interaction with the essential glutamate residue in all 3 cytokines. This may involve formation of a hydrogen bond between the glutamate residue and Tyr365 or His367 or the cooperative formation of part of the cytokine-binding surface. In this context it is worth noting that in the IL-4 receptor system, the homologous Tyr127 is close to Glu9 but does not form a hydrogen bond with it.31 All other residues of the B-C loop are orientated in a different direction from the binding triad, which is consistent with their not taking part in binding cytokines. On the other hand, Tyr421 in the neighboring F-G loop is positioned to contribute directly or indirectly to binding cytokines. Directly, through its hydroxyl group, Tyr421 may form a hydrogen bond with the conserved glutamate. This would be akin to the known interaction of Tyr183 in a homologous position in IL-4Rα-chain and Glu9 of IL-4.31 Indirectly, Tyr421 may interact with the A-B loop of domain 3 of βc, as seen with Phe205 of EPOR,28 and thus support an appropriate orientation of this domain, or Tyr421 may facilitate receptor assembly. Given that the active receptor probably has a stoichiometry of 2 α-chains : 2 βc : 2 ligands, Tyr421 may directly interact with either a second βc subunit or α-chain subunit in the hexameric complex.3
The separate and combined mutagenesis of the B-C loop and Tyr421 revealed that both sites are involved in high-affinity binding and receptor activation of all 3 cytokines, GM-CSF, IL-3, and IL-5, albeit in subtly different ways. The observation that the tetra-alanine substitution of the B-C loop abrogated IL-3 high-affinity binding (Figure 3A) is particularly interesting because single or paired alanine substitutions along this loop have marginal or no effect on IL-3 high-affinity binding.8 The latter is in contrast to GM-CSF and IL-5, where substitution of either Tyr365, His367, or Ile368 completely eliminates high-affinity binding.7,8 Conversely, substitution of Tyr421, while abrogating high-affinity binding of all 3 cytokines, has a profound effect on IL-3 receptor activation, as measured by phosphorylation of cytoplasmic tyrosine residues of βc(Figure 3B), but a minor effect on the activation of the GM-CSF receptor.9 These differences in receptor activation probably reflect the different abilities of mutated βc to be recruited to IL-3/IL-3Rα-chain complexes and suggest that, in the case of Y421A, the stability of the active IL-3/IL-3Rα-chain/βc complex is considerably lower than even that of an IL-3/IL-3Rα-chain complex.
The combination of substitutions within the B-C loop and Tyr421 caused a complete loss of tyrosine-phosphorylation of βc. This suggests that even in the presence of concentrations of IL-3 that saturate the IL-3Rα-chain, βc is not recruited significantly (Figure 3B). These functional observations in the context of the structure support the notion of a central cytokine-binding “hot spot” in D4βc, with a structural plasticity that allows it to accommodate 3 cytokines of significant diversity as well as monomeric (GM-CSF and IL-3) and dimeric (IL-5) structure. In this model, GM-CSF may interact more closely with the B-C loop, while the orientation of IL-3 may be slightly different and more dependent on Tyr421for its interaction with βc. Comparison of the cytokine-binding surface of D4βc with the corresponding surface of GHR reveals substantial similarity in terms of the parts of the B-C and F-G loops involved (Figure 5), although the contributions to cytokine binding of these GHR residues have not been assessed by mutational analysis for this homodimeric receptor. The location of these cytokine-binding residues of βc relative to the hydrophobic patch, H2, which may interact with α-chains, is similar to the intermolecular contacts seen in the GH:GHR complex.26 Ultimately, solving the structures of the GM-CSF and IL-3 receptor complexes may provide a definitive answer to the relative positioning of GM-CSF, IL-3, and their α-chains in respect to βc.
The epitope of D4βc that interacts with cytokines, largely overlaps the surface that is recognized by BION-1. Although several residues that are required for affinity-conversion (Tyr365 and His367 and others such as Lys362 make intimate contact) with BION-1, they are not required for binding of the mAb. Rather, a set of adjacent residues, including Met363 or Arg364, Glu366and Arg418, provides the key determinant for binding BION-1 (Figure 4). While the basis for the roles of these residues can be seen clearly from the structure, the absence of productive contributions to binding from other residues, especially Tyr365 and His367, suggests that their corresponding contacts in BION-1 may be targets for mutagenesis. This may lead to improved forms of the mAb or derivatives of it, which may be higher affinity antagonists. Because BION-1 has been shown to inhibit the GM-CSF/IL-3/IL-5–induced proliferation of eosinophils in vitro,10 this highlights the possibility of developing single-molecule antagonists of several cytokines. This approach of targeting a common receptor subunit may also be extended to other receptor chains, such as the common subunit of the IL-4/IL-13 receptors, which mediates allergen-induced asthma induced by IL-4 and IL-13.36
Note added in proof: The coordinates have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank, code 1EGJ.
Acknowledgments
We thank Craig Gaunt, Betty Zacharakis, and Frosa Katsis for technical assistance and Elspeth Garman for advice on flash freezing of crystals. We also thank Harry Tong and other staff at BioCARS for their help with data collection during our visit to Advanced Photon Source.
Supported in part by a grant from the National Health and Medical Research Council of Australia, Canberra, Australia, and a grant from the Australian Synchrotron Research Program, Australian Nuclear Science and Technology Organisation, Menai, Australia, which is funded by the Commonwealth of Australia under the Major National Research Facilities Program, Australia. Use of the Advanced Photon Source was supported under contract W-31-109-Eng-38 from the U.S. Department of Energy, Basic Energy Sciences, Office of Science, Washington DC. Use of the BioCARS Sector 14 (Advanced Photon Source, Argonne National Laboratory, 9700 South Cass Ave, Argonne, IL) was supported by grant RR07707 from the National Institutes of Health, National Center for Research Resources, Bethesda, MD.
J.R. and W.J.M. contributed equally to the structural biology aspects of this work. J.R. is a postdoctoral fellow of the Australian Research Council, Canberra, Australia. J.M.W. is a fellow of the Anti-Cancer Foundation of South Australia, Adelaide, South Australia, Australia; M.W.P. is a senior research fellow of the Australian Research Council, Canberra, Australia; and C.J.B. is a Florey fellow of the Royal Adelaide Hospital, Adelaide, South Australia, Australia.
Reprints: Angel F. Lopez, Cytokine Receptor Laboratory, Hanson Centre for Cancer Research, Institute of Medical and Veterinary Science, Frome Rd, Adelaide, South Australia 5000, Australia; e-mail: angel.lopez@imvs.sa.gov.au; or Michael W. Parker, The Ian Potter Foundation Protein Crystallography Laboratory, St Vincent's Institute of Medical Research, 41 Victoria Parade, Fitzroy, Victoria 3065, Australia; e-mail: mwp@rubens.its.unimelb.edu.au.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal