Abstract
Antigen (Ag) immunization induces formation of the germinal center (GC), with large, rapidly proliferating centroblasts in the dark zone, and small, nondividing centrocytes in the light zone. We identified a novel nuclear protein, GANP, that is up-regulated in centrocytes. We found that GANP was up-regulated in GC B cells of Peyer's patches in normal mice and in spleens from Ag-immunized mice. GANP-positive cells appeared in the light zone of the GC, with coexpression of the peanut agglutinin (PNA) (PNA)-positive B220-positive phenotype. The expression of GANP was strikingly correlated with GC formation because Bcl6-deficient mice did not show the up-regulation of GANP. GANP-positive cells were mostly surrounded by follicular dendritic cells. Stimulation with anti-μ and anti-CD40 induced up-regulation of ganp messenger RNA as well as GANP protein in B220-positive B cells in vitro. GANP is a 210-kd protein localized in both the cytoplasm and nuclei, with a homologous region to Map80 that is associated with MCM3, a protein essential for DNA replication. Remarkably, GANP is associated with MCM3 in B cells and MCM3 is also up-regulated in the GC area. These results suggest that the up-regulation of GANP might participate in the development of Ag-driven B cells in GCs through its interaction with MCM3.
Antigen (Ag) binding to B-cell antigen receptor (BCR) initiates activation and maturation of Ag-specific B cells in the peripheral lymphoid organs.1,2 B cells enter the outer periarterial lymphoid sheath and initiate costimulus-dependent interactions with specific Th cells and interdigitating dendritic cells within 48 hours after immunization.3,4 Ag-driven B cells proliferate in the outer periarterial lymphoid sheath and then undergo further activation in lymphoid follicles to establish germinal centers (GCs).5-7 Such B cells develop into large centroblasts that move rapidly through the cell cycle to form the dark zone and further differentiate into centrocytes (PNA+B220+sIgM+sIgD−CD38−) in the light zone of GCs.8-12 Centrocytes presumably undergo either apoptosis or differentiate into B cells, with affinity-maturated BCRs and class switching toward IgG-class antibodies (Abs). Ag-specific B cells selected in GCs migrate further into the red pulp of the spleen and differentiate into Ab-secreting plasma cells.
Several studies have demonstrated various differentiation Ags in GCs as molecules identified with monoclonal antibodies (mAbs) and their complementary DNAs (cDNAs) have been cloned.13-15 These molecules appear mostly in cells of the whole GC area; for example, GC kinase is up-regulated in GC B cells,14 whereas 8-oxoguanine DNA glycosylase is expressed in the dark zone.15 Ag-driven B cells appear as centroblasts that proliferate at a very high rate, accompanied by random somatic mutations in the V-region genes. The affinity maturation of BCRs on centroblasts potentially generates clones that require subsequent selection. Such mutated B cells will presumably migrate into the light zone and differentiate into centrocytes. Studies in several kinds of gene-targeted mice, including investigations of genes in the tumor necrosis factor (TNF)- or TNF-receptor families, demonstrated absence or impairment of GC formation after Ag immunization.16-18It is possible that the follicular dendritic cell (FDC) network or immune complexes trapped on FDCs are required for GC formation. FDCs sequester Ags in a form of immune complexes through the complement receptor (CR). It is generally assumed that the immune complexes trapped on the surface of FDCs compete for binding to Ags with the BCR expressed at low levels on the surface of centrocytes. Only centrocytes with highest affinity for the Ags will succeed in binding to Ags on FDCs. Centrocytes with low affinity will die from apoptosis, whereas those with the highest affinity will survive. In support of this idea, histochemical studies found that centrocytes are mostly surrounded by CR1-positive (CR+) FDCs, with tight contact.3FDC-mediated inhibition of apoptosis in GC B cells has been suggested, but the molecular mechanism remains largely unknown.
Studies have shown that RAG-1 and RAG-2 proteins are expressed selectively in centrocytes in the light zone.19,20 It is known that mature B cells undergo secondary Ig gene rearrangements that will create a variety of BCR specificity triggered by the binding of Ags. The GC area probably provides signals leading to receptor editing through T-cell–dependent activation of B cells, as described by Papavasiliou et al21 and Han et al.22Centrocytes expressing RAG proteins will undergo exchange of V-region segments, which would further generate new combinatory Ab specificities. The secondary repertoire generated in centrocytes would be selected, presumably in the microenvironment provided by the FDC network.
Although much information is available on GCs, the molecules involved in the differentiation and activation of GC B cells remain unknown. To study such molecules, we prepared mAbs against intracellular components of a murine B-cell line, WEHI-231. An mAb, 29-15, recognizes a differentiation Ag whose expression is selectively augmented in centrocytes of GCs in the spleens of Ag-immunized mice. The 29-15–positive (29-15+) B cells are surrounded by CR1+ FDCs, suggesting that there is a functional association between the 29-15 Ag and the FDCs. With use of cDNA cloning, we determined that the 29-15 Ag is a 210-kd phosphoprotein, and we named it GANP. GANP shows regional homology to Map80, a protein that binds to MCM3, which is a protein essential for the initiation of DNA replication.23 We found that GANP was associated with MCM3 in B cells and studied the possible relation between molecular events within the GC and DNA replication.
Materials and methods
Mice and cells
BALB/c mice and Lewis rats were purchased from Seac Yoshitomi (Fukuoka, Japan) and maintained in the Center for Animal Resources and Development at Kumamoto University. Mice were immunized by multiple intraperitoneal injections with sheep red blood cells (SRBC; Nippon Bio-Test Laboratories, Tokyo, Japan). Spleen B cells from the mice were enriched and cultured in RPMI-1640 medium as described previously.24
Establishment of 29-15 mAb
Antibodies and reagents
Reagents were the F(ab′)2 fragment of the affinity-purified goat antimouse μ Ab (anti-μ Ab; ICN Pharmaceutical, Costa Mesa, CA), biotin-conjugated PNA (Vector Laboratories, Burlingame, CA), biotin-conjugated anti-CD35 mAb (anti-CR1; PharMingen, San Diego, CA), alkaline phosphatase (ALP)-conjugated goat antirat Ig Ab (ALP-antirat Ig [59 301]; ICN Pharmaceutical), horseradish peroxidase (HRP)-conjugated goat antirat Ig Ab (HRP-antirat Ig; ICN Pharmaceutical), HRP-conjugated streptavidin (HRP-ST; Kirkegaard & Perry Laboratories, Gaithersburg, MD), mouse anti-bromodeoxyuridine (BrdU) mAb (Novocastra Laboratories, Newcastle, UK), ALP-conjugated goat antimouse Ig Ab (ALP-antimouse Ig; Sigma Chemical, St Louis, MI), fluorescein isothiocyanate-conjugated (FITC) mouse antirat κ mAb (FITC-antirat κ; ICN Pharmaceutical), phycoerythrin (PE)-conjugated anti-B220 mAb (PharMingen), and ALP-conjugated goat antirabbit Ig Ab (Zymed Laboratories, South San Francisco, CA). Biotin-conjugated mAbs, such as anti-B220 (RA3-6B2) and anti-δ (CS/15), and purified anti-CD40 mAb (LB429) were prepared as described previously.24 Rabbit antimouse MCM3/P1 Ab was prepared as described previously.27
Immunohistochemical studies
Six-micrometer cryosections of organs were placed on gelatin-coated slides, dried, and fixed in acetone.28,29 After rehydration in PBS, the sections were incubated with the 29-15 mAb, washed, incubated with ALP-antirat Ig, and developed with Vector Blue (Vector Laboratories). Double staining was carried out using biotin-labeled mAbs in combination with HRP-ST and development with 3,3′-diaminobenzidine tetrahydrochloride (Dojindo, Kumamoto, Japan). BrdU incorporation after labeling in vivo for 1 hour was detected with anti-BrdU mAb in combination with ALP-antimouse Ig and Vector Red (Vector Laboratories).30 Sections from RAG-1–deficient mice reconstituted with bone marrow cells from Bcl6-deficient (Bcl6−/−) mice (reconstitution marrow [RM]) were prepared as described previously.31 For cell smears, spleen B cells were attached to gelatin-coated slides, fixed with acetone, and stained by anti-GANP mAb in combination with ALP-antirat Ig. After development with Vector Red, sections were lightly counterstained with hematoxylin.
Molecular cloning of a cDNA clone and in situ RNA hybridization on tissue sections
A λgt-11 cDNA library of WEHI-231 was screened with the 29-15 mAb in combination with sheep antirat Ig Ab labeled with iodine 125 (Amersham, Buckinghamshire, UK).32 With the initial cDNA clone (280-base pair [bp] insert), we obtained longer cDNA clones and determined the coding sequence. In situ RNA hybridization was carried out as described previously.33 Paraffin-embedded sections mounted on silanized slides were hybridized withganp 280-bp riboprobe labeled with digoxigenin. Slides were washed with TNE buffer (10 mmol/L of Tris-hydrochloric acid (HCl) [pH 7.6], 500 mmol/L of sodium chloride (NaCl), and 1 mmol/L of EDTA) at 37 °C and with 1 × SSC solution at 50 °C and developed with anti-digoxigenin Ab followed by ALP substrate.
Preparation of the glutathione-S-transferase–cDNA fusion protein
The glutathione-S-transferase (GST)-GANP fusion protein was prepared from bacteria harboring a pGEX-4T-1 vector (Pharmacia Biotech, Piscataway, NJ)-based plasmid containing the ganp cDNA encoding amino acids 579 to 1028 with use of glutathione-Sepharose (Pharmacia Biotech) chromatography.32 Another hybridoma secreting the anti-GANP mAb 42-23, used for the immunoprecipitation, was established as described above by immunizing rats with the GST-GANP protein.
Immunoprecipitation-Western blot analysis
Proteins obtained by subcellular fractionation as described previously34 were immunoprecipitated with the anti-GANP mAb in combination with protein G-Sepharose and analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The Western blot filter was incubated with the anti-GANP mAb followed by HRP-antirat Ig. An enhanced-chemiluminescence (ECL) detection kit (Amersham) was used for development.
Immunostaining of spleen B cells
Spleen B cells were fixed with 2.5% paraformaldehyde in PBS and subjected to permeabilization with 70% ethanol. The cells were incubated with the 29-15 mAb in combination with FITC-antirat κ and were analyzed using a flow cytometer (FACScan; Becton Dickinson, Mountain View, CA) as described previously.24
Reverse transcriptase-polymerase chain reaction
Total RNA (1 μg each) purified from cultured B cells by using Trizol (Gibco BRL, Rockville, MD) was used as a template for cDNA synthesis (100-μL volume) with Superscript (GIBCO BRL). Polymerase chain reaction (PCR) amplification was carried out by using 2 μL of each cDNA solution with Taq Gold (Perkin-Elmer, Foster City, CA) and the primers for ganp or HPRT.20 Theganp transcripts were amplified by 5′-CCGTGGGATGACATCATCAC-3′ (the forward primer) and 5′-CATGTCCACCATCTCCAGCA-3′ (the reverse primer).
In vitro kinase assay and V8 cleavage mapping
Kinase reactions were carried out in vitro with immunoprecipitates as described previously.25 Spleen B cells were stimulated in vitro for 48 hours with anti-μ Ab and anti-CD40 mAb.24After harvesting and washing, cells were lysed with TNE buffer (10 mmol/L of Tris-HCl [pH 7.8], 150 mmol/L of NaCl, 1 mmol/L of EDTA, 1% NP-40, 10 μg/mL of aprotinin, and 1 mmol/L of phenylmethylsulfonyl fluoride [PMSF]), and immunoprecipitated with the anti-GANP mAb. The immunoprecipitates were incubated with γ-adenosine triphosphate labeled with phosphorous 32 (Amersham) and radiolabeled proteins, separated by SDS-PAGE, and detected with use of autoradiography. In addition to the TNE buffer, we used the more harsh washing buffer, radioimmunoprotein-assay (RIPA) buffer (10 mmol/L of Tris-HCl [pH 7.4], 150 mmol/L of NaCl, 1 mmol/L of EDTA, 1% NP-40, 0.1% sodium deoxycholate, 0.1% SDS, 10 μg/mL of aprotinin, and 1 mmol/L of PMSF) to detect the target molecule after removing other coprecipitated molecules. V8 cleavage mapping of the indicated protein was performed as described previously.25
cDNA transfection and immunoprecipitation
The expression vector of influenza hemagglutinin (HA)-tagged mouse MCM3 was described previously.35 An expression vector for FLAG-taggedganp cDNA was prepared with pCXN2 vector.36 COS7 cells were transfected with 4 μg of each plasmid DNA by LipofectAMINE Plus (GIBCO BRL) according to the manufacturer's instructions. After 48 hours, cells were harvested, washed, and lysed in TNE buffer. The lysates were immunoprecipitated with rabbit anti-HA Ab (Santa Cruz Biotechnology, Santa Cruz, CA) in combination with protein A-Sepharose. After SDS-PAGE and electrotransfer, the blot filter was incubated with mouse anti-FLAG mAb (M2; Stratagene, La Jolla, CA). For ECL detection, we used HRP-conjugated goat antimouse Ig Ab (Zymed) as the secondary Ab.
Results
Expression of the 29-15 Ag in lymphoid organs
We prepared an mAb that recognizes a differentiation Ag expressed in peripheral B cells by immunizing rats with the lysate of WEHI-231. Immunohistochemical analysis with the 29-15 mAb on normal lymphoid organs of BALB/c mice did not detect expression of the Ag in the bone marrow, but lymphoid organs such as the thymus, spleen, and lymph nodes were stained weakly (data not shown). Interestingly, expression of the 29-15 Ag was very high in the central area of follicles of the Peyer's patches (Figure 1A). The Ag was expressed in B220-positive (B220+) IgD-negative B cells. In the study with lymphoid follicles from Ag-immunized mice, 29-15+ cells appeared in the GC area of the spleen (Figure1B) as well as in GCs of the Peyer's patches (Figure 1A). In secondary lymphoid follicles, nearly half of the PNA-positive (PNA+) GC B cells were positive with the 29-15 mAb but were almost all negative with the anti-BrdU mAb. Interestingly, expression of the 29-15 Ag was up-regulated in the centrocyte area at the distal region of the central artery. To confirm this, we used the Bcl6−/− RM, which does not show formation of GCs after Ag immunization, while the extrafollicular responses seem to be intact.31 We compared the expression of the 29-15 Ag on follicular regions in Bcl6−/− RM and Bcl6-positive (Bc16+/+) RM. Ag immunization induced the formation of GCs in Bcl6+/+ RM but not in Bcl6−/−RM (Figure 1C). The 29-15 Ag was up-regulated in PNA+ cells in Bcl6+/+ RM.
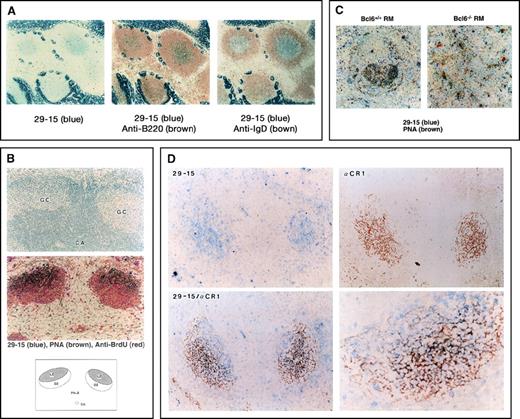
Detection of 29-15–positive (29-15+) germinal center (GC) B cells.
(A) The immunohistochemical analysis was carried out on Peyer's patches with the 29-15 monoclonal antibody (mAb) and alkaline phosphatase (ALP) antirat Ig antibody (Ab). Positive cells, stained with Vector Blue ALP substrate, appear in the central area . The strong signal in the surrounding area is in intestinal villi containing nonspecific endogenous ALP activity. For 2-color staining, the sections were further stained with either biotin–anti-B220 mAb or biotin–anti-IgD mAb followed by horseradish peroxidase-streptavidin in combination with 3,3′-diaminobenzidine tetrahydrochloride. (B) The sections of the GC area from mice immunized with sheep red blood cells (SRBC) were stained with PNA, anti-bromodeoxyuridine, and the 29-15 mAb in combination with the individual colors. The upper panel shows hematoxylin staining of the GC area and the central artery (CA). The middle panel shows 3-color staining, indicating 29-15+PNA-positive (PNA+) cells. The lower panel indicates the location of 29-15+PNA+ cells. (C) The sections from Bcl6-positive (Bc6+/+) reconstitution marrow (RM) and Bcl6-negative (Bc5−/−) RM after immunization were doubly stained with the 29-15 mAb and PNA. No 29-15+PNA+ cells were observed in Bcl6−/− RM. (D) Sections from SRBC-immunized mice were analyzed with the 29-15 mAb (blue) and anti-CR1 mAb (brown). Two-color staining shows the close contact of 29-15+ cells with CR1-positive follicular dendritic cells (FDC). The magnification in the photomicrograph at the lower left is × 125; that in the photomicrograph at the lower right is × 320.
Detection of 29-15–positive (29-15+) germinal center (GC) B cells.
(A) The immunohistochemical analysis was carried out on Peyer's patches with the 29-15 monoclonal antibody (mAb) and alkaline phosphatase (ALP) antirat Ig antibody (Ab). Positive cells, stained with Vector Blue ALP substrate, appear in the central area . The strong signal in the surrounding area is in intestinal villi containing nonspecific endogenous ALP activity. For 2-color staining, the sections were further stained with either biotin–anti-B220 mAb or biotin–anti-IgD mAb followed by horseradish peroxidase-streptavidin in combination with 3,3′-diaminobenzidine tetrahydrochloride. (B) The sections of the GC area from mice immunized with sheep red blood cells (SRBC) were stained with PNA, anti-bromodeoxyuridine, and the 29-15 mAb in combination with the individual colors. The upper panel shows hematoxylin staining of the GC area and the central artery (CA). The middle panel shows 3-color staining, indicating 29-15+PNA-positive (PNA+) cells. The lower panel indicates the location of 29-15+PNA+ cells. (C) The sections from Bcl6-positive (Bc6+/+) reconstitution marrow (RM) and Bcl6-negative (Bc5−/−) RM after immunization were doubly stained with the 29-15 mAb and PNA. No 29-15+PNA+ cells were observed in Bcl6−/− RM. (D) Sections from SRBC-immunized mice were analyzed with the 29-15 mAb (blue) and anti-CR1 mAb (brown). Two-color staining shows the close contact of 29-15+ cells with CR1-positive follicular dendritic cells (FDC). The magnification in the photomicrograph at the lower left is × 125; that in the photomicrograph at the lower right is × 320.
As an essential component of the GC area, FDCs appear in the dark and light zones. It is known that Ag-driven B cells are surrounded closely by the FDC network in the light zone. To investigate the close contact with FDCs, the locations of staining in the 29-15+ and CR1+ cells were compared. Anti-CR1 staining showed a net-like appearance of FDCs surrounding 29-15+ B cells (Figure 1D). The staining profiles were mostly superimposed on the same area with the individual colors (blue for 29-15 and brown for CR1), but there were few doubly stained cells (lower panels). This staining pattern confirmed that 29-15+ cells are the centrocytes surrounded by FDCs. The 29-15+ B cells appeared in a more proximal area of the GC than the CR1+ cells, suggesting that up-regulation of the 29-15 Ag begins during the transition from centroblasts to centrocytes. The 29-15 Ag was observed mainly in centrocytes trapped in the FDC network.
Identification of a cDNA clone encoding GANP Ag
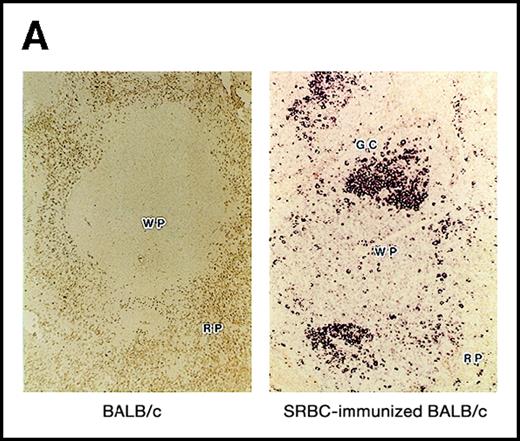
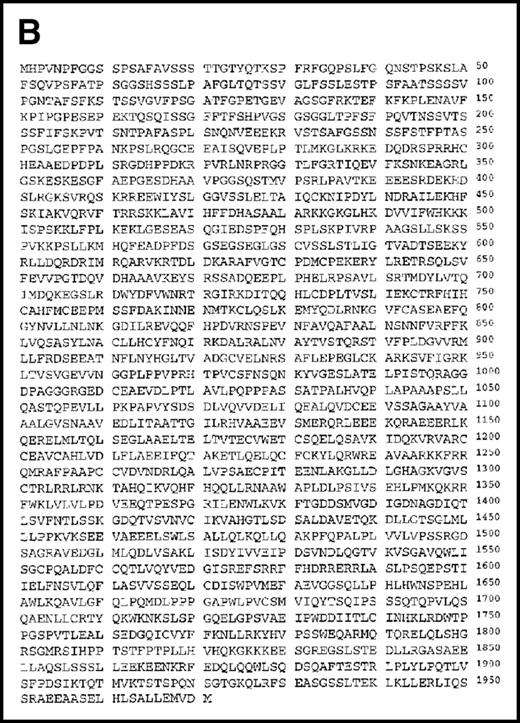
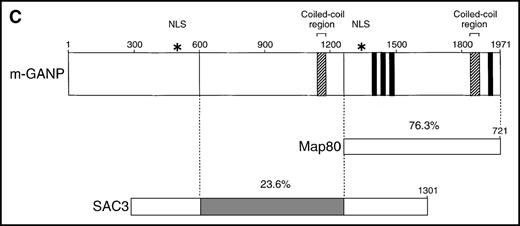
Using the 29-15 mAb, we isolated a ganp cDNA clone. We studied whether the ganp messenger RNA (mRNA) was up-regulated in the same area detected on sections with the 29-15 mAb. In situ RNA hybridization analysis revealed that the ganp mRNA was expressed abundantly in the central area of GCs of spleens from mice immunized with SRBC but not in spleens (Figure2A), thymuses, or lymph nodes from nonimmunized mice (data not shown). The ganp mRNA was up-regulated in GC B cells of immunized mice. This expression pattern was quite similar to the results with the 29-15 mAb on similar sections stained with hematoxylin. Further characterization of overlapping cDNA clones revealed a full-length nucleotide sequence (6429 bp). Theganp cDNA encoded a putative polypeptide composed of 1971 amino acids with a predicted molecular size of 210 kd (Figure 2B). GANP showed a regional homology to SAC3, a possible nuclear transcription regulator characterized in temperature-mutantSaccharomyces cerevisiae (Figure 2C),37 and to human Map80 protein.23 GANP has 2 nuclear localization sequences (497HKKK and 1344PMKQKRR), which could potentially support the expression of GANP in the nucleus, as suggested by the PSORT computer program. GANP also has 2 coiled-coil motifs but no zinc-finger, leucine-zipper, or homeo-domain motifs. There are 4 LXXLL motifs that are observed in nuclear transcription coactivator molecules, including CBP/p300 and p/CIP,38 39 but we did not identify the associated molecules through these motifs.
Structure of the GANP protein.
(A) Sections of spleens from SRBC-immunized and nonimmunized BALB/c mice were hybridized with the ganp antisense probe. The white pulp area (WP), red pulp area (RP), and GC area are indicated. (B) Deduced amino acid sequence of the ganp-encoded protein (sequence data available from the European Molecular Biology Laboratory; accession number AJ006590). (C) Schematic representation of the murine GANP protein. The region homologous to SAC3 and Map80, nuclear localization sequences (NLSs), and coiled-coil regions are indicated. The 4 LXXLL motifs are indicated by black.
Structure of the GANP protein.
(A) Sections of spleens from SRBC-immunized and nonimmunized BALB/c mice were hybridized with the ganp antisense probe. The white pulp area (WP), red pulp area (RP), and GC area are indicated. (B) Deduced amino acid sequence of the ganp-encoded protein (sequence data available from the European Molecular Biology Laboratory; accession number AJ006590). (C) Schematic representation of the murine GANP protein. The region homologous to SAC3 and Map80, nuclear localization sequences (NLSs), and coiled-coil regions are indicated. The 4 LXXLL motifs are indicated by black.
Regulation of GANP expression by BCR- and CD40-mediated signals
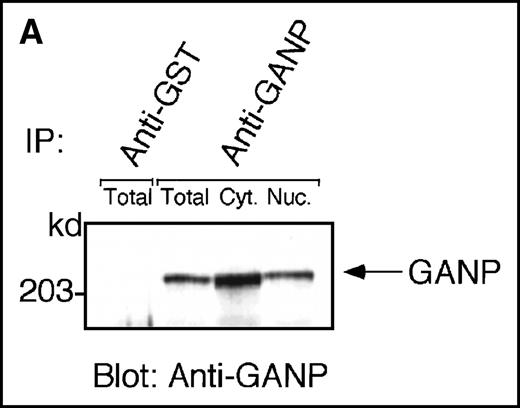
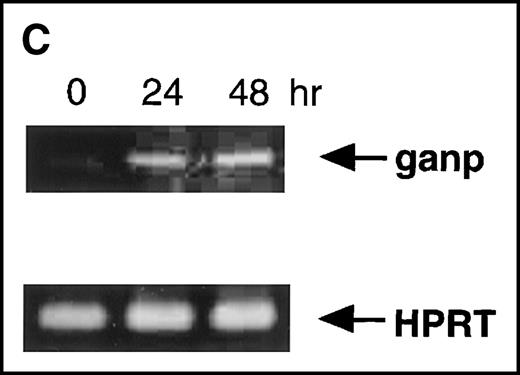
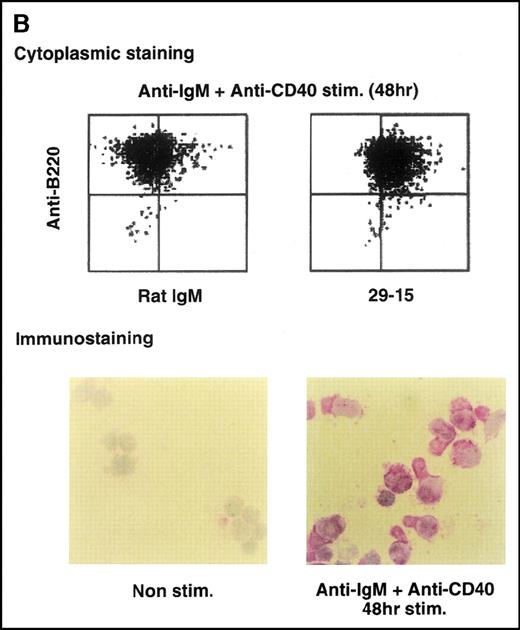
Anti-GANP mAb 42-23 prepared with the gene product detected a protein band at 210 kd in both the nuclear and cytoplasmic compartments of WEHI-231 (Figure 3A) The GANP in the cytoplasmic fraction might result from the transition from the nucleus in WEHI-231. Because the signals through BCR and CD40 are crucial for activation of GC B cells, we examined whether either or both signals caused up-regulation of GANP expression in vitro. Flow cytometric analysis and cytostaining on the smears (Figure 3B) found that B220+ B cells showed up-regulation of GANP in comparison to the control with irrelevant mAb. Stimulation with both anti-μ Ab and anti-CD40 mAb produced a maximal response, but use of either of these reagents alone resulted in only a marginal response (data not shown). The up-regulation was also detected by the increase in ganpmRNA in B cells stimulated by anti-μ and anti-CD40 coligation in vitro (Figure 3C). Reverse transcriptase (RT-PCR) demonstrated that the amount of ganp mRNA increased at 24 and 48 hours after stimulation in comparison with the control HPRT mRNA.
Characterization of the GANP protein.
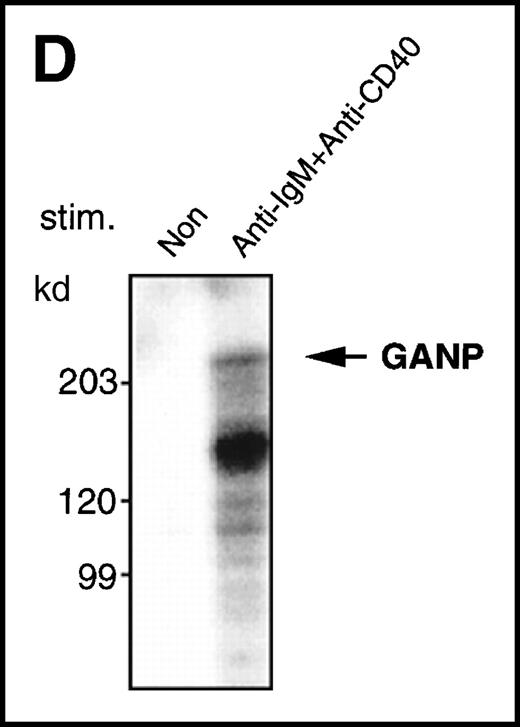
(A) The GANP protein is detected as a 210-kd protein in cytoplasmic and nuclear fractions from WEHI-231 by Western blot analysis after immunoprecipitation with the anti-GANP 42-23 mAb. Anti-glutathione-S-transferase (GST) mAb was used as an isotype-matched control mAb. (B) Spleen B cells from normal BALB/c mice were stimulated with F(ab′)2 of goat anti-μ Ab (10 μg/mL) and anti-CD40 mAb (10 μg/mL) for 48 hours and stained with the anti-GANP 29-15 mAb. The upper panel shows the profile of the cytoplasmic staining analyzed by the flow cytometry (FACScan). After permeabilization, cells were stained with the 29-15 mAb followed by fluorescein isothiocyanate-conjugated anti-κ. Two-color staining with phycoerythrin-B220 mAb showed the increase of GANP in B220-positive B cells. The lower panel shows cytostaining on the smears with the 29-15 mAb in combination with ALP-antirat Ig. (C) Reverse transcriptase-polymerase chain reaction assay showed the up-regulation of ganp messenger RNA in B cells stimulated with antimouse μ Ab and anti-CD40 in vitro. HPRT was used as a control to confirm the amount of each template. (D) The cell lysate was prepared from unstimulated (left) or stimulated (right) cells and subjected to anti-GANP immunoprecipitation. In vitro kinase reaction was carried out for 10 minutes with the anti-GANP 42-23 immunoprecipitates in the presence of γ-adenosine triphosphate labeled with phosphorous 32. Phosphorylation on the proteins was detected by autoradiography after separation with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Arrow indicates the position of phosphorylated GANP.
Characterization of the GANP protein.
(A) The GANP protein is detected as a 210-kd protein in cytoplasmic and nuclear fractions from WEHI-231 by Western blot analysis after immunoprecipitation with the anti-GANP 42-23 mAb. Anti-glutathione-S-transferase (GST) mAb was used as an isotype-matched control mAb. (B) Spleen B cells from normal BALB/c mice were stimulated with F(ab′)2 of goat anti-μ Ab (10 μg/mL) and anti-CD40 mAb (10 μg/mL) for 48 hours and stained with the anti-GANP 29-15 mAb. The upper panel shows the profile of the cytoplasmic staining analyzed by the flow cytometry (FACScan). After permeabilization, cells were stained with the 29-15 mAb followed by fluorescein isothiocyanate-conjugated anti-κ. Two-color staining with phycoerythrin-B220 mAb showed the increase of GANP in B220-positive B cells. The lower panel shows cytostaining on the smears with the 29-15 mAb in combination with ALP-antirat Ig. (C) Reverse transcriptase-polymerase chain reaction assay showed the up-regulation of ganp messenger RNA in B cells stimulated with antimouse μ Ab and anti-CD40 in vitro. HPRT was used as a control to confirm the amount of each template. (D) The cell lysate was prepared from unstimulated (left) or stimulated (right) cells and subjected to anti-GANP immunoprecipitation. In vitro kinase reaction was carried out for 10 minutes with the anti-GANP 42-23 immunoprecipitates in the presence of γ-adenosine triphosphate labeled with phosphorous 32. Phosphorylation on the proteins was detected by autoradiography after separation with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Arrow indicates the position of phosphorylated GANP.
Because the 210-kd GANP has many possible phosphorylation sites, we examined the induction of phosphorylation by an in vitro kinase reaction with anti-GANP immunoprecipitates after lysis of cells with TNE buffer containing NP-40. As shown in Figure 3D, phosphorylation of the 210-kd protein was found in the anti-GANP immunoprecipitates from spleen B cells stimulated by anti-μ and anti-CD40 coligation. This result indicates that a kinase activity is coprecipitated with GANP, which could be a phosphoprotein.
Association of GANP with MCM3 protein
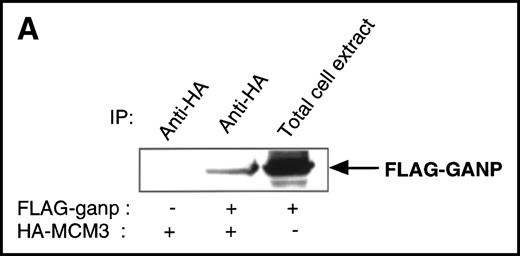
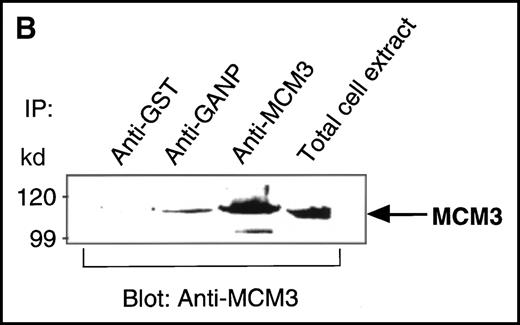
We found a Map80-homologous region (76.3% identity at the amino acid level) in the carboxyl-terminal part of GANP. Map80 is an 80-kd nuclear protein that is involved in the translocation of MCM3, a protein essential for DNA replication, between the cytoplasm and the nuclei.23,27 40-44 Therefore, we examined the interaction between GANP and MCM3 in cells. First, we used COS7 cells transfected with cDNA of HA-MCM3 and FLAG-ganp. GANP coprecipitated with MCM3 in the double cDNA transfectants detected by Western blot analysis (Figure 4A). Next, we used WEHI-231 cells in experiments to determine whether MCM3 would coprecipitate with GANP. Anti-GANP immunoprecipitates also coprecipitated with the 100-kd protein of MCM3 (Figure 4B). These results indicate that GANP is associated with MCM3 in vivo.
Physical association between GANP and MCM3.
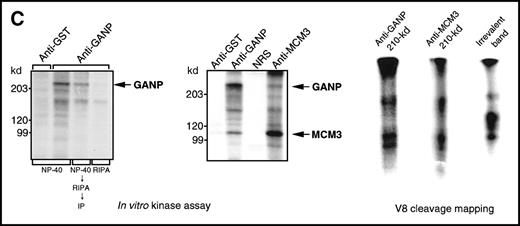
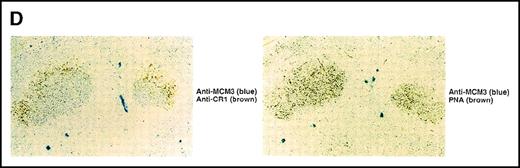
(A) Association of MCM3 and GANP in cells in vivo. COS7 cells were transfected with complementary DNA (cDNA) constructs of both HA-MCM3 and FLAG-ganp. The NP-40 cell lysates were immunoprecipitated with anti-HA Ab, and Western blot analysis was carried out with anti-FLAG mAb to detect GANP. The transfectants with the single cDNA constructs were used as the negative control. (B) The cell lysate from WEHI-231 was immunoprecipitated with anti-GST, anti-GANP 42-23, or anti-MCM3 Ab. After separation by SDS-PAGE, the proteins were electrophoretically transferred to a membrane and probed with anti-MCM3 Ab. (C, left panel) In vitro kinase reactions were carried out with the anti-GANP immunoprecipitates washed with either NP-40 washing buffer or radioimmunoprotein-assay (RIPA) buffer. After the kinase reaction, the phosphorylated sample was washed further with the RIPA buffer and reimmunoprecipitated. The anti-GST immunoprecipitate was used as the negative control. (C, middle panel) Anti-GST, anti-GANP 42-23, and anti-MCM3 immunoprecipitates from WEHI-231 cell lysates were subjected to in vitro kinase assay. Normal rabbit serum was used as the control for anti-MCM3 Ab. The samples were separated by 7% SDS-PAGE. The bands corresponding to the sizes of GANP and MCM3 are indicated by arrows. (C, right panel) V8 cleavage mapping of 210-kd bands showed an identical cleavage pattern. As the control, an irrelevant V8-digested protein was separated in parallel. (D) Double staining with anti-MCM3 Ab and anti-CR1 mAb, or with PNA, was performed. The expression of MCM3 was up-regulated in the GC area.
Physical association between GANP and MCM3.
(A) Association of MCM3 and GANP in cells in vivo. COS7 cells were transfected with complementary DNA (cDNA) constructs of both HA-MCM3 and FLAG-ganp. The NP-40 cell lysates were immunoprecipitated with anti-HA Ab, and Western blot analysis was carried out with anti-FLAG mAb to detect GANP. The transfectants with the single cDNA constructs were used as the negative control. (B) The cell lysate from WEHI-231 was immunoprecipitated with anti-GST, anti-GANP 42-23, or anti-MCM3 Ab. After separation by SDS-PAGE, the proteins were electrophoretically transferred to a membrane and probed with anti-MCM3 Ab. (C, left panel) In vitro kinase reactions were carried out with the anti-GANP immunoprecipitates washed with either NP-40 washing buffer or radioimmunoprotein-assay (RIPA) buffer. After the kinase reaction, the phosphorylated sample was washed further with the RIPA buffer and reimmunoprecipitated. The anti-GST immunoprecipitate was used as the negative control. (C, middle panel) Anti-GST, anti-GANP 42-23, and anti-MCM3 immunoprecipitates from WEHI-231 cell lysates were subjected to in vitro kinase assay. Normal rabbit serum was used as the control for anti-MCM3 Ab. The samples were separated by 7% SDS-PAGE. The bands corresponding to the sizes of GANP and MCM3 are indicated by arrows. (C, right panel) V8 cleavage mapping of 210-kd bands showed an identical cleavage pattern. As the control, an irrelevant V8-digested protein was separated in parallel. (D) Double staining with anti-MCM3 Ab and anti-CR1 mAb, or with PNA, was performed. The expression of MCM3 was up-regulated in the GC area.
Because the phosphorylation states of MCM proteins seem crucial in the regulation of cell-cycle progression,27 40-44 we studied whether the phosphorylation reaction of the anti-GANP immunoprecipitates was similar to that of the anti-MCM3 immunoprecipitate (Figure 4C; left panel). The in vitro kinase reaction revealed active protein kinase activity in the GANP precipitates when we used NP-40 lysis and washing buffer (TNE buffer). The 210-kd phosphorylated band, however, was not visible when the immunoprecipitate was prewashed with the more harsh RIPA buffer, suggesting that the weakly coprecipitated molecules had been removed. Rewashing after the kinase reaction and reimmunoprecipitation revealed the 210-kd protein, which suggests that phosphorylation occurs in 210-kd GANP. Immunoprecipitation of MCM3 also coprecipitated a similar 210-kd phosphorylatable protein (Figure 4C; middle panel). These 210-kd bands from anti-GANP and anti-MCM3 immunoprecipitates showed an identical pattern in the V8 cleavage mapping (Figure 4C; right panel), suggesting that GANP and MCM3 are associated with the common molecules for phosphorylation in vitro.
We next studied whether MCM3 is up-regulated in GC B cells by Ag immunization of mice in vivo. Sections contiguous to those used in the experiments described above were stained with the anti-MCM3 Ab (Figure4D). MCM3 was also up-regulated in GCs. Double staining showed colocalization of MCM3 and PNA. Part of the GC area was surrounded closely by FDCs. These results demonstrated that MCM3 is up-regulated in GC B cells, including centroblasts and the GANP-positive (GANP+) centrocytes mostly surrounded by FDCs (Figure 1D and Figure 4D).
Discussion
We found a novel protein, GANP, that is expressed in GC B cells localized in the light zone of secondary follicles in the spleens of Ag-immunized mice. Although a trace amount of the ganp mRNA was detectable in many kinds of cells under normal conditions (data not shown), GANP appears to be up-regulated in the specified GC area in immunized mice. GANP might be required for maturation of Ag-specific B cells at the centrocyte stage.
Ag immunization induces the formation of GCs that are assumed to be the activation platforms for effective Ab responses. The dark and light zones are divided into 2 compartments, each of which contain B cells in different activation states. Ag-driven B cells are triggered initially for proliferation to enlarge the clone size, accompanied by a high rate of somatic mutation of the BCR genes. Such B cells inevitably involve the altered BCR specificity presumably associated with the generation of self-reactive B-cell clones. After sufficient cell divisions, centroblasts arrest cell cycling at the level of little somatic mutation and become centrocytes. Centrocytes expressing low amounts of BCR are selected through the differentiation in the FDC network of the light zone.3 GANP+ B cells showed mostly close contact with CR1+ FDCs that were occupying the tight interspace of the light-zone area. The role of FDCs has been characterized as the major support for the activation and maturation of Ag-driven centrocytes. Histologic studies showed that Ag-driven B cells interact through binding of LFA-1 to ICAM-1 and VLA-4 to VCAM-1 on FDCs.45 These interactions are thought to stimulate FDCs to prevent the apoptotic pathway or to lead to the assembly of molecular machinery for B-cell selection.45
We found that the carboxyl-terminal portion of GANP has a marked similarity to human Map80, which facilitates the nuclear transport of MCM3.23 Immunoprecipitation experiments demonstrated that GANP also binds to MCM3 in WEHI-231. MCM3 is a member of the MCM protein family essential for the initiation of DNA replication.27,40-44 The major fractions of nuclear MCM proteins bind to chromatin at the beginning of the S phase but dissociate during replication and accumulate as free proteins in the nuclei. The release of MCMs from chromatin is accompanied by the phosphorylation of several MCM proteins, and their reassociation after mitosis is concomitant with their dephosphorylation. It has been suggested that MCM proteins are no longer synthesized in growth-arrested differentiating cells and disappear, with kinetics related to their half-life.46 The MCM3 protein was shown to be an early target in apoptotic proteolysis.47 Schwab et al47 proposed that active destruction of MCM3 inactivates the MCM complex and serves to prevent untimely DNA replication events during execution of the cell-death program.
We found that GC B cells express high levels of MCM3, some of which is associated with GANP. However, it is curious that a protein that is up-regulated in differentiated cells that arrest the cell cycle binds to a protein essential for progression of the S phase. One possible explanation for this finding is that a function of GANP is inactivation of MCM3 by means of binding. Our immunohistochemistry data are consistent with this idea: GANP was up-regulated in growth-arrested centrocytes, whereas MCM3 was expressed in both rapid-cycling centroblasts and centrocytes in GCs. Although the amount of MCM3 would decrease if gene expression and active destruction ceased,46,47 inactivation of MCM3 (which is still expressed in centrocytes) through the interaction with GANP might be another mechanism to prevent DNA replication. In addition, both GANP and MCM3 became phosphorylated with the coprecipitated kinase (Figure 4B). Because highly phosphorylated MCM3 is thought to be an inactivated form,27 the association with GANP may stimulate phosphorylation of MCM3.
Additionally, GANP has a close similarity to the suppressor of actin 3 (SAC3) gene of Saccharomyces cerevisiae, which was isolated in a genetic screen for suppressors of a temperature-sensitive mutation (act1-1) in the essential actin gene (Figure3C).37 SAC3 protein is expressed in nuclei and is required for normal progression of mitosis and protection against chromosome loss.48 Null mutants of SAC3 grow slowly and are larger than wild-type cells.37 SAC3 participates in a process that affects both the actin cytoskeleton and mitosis, which suggests that SAC3 regulates gene expression of actin or actin-binding proteins. LEP-1, a gene that augments the transcription of the leucine permease activity in Saccharomyces organisms, was identical to SAC3.49 The LEP-1 gene induces up-regulation of the yeast leucine permease involved in selective amino acid transport. Our knowledge of amino acid transport in eukaryotic cells is insufficient, especially with regard to the molecules involved in amino acid permeation.50 Although the SAC3/LEP-1sequence did not show motifs homologous to a number of transcription factors, the biologic functions observed previously49suggest its regulatory activity of various target genes in the nucleus. Mouse ganp did not show consensus motifs typical for nuclear transcription factors, but it has structural similarity to the ancestorSAC3, possible phosphorylation sites, 2 nuclear localization sequences, and 2 coiled-coil structures that might interact with other transcription molecules.
We found that GANP is selectively up-regulated in centrocytes of spleens from Ag-immunized mice. It is also useful as the differentiation marker to define the centrocyte subset that is interacting closely with FDCs in the GC area. Our study showed that the BCR signal and the CD40 costimulation together cause the up-regulation of GANP and lead to the signal transduction mediated through the GANP/MCM3 complex. Studies of GANP expression and its function in B-cell activation would improve our understanding of immune responses.
Acknowledgments
We thank Mitsuru Matsumoto (University of Tokushima) and Kensuke Miyake (Saga Medical School) for valuable discussions and useful mAbs, respectively.
Supported in part by a grant-in-aid from the Ministry of Education, Science, Sports, and Culture in Japan. K.K. was supported by research fellowships from the Japanese Society for the Promotion of Science for Young Scientists.
Reprints:Nobuo Sakaguchi, Department of Immunology, Kumamoto University School of Medicine, 2-2-1, Honjo, Kumamoto 860-0811, Japan; e-mail: nobusaka@kaiju.medic.kumamoto-u.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal