The autoimmune lymphoproliferative syndrome (ALPS) is a rare inherited disorder associated with lymphadenopathy, accumulation of CD4/CD8 double-negative T cells, and a broad range of autoimmune phenomena.1 The clinical phenotype can, in all cases, be ascribed to defective lymphocyte apoptosis caused by defects in the Fas signaling pathway. However, recent work has suggested that at least 3 genes may account for the inherited component of the disease. The most common form of the disease, designated ALPS type Ia, is associated with inherited mutations in the Fas receptor gene,APT1,2 whereas ALPS type Ib is associated with mutations in the gene encoding the Fas ligand.3 The remaining cases with documented lack of mutations in either of the 2 above-mentioned genes have been collectively assigned as ALPS type II.4
In a recent issue of Cell, Wang et al5 reported on inherited missense mutations in the caspase-10 gene in two kindreds with ALPS type II. One of the probands carried a heterozygous mutation,L285F (nucleotide change C853T; nomenclature referring to the long caspase-10 isoform; Genbank accession number AF111345), that was inherited from the mother who also showed features consistent with ALPS. The other proband, an Ashkenazi Jew, was homozygous for a G to A transition at position 1228, predicted to cause the substitution of valine with isoleucine at codon 410. Both heterozygous parents were clinically normal. None of the caspase-10 variants was found on 440 normal alleles, including 200 alleles of Ashkenazi Jewish origin. The different modes of inheritance in the 2 kindreds are compatible with the investigators' in vitro expression studies, showing that the L285F mutation severely impairs caspase-10-mediated apoptosis and acts in a dominant-negative fashion, whereas the V410I mutation has a significant, albeit less severe, effect on apoptosis. The mutant caspase-10 alleles were shown to affect apoptosis induced byvarious death receptors including Fas, TNFR1, DR3, DR4(TRAIL-R1) and DR5 (TRAIL-R2), presumably by interaction with caspase-8/FADD in the death-inducing signaling complexes(DISCs).
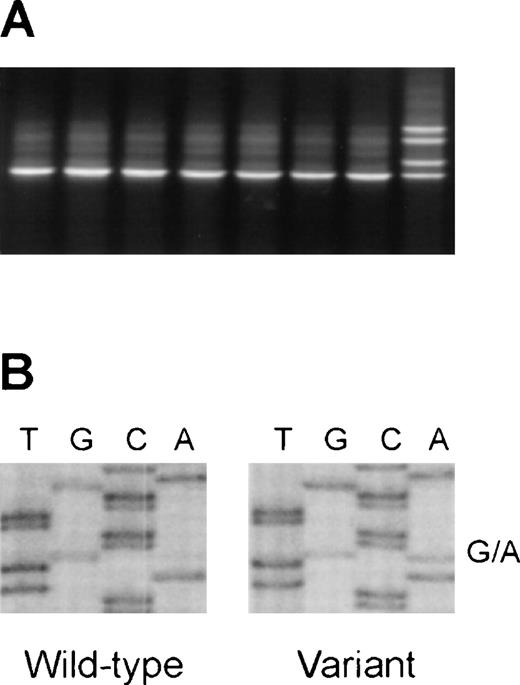
Detection of the V410I (G1228A) variant of the caspase-10 gene in DNA from healthy blood donors.
(A) Representative denaturing gradient gel electrophoresis (DGGE)analysis of a region encompassing nucleotides 1186-1322 (codons 396-441) of the caspase-10 gene. (B) Direct sequence analysis of a sample with aberrant DGGE pattern, showing the G1228A variant.
Detection of the V410I (G1228A) variant of the caspase-10 gene in DNA from healthy blood donors.
(A) Representative denaturing gradient gel electrophoresis (DGGE)analysis of a region encompassing nucleotides 1186-1322 (codons 396-441) of the caspase-10 gene. (B) Direct sequence analysis of a sample with aberrant DGGE pattern, showing the G1228A variant.
We have recently demonstrated somatic APT1 mutations in a subset of non-Hodgkin's lymphomas (NHL) associated with extranodal disease and autoimmune features.6 Because of the proposed association between ALPS and caspase-10, an obvious extension of our work would be to examine whether somatic caspase-10 mutations might also be involved in the pathogenesis of NHL. We initially established a mutation detection assay based on polymerase chain reaction (PCR) and denaturing gradient gel electrophoresis (DGGE) to analyze a 137-bp genomic region of caspase-10 encompassing nucleotides 1186-1322 (codons 396-441). During the establishment of this assay, we observed an altered DGGE pattern in 1 of 10 normal control DNA samples(Figure 1A). Surprisingly, direct sequence analysis of this sample demonstrated the G1228A substitution in the heterozygous constellation(Figure 1B). Subsequent analysis of DNA samples from 85 healthy Danish blood donors (170 alleles) identified 12 additional heterozygotes,corresponding to allele frequencies of 93.2% (for the G1228 allele)and 6.8% (for the A1228 allele). Our preliminary analysis of 96 NHL cases shows a similar allele distribution in NHL.
Because we did not identify V410I homozygotes among our panel of normal controls, we cannot formally exclude that this variant is associated with severe impairment of Fas-mediated apoptosis in vivo.Nevertheless, the predicted homozygosity frequency of approximately 1 in 200 in the Danish population strongly argues against a major contribution to the ALPS phenotype. Further documentation is required to solidify the association between caspase-10 mutations and ALPS type II, and to establish whether the V410I variant is a benign polymorphism or may contribute to the ALPS phenotype together with other yet undefined factors involved in the control of apoptosis and lymphoproliferation.
Acknowledgment
We thank V. Ahrenkiel and B. Rasmussen for technical assistance. The study was supported by grants from The Danish Cancer Society, the E. Willumsen Foundation, and the Danish Cancer Research Foundation.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal