Abstract
A new monoclonal antibody (MUM1p) was used to study the cell/tissue expression of human MUM1/IRF4 protein, the product of the homologous gene involved in the myeloma-associated t(6;14) (p25;q32). MUM1 was expressed in the nuclei and cytoplasm of plasma cells and a small percentage of germinal center (GC) B cells mainly located in the “light zone.” Its morphologic spectrum ranged from that of centrocyte to that of a plasmablast/plasma cell, and it displayed a phenotype (MUM1+/Bcl-6−/Ki67−) different from that of most GC B cells (MUM1−/Bcl-6+/Ki67+) and mantle B cells (MUM1−/Bcl-6−/Ki67−). Polymerase chain reaction (PCR) analysis of single MUM1+cells isolated from GCs showed that they contained rearranged Ig heavy chain genes with a varying number of VHsomatic mutations. These findings suggest that these cells may represent surviving centrocytes and their progeny committed to exit GC and to differentiate into plasma cells. MUM1 was strongly expressed in lymphoplasmacytoid lymphoma, multiple myeloma, and approximately 75% of diffuse large B-cell lymphomas (DLCL-B). Unlike normal GC B cells, in which the expression of MUM1 and Bcl-6 were mutually exclusive, tumor cells in approximately 50% of MUM1+ DLCL-B coexpressed MUM1 and Bcl-6, suggesting that expression of these proteins may be deregulated. In keeping with their proposed origin from GC B cells, Hodgkin and Reed–Sternberg cells of Hodgkin's disease consistently expressed MUM1. MUM1 was detected in normal and neoplastic activated T cells, and its expression usually paralleled that of CD30. These results suggest that MUM1 is involved in the late stages of B-cell differentiation and in T-cell activation and is deregulated in DLCL-B.
Chromosomal translocations (14q+) affecting band 14q32 and unidentified partner chromosomes are common in multiple myeloma, suggesting that they may cause the activation of novel oncogenes.1,2 Recently, Iida et al1 reported that the 14q+ translocation occurring in multiple myeloma is a cryptic translocation (6;14) (p25;q32) that causes juxtaposition of the immunoglobulin heavy-chain (IgH) locus to the multiple myeloma oncogene 1 (MUM1)/IRF4 gene.3 It has been suggested that as consequence of this translocation, theMUM1/IRF4 gene is overexpressed, an event that may contribute to tumorigenesis because MUM1/IRF4 has oncogenic activity in vitro.1
The product of the MUM1/IRF4 gene (also called PIP, LSIRF, ICSAT)3-6 is a member of the interferon regulatory factor (IRF) family of transcription factors, known to play an important role in the regulation of gene expression in response to signaling by interferons and by other cytokines.6 By Northern blot analysis, strong expression of MUM1 mRNA has been detected in mature B cell–derived lymphoma and myeloma cell lines1 and in human T-cell leukemia virus type 1 (HTLV-1)–infected T cells.6 Moreover, IRF4-deficient mice (IRF4−/−) were unable to form germinal centers (GCs), lacked plasma cells in the spleen and lamina propria, and exhibited a profound reduction of serumimmunoglobulin levels and an inability to mount detectable antibody responses or to generate T-lymphocyte cytotoxic or antitumor responses.7 These findings provide evidence that the MUM1/IRF4 gene is essential for the function of both mature B cells and cytotoxic T lymphocytes.7 At molecular level, MUM1/IRF4 acts by forming a cooperative ternary complex with the transcription factor PU.1 at immunoglobulin enhancer elements, such as λB and κE3′ sites.8-11
In spite of its recognized importance in the development of the immune system, the expression of MUM1/IRF4 protein in normal and neoplastic lymphohematopoietic tissues is unknown. To gain further insights into this issue, we produced a monoclonal antibody (MUM1p) specifically directed against a fixative-resistant epitope of the human MUM1 protein. The antibody was used to detect by immunohistochemistry expression of the MUM1 protein in human cell lines and in paraffin sections from normal and neoplastic lymphohematopoietic tissues. The results presented in this article indicate that the MUM1 protein is more strongly expressed in late plasma cell–directed stages of B-cell differentiation and in activated T cells and suggest that the MUM1p monoclonal antibody may serve as a marker for lymphohematopoietic neoplasms thought to be derived from these cells.
Materials and methods
Generation of the recombinant glutathione S-transferase–MUM1 protein
A cDNA fragment corresponding to amino acids 144 to 451 of the human MUM1 protein was subcloned to BamHI and EcoRI cloning sites of pGEX 3X (Pharmacia Biotech, Piscataway, NJ) bacterial expression vector.1 The insert was cloned in frame to glutathione S-transferase (GST) coding sequences and confirmed by sequencing. N-terminal sequences encoding DNA-binding motifs were eliminated because they have extremely high homology with other family proteins. The GST–MUM1 fusion protein was then expressed in BL21-competent bacteria and purified by affinity chromatography following the manufacturer's instructions.
Production of a monoclonal antibody (MUM1p) against the MUM1 protein
BALB/c mice were injected intraperitoneally (3 times at 10-day intervals) with 150 μg GST–MUM1 fusion protein (amino acids 144 to 451) plus Freund's adjuvant. A 150-μg booster of the recombinant GST–MUM1 protein was injected intraperitoneally, and fusion was carried out 3 days later, as described previously.12Hybridoma supernatants were screened by the immuno-alkaline phosphatase (APAAP) technique13 on cytocentrifuge preparations of the IM9 human myeloma cell line and on paraffin sections of normal human tonsil. Two of 1000 hybridoma supernatants (MUM1p and MUM97) that reacted strongly with the IM9 myeloma cells and with normal plasma cells in tonsil paraffin sections were cloned by a limiting dilution technique, and 1 of them (MUM1p) was selected for further study.
Other antibodies
The reactivity pattern of the MUM1p monoclonal antibody was compared with that of a goat polyclonal antibody directed against the carboxy terminus of the murine IRF4/ICSAT molecule that is marketed by the manufacturer (Santa Cruz Biotechnology, Heidelberg, Germany) as cross-reacting with its human homologue.
Double immuno-enzymatic stainings on frozen and paraffin tonsil sections were performed using the MUM1p monoclonal antibody in combination with antibodies directed against the following antigens: κ and λ light chains, IgD, CD19, CD20, CD3, the follicular dendritic cell markers CD21 and CD23, and the intermediate cytokeratin filaments (antibody MNF116) (all purchased from DAKO A/S, Glostrup, Denmark); against the following plasma cell markers VS38 (DAKO A/S), CD138/syndecan clone BB4 (Serotec, Oxford, UK), and CD38 (kindly provided by Prof Fabio Malavasi, Turin, Italy); and against CD30 and Ki67 (both generated in the laboratory of H.S.), CD68/PG-M1,14 and Bcl-612 (both produced in the laboratory of B.F.).
Transfected cells
A pHeBo-CMV-MUM1-HA and a pHeBo-CMV (as control) were used for the transient transfection of HeLa cells by a calcium chloride HEPES-buffered-saline method. MUM1-transfected and control cells were grown in Dulbecco modified essential medium containing 10% fetal calf serum, penicillin (100 IU/mL), and streptomycin (100 μg/mL). Cells were lysed and analyzed by Western blotting (see below). Cells were also grown exponentially on slides, air dried overnight, fixed in acetone for 10 minutes, and immunostained by the APAAP technique.
Cell lines
MUM1 expression was studied on the following human cell lines: IM9 (myeloma); Namalwa, Bjab, Ramos (B-lymphoid); MOLT-4, Jurkat (T-lymphoid); Karpas 299 (CD30+ anaplastic large-cell lymphoma with 2;5 translocation); U937 and HL60 (myeloid); and HeLa (epithelial). Cell lines were cultured in RPMI 1640 containing 10% fetal calf serum (Life Technologies, Grand Island, NY). Cytospin was prepared from exponentially growing cells, fixed in acetone for 10 minutes at room temperature, and then used for immunocytochemical studies.
Phytohemagglutinin stimulation
Ficoll-separated normal peripheral blood mononuclear cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum and phytohemagglutinin (PHA) (2.5 μg/mL) at a concentration of 1 × 106/mL in a humidified incubator at 37°C with 5% CO2 atmosphere. The proliferative response of the cultured cells was assessed in cytospin with the monoclonal antibody Ki67. Basal and stimulated cells were cyto-centrifuged and subjected to immunostaining. An aliquot of cells was tested for MUM1 protein expression by Western blotting.
Western blotting
Western blotting was performed on cell lysates from the MUM1-transfected and control HeLa cells and from IM9, U937, and HeLa cell lines. Unfractionated tonsil cell suspensions and Ficoll-separated peripheral blood T cells (both in basal condition and after stimulation with PHA) were also studied. Cells were lysed with sodium dodecyl sulfate (SDS) loading buffer, and an aliquot of each lysate was loaded onto an 8% SDS polyacrylamide gel and electrotransferred to nitrocellulose sheets, as previously described.12
Immunoprecipitation and Western blotting
IM9 myeloma cells were lysed with a single detergent lysis buffer (150 mmol/L NaCl, 50 mmol/L Tris HC1, pH 8, 1% NP40, 1 mmol/L EDTA) containing a protease inhibitor cocktail (leupeptin, aprotinin, pepstatin A, and phenylmethylsulfonyl fluoride) and immunoprecipitated with the MUM1p monoclonal antibody (supernatant at 1:5 dilution). The immunoprecipitates were Western blotted and then incubated overnight at 4°C with the goat anti-MUM1 polyclonal antibody (Santa Cruz Biotechnology). Finally, the blots were incubated for 1 hour at room temperature with a horseradish peroxidase–conjugated antigoat antibody and stained by the ECL system (Amersham, Arlington Heights, IL).
Tissue specimens
Expression of the MUM1 protein was studied in normal lymphohematopoietic tissues (tonsil, n = 10; spleen, n = 5; bone marrow, n = 6) and lymphomas (n = 150) representative of most categories of the Revised European American Lymphoma Classification.15 Normal and neoplastic samples were fixed either in 10% buffered formalin or in B5 (followed by 2 hours' decalcification in EDTA for bone marrow biopsies) and routinely processed for paraffin embedding. Immunohistologic analysis was also performed on cryostat sections cut from tonsil specimens that had been previously snap-frozen in liquid nitrogen.
Tissue processing for immunohistochemistry
Paraffin sections (5-μm thick) were attached on silane-coated slides, rehydrated, and subjected to microwaving (750 W × 3 cycles at 5 minutes each) using 1-mmol/L EDTA buffer, pH 8, as the antigen retrieval solution.12 16 Tonsil frozen sections (5-μm thick) were air dried overnight and fixed in acetone for 10 minutes before immunostaining.
Single immunoenzymatic labeling
Double-staining procedures
Paraffin and tonsil frozen sections were double stained for MUM1/κ and MUM1/λ light chains, MUM1/IgD, MUM1/CD21, MUM1/CD23, MUM1/cytokeratin, MUM1/Bcl-6, MUM1/VS38, MUM1/CD138, MUM1/CD38, MUM1/CD3, MUM1/CD30, and MUM1/Ki67. The MUM1 protein was usually detected by a biotin–avidin peroxidase technique using diaminobenzidine (Sigma-Aldrich, Milan, Italy)/hydrogen peroxide as substrate.18 The second pair of antigens was revealed by the APAAP procedure,13 using naphthol AS-MX plus Fast Red TR (both purchased from Sigma-Aldrich) or naphthol AS-MX plus Fast Blue BB salt (both purchased from Sigma-Aldrich) as substrate.18Because the Bcl-6 and CD20 proteins and cytokeratin proved to be partially or totally denatured or masked by the reaction product of peroxidase substrate, a reverse procedure (immunoperoxidase detection of Bcl-6, CD20, and cytokeratin followed by APAAP labeling of MUM1) was used for double staining of MUM1/Bcl-6, MUM1/CD20, and MUM1/cytokeratin. Slides were mounted in Kaiser gelatin after counterstain for 30 seconds in Gill hematoxylin or without counterstain.
Double immunofluorescence labeling19 for MUM1/Bcl-6 was also performed on tonsil frozen sections by 30-minute incubation with a mixture of rabbit polyclonal anti-Bcl-6 (Santa Cruz Biotechnology) and MUM1p monoclonal antibody. After extensive washing, the primary antibodies were detected with fluorescein-conjugated antirabbit and rhodaminated antimouse secondary antibodies.
Polymerase chain reaction (PCR) analysis of single MUM1+cells
Immunostaining and isolation of single cells.
Tonsil frozen sections immunostained for MUM1 were overlaid with phosphate-buffered saline. Single MUM1+ cells, totaling 30 cells per section, were isolated from GCs using a hydraulic micromanipulator as previously described20 and were transferred to PCR tubes containing 10 μL proteinase K (1 mg/mL). In each PCR tube, 5 single MUM1+ cells were pooled together. From each section, aliquots of the overlaying buffer were aspirated and used as negative controls. B cells isolated from the mantle zone served as positive controls.
PCR and cloning.
After proteinase K digestion (1 hour at 55°C), fully nested PCR was performed for the detection of the rearranged VHgene using family-specific framework (FW)1 primers for the first amplification, as previously described.20 In the second round of amplification, we further amplified 2-μL aliquots from the first round with family-specific FW2 primers. Both FW primer sets were used in conjunction with 2 nested primers for the joining region (JH). The PCR products were analyzed on an ethidium bromide–stained polyacrylamide gel (6%). Visualized PCR products were then gel-purified, cloned to plasmid, and sequenced on an automatic fluorescence DNA sequencer (377A; Applied Biosystem, Weiterstadt, Germany) by using the dye deoxy terminator method. At least 6 clones derived from each amplified portion were analyzed by sequencing in each direction using SP6 and T7 primers in 2 separate reactions.
Sequence analysis
Sequences were compared with the corresponding germ line VH(VBASE databank)21 to determine the VHfamily usage and to demonstrate the number of somatic mutations. Furthermore, all sequences were compared with each other to detect intraclonal diversities and with our own and published databank sequences.
Results
Western blotting
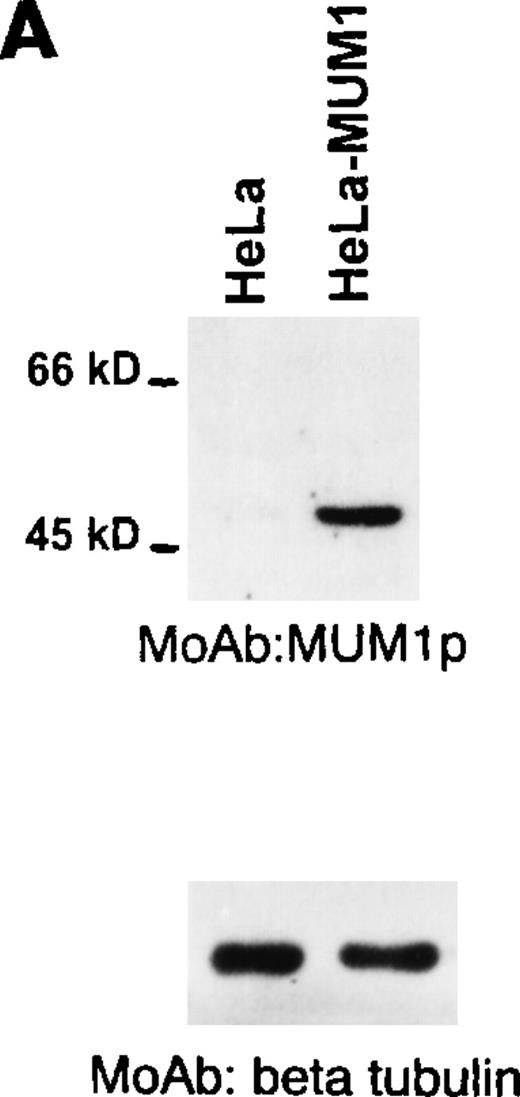
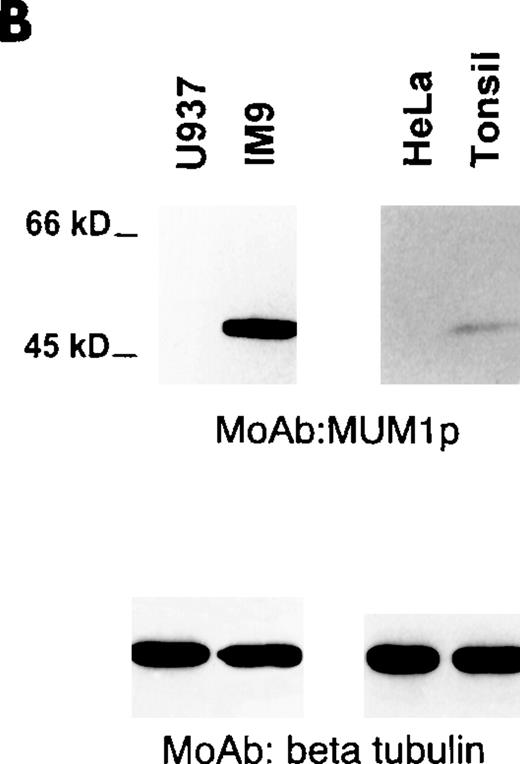
A single band of 52 kd corresponding to the molecular weight of the MUM1-HA protein was observed on lysates from MUM1-transfected, but not control HeLa, cells (Figure 1A, line 2). A 50-kd band corresponding to the expected molecular weight of the MUM1 protein was detected with the MUM1p antibody on Western blot lysates from IM9 myeloma cells (Figure 1B, line 2) and on unfractionated lysates from normal tonsil (Figure 1B, line 4). In the latter lysates, the 50-kd band was weaker probably because of the dilution of plasma cells with other tonsil cell populations. No bands were detected by MUM1p in lysates of the U937 and HeLa cell lines that served as negative controls because they did not express MUM1 RNA (Figure 1B, lines 1 and 3).
Western blotting with the MUM1p monoclonal antibody.
(A) A band of 52-kd of the expected size of the MUM1-HA protein is seen in line corresponding to pHeBo-CMV-MUM1-HA HeLa-transfected cells but not in negative control HeLa cells. (B) A 50-kd band of the expected molecular size of the MUM1 protein is seen in lanes 2 and 4, corresponding to the IM9 myeloma cell line and normal tonsil. No bands are detected in lanes 1 and 3, corresponding to U937 and HeLa cell lines. Identical results (not shown) were obtained with the monoclonal (clone MUM97) and polyclonal anti-IRF4/ICSAT antibody. In both experiments, β tubulin levels are shown below to control for the integrity and amount of the loaded protein.
Western blotting with the MUM1p monoclonal antibody.
(A) A band of 52-kd of the expected size of the MUM1-HA protein is seen in line corresponding to pHeBo-CMV-MUM1-HA HeLa-transfected cells but not in negative control HeLa cells. (B) A 50-kd band of the expected molecular size of the MUM1 protein is seen in lanes 2 and 4, corresponding to the IM9 myeloma cell line and normal tonsil. No bands are detected in lanes 1 and 3, corresponding to U937 and HeLa cell lines. Identical results (not shown) were obtained with the monoclonal (clone MUM97) and polyclonal anti-IRF4/ICSAT antibody. In both experiments, β tubulin levels are shown below to control for the integrity and amount of the loaded protein.
Lysates of the MUM1p–immunoprecipitated IM9 myeloma cells revealed by Western blotting with an anti-MUM1 polyclonal antibody gave the expected 50-kd band of MUM1.
These results demonstrated that MUM1p reacted specifically with the MUM1 protein and that this antibody was suitable for both Western blotting and immunoprecipitation studies. Identical results (not shown) were obtained with the monoclonal antibody MUM97 generated in the same fusion.
Expression of the MUM1 protein in human cell lines
The MUM1p antibody strongly reacted with the nuclei of MUM1-transfected HeLa cells but not those of control cells (data not shown), and this further supported the specificity of the antibody (see above).
MUM1p reacted with the nucleus and with the cytoplasm of the IM9 myeloma cells; nuclear positivity was stronger than that observed in the cytoplasm. Both nucleus and cytoplasm showed a microgranular positivity (Figure 2A), but the nucleoli were consistently MUM1− (Figure 2A). A similar reactivity (not shown) was observed with the cell line Karpas 299, derived from a T-cell anaplastic large-cell lymphoma with t(2;5). Only a small percentage of the Namalwa cells was labeled for MUM1. The Burkitt cell line Daudi was MUM1− but strongly expressed the Bcl-6 protein. In contrast, the Burkitt cell line Ramos coexpressed strongly the MUM1 and the Bcl-6 proteins. In general, nuclear reactivity of the MUM1-expressing cell lines was stronger with the MUM1p monoclonal antibody than with the polyclonal anti-IRF4/ICSAT. The myeloid-derived (U937 and HL60) and the epithelial-derived (HeLa) cell lines were consistently MUM1−.
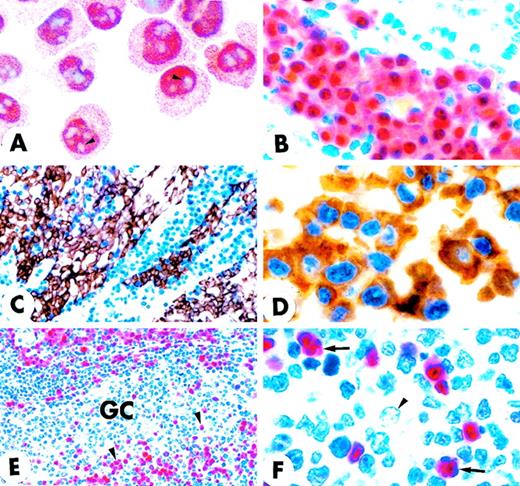
MUM1 expression in myeloma cell line and normal lymphoid tissues.
(A) IM9 human myeloma cell line. Microgranular positivity for the MUM1 protein is observed in the nucleus (stronger) and in the cytoplasm (weaker) of tumor cells. Arrowheads indicate negative nucleoli (APAAP; × 1000). (B) MUM1 protein expression in the nucleus (stronger) and in the cytoplasm (weaker) of plasma cells in a lymph node involved by a plasmacellular variant of Castleman disease (paraffin section; APAAP; × 800). (C) Paraffin section from normal tonsil double stained for cytokeratins (tonsil epithelium labeled in brown) and MUM1 (plasma cells labeled in blue) (APAAP; × 250). (D) All plasma cells in the tonsil epithelium double stain for intracytoplasmic light chains (brown) and nuclear MUM1 (blue) (× 1000). (E) Isolated or small clusters of MUM1+ cells (arrowheads) are present within GC (APAAP; × 250). (F) At higher magnification, some of the MUM1+ cells in the GC show a markedly irregular (often multilobated) nucleus (arrows). Arrowhead points to a MUM1− macrophage (APAAP; × 1000). (A-F) Immunostaining with MUM1p monoclonal antibody; hematoxylin counterstain.
MUM1 expression in myeloma cell line and normal lymphoid tissues.
(A) IM9 human myeloma cell line. Microgranular positivity for the MUM1 protein is observed in the nucleus (stronger) and in the cytoplasm (weaker) of tumor cells. Arrowheads indicate negative nucleoli (APAAP; × 1000). (B) MUM1 protein expression in the nucleus (stronger) and in the cytoplasm (weaker) of plasma cells in a lymph node involved by a plasmacellular variant of Castleman disease (paraffin section; APAAP; × 800). (C) Paraffin section from normal tonsil double stained for cytokeratins (tonsil epithelium labeled in brown) and MUM1 (plasma cells labeled in blue) (APAAP; × 250). (D) All plasma cells in the tonsil epithelium double stain for intracytoplasmic light chains (brown) and nuclear MUM1 (blue) (× 1000). (E) Isolated or small clusters of MUM1+ cells (arrowheads) are present within GC (APAAP; × 250). (F) At higher magnification, some of the MUM1+ cells in the GC show a markedly irregular (often multilobated) nucleus (arrows). Arrowhead points to a MUM1− macrophage (APAAP; × 1000). (A-F) Immunostaining with MUM1p monoclonal antibody; hematoxylin counterstain.
Expression of the MUM1 protein in normal and reactive lymphoid tissues
In tissue paraffin sections and cytospins, the MUM1p monoclonal antibody gave stronger positivity and lower background than the polyclonal reagent. There was no difference in terms of MUM1p specificity in tissue samples processed as frozen or paraffin sections (B5 or formalin fixed) and immunostained either with peroxidase or APAAP procedures, by hand or by an automatic DAKO immunostainer (Techmate 500). However, the intensity of MUM1 labeling was usually stronger in paraffin sections than in frozen sections.
The immunostaining results on paraffin sections from normal lymphohematopoietic tissues are summarized in Table1. The most striking reactivity of MUM1p in sections of normal tonsil and reactive lymph nodes was with plasma cells (Figures 2B-2D) that showed strong MUM1 nuclear positivity in addition to weaker labeling of cytoplasm (Figure 2B). Negativity of nucleoli, clearly evident in the cytospin of unfractionated tonsil cells (not shown), was not detectable in paraffin sections from tonsil (or other tissues) in which the nucleus of MUM1+ cells appeared to be homogeneously stained (Figures 2B-2D). Diffuse nuclear positivity with the inability to recognize in paraffin sections the reactivity pattern of specific nuclear structures (nucleoli, nuclear bodies) was probably a fixation artifact because it is also observed with monoclonal antibodies directed against other nuclear-located antigens (PML, Bcl-6, NPM-ALK, NPM).22-24 In double-stained sections, most plasma cells were usually found to coexpress MUM1 and other plasma cell–associated markers (intracytoplasmic Ig light chains, CD138/syndecan, VS38, CD38) (Figure 2D). However, occasional MUM1+ plasma cells that failed to express 1 or more of the plasma cell markers, and vice versa, were also present.
MUM1 protein expression in normal lymphohematopoietic tissues
| Tissue . | MUM1 Expression . |
|---|---|
| Tonsil/spleen | |
| Germinal center B cells | −* |
| Mantle B cells | −† |
| Marginal zone B cells | − |
| Plasma cells | + |
| T cells | −‡ |
| Macrophages | − |
| Follicular dendritic cells | − |
| Endothelia | − |
| Epithelia | − |
| Thymus | |
| Cortex | − |
| Medulla | − |
| Hassal corpuscle | − |
| Bone marrow | |
| Erythroid precursors | − |
| Myeloid precursors | − |
| Megakaryocytes | − |
| Osteoblasts | − |
| Osteoclasts | − |
| Tissue . | MUM1 Expression . |
|---|---|
| Tonsil/spleen | |
| Germinal center B cells | −* |
| Mantle B cells | −† |
| Marginal zone B cells | − |
| Plasma cells | + |
| T cells | −‡ |
| Macrophages | − |
| Follicular dendritic cells | − |
| Endothelia | − |
| Epithelia | − |
| Thymus | |
| Cortex | − |
| Medulla | − |
| Hassal corpuscle | − |
| Bone marrow | |
| Erythroid precursors | − |
| Myeloid precursors | − |
| Megakaryocytes | − |
| Osteoblasts | − |
| Osteoclasts | − |
A small percentage of strongly MUM1+ cells was present within germinal centers (see “Results”).
Mantle B cells were usually negative but occasionally showed very faint MUM1 expression.
A small percentage (1%-5%) of MUM1+ T cells was present in the GC and the interfollicular area.
In the B-cell follicles, IgD+ mantle lymphocytes were usually MUM1− but occasionally showed faint nuclear MUM1 positivity. Rare cells strongly expressing the MUM1 protein were sometimes observed in the mantle zones, and they probably represented MUM1+ cells exiting the GC and transiting through the follicle mantle. This view is supported by the finding that PCR analysis on such isolated cells revealed mutations in their rearranged VH region genes (Table 2). Most GC cells were MUM1−, but a percentage of them (ranging from 3% to 10%, depending on the GC) strongly expressed the MUM1 protein (Figure 2E). These MUM1+ cells were morphologically heterogeneous. Some had markedly irregular nuclei (Figure 2F), whereas others had an immunoblast-like or a plasma cell–like appearance and were predominantly located in the light zone of the GCs (the centroblasts of the dark zone were usually MUM1−) (Figure 3A). This topographic distribution was clearly evident in sections double stained for MUM1/CD23 or MUM1/CD21 that showed the intimate contact of MUM1+ elements with the meshwork of follicular dendritic cells (Figures 3A and 3B). Double staining for MUM1/CD19 and MUM1/CD20 was difficult to interpret because the few MUM1+ cells within the GCs were surrounded by MUM1−/CD19+/CD20+ GC B-cells, and it was impossible to establish whether the membrane positivity for CD19 and CD20 belonged to the MUM1+ elements or the adjacent MUM1− B cells. Thirty percent to 50% of MUM1+ cells in the GCs contained intracytoplasmic Ig light chains (data not shown). Many MUM1+ cells in the GC were negative for the plasma cell marker CD138/syndecan (not shown), suggesting that MUM1 expression most likely precedes that of CD138. Double staining for MUM1/VS38 and MUM1/CD38 was difficult to interpret. These findings, together with the results of MUM1/CD3 double staining and of single-cell PCR (see below), provide evidence that more then 95% of MUM1+ cells within the GC are B cells.
IgH polymerase chain reaction single cell analysis of MUM1+ and MUM1− cells
| . | IgH-R . | VH Family . | Range of Somatic Mutations (R + S) FW3 and CDR2 . | Coding Capacity . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VH1 . | VH2 . | VH3 . | VH4 . | VH5 . | VH6 . | VH7 . | ||||
| MUM1+ | ||||||||||
| Germinal center cells | 10/74 | 7 | 3 | 1-11 | ±* | |||||
| Intramantle cells | 5/35 | 3 | 5-8 | + | ||||||
| Intraepithelial plasma cells | 5/62 | 5 | 2-16 | + | ||||||
| MUM1− | ||||||||||
| Mantle cells | 12/82 | 3 | 1 | 0-2 | + | |||||
| . | IgH-R . | VH Family . | Range of Somatic Mutations (R + S) FW3 and CDR2 . | Coding Capacity . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VH1 . | VH2 . | VH3 . | VH4 . | VH5 . | VH6 . | VH7 . | ||||
| MUM1+ | ||||||||||
| Germinal center cells | 10/74 | 7 | 3 | 1-11 | ±* | |||||
| Intramantle cells | 5/35 | 3 | 5-8 | + | ||||||
| Intraepithelial plasma cells | 5/62 | 5 | 2-16 | + | ||||||
| MUM1− | ||||||||||
| Mantle cells | 12/82 | 3 | 1 | 0-2 | + | |||||
In one cell, the VH rearrangement was out of frame.
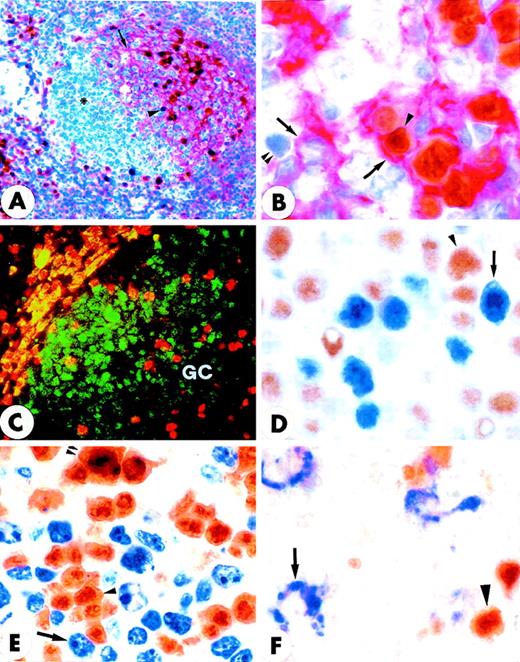
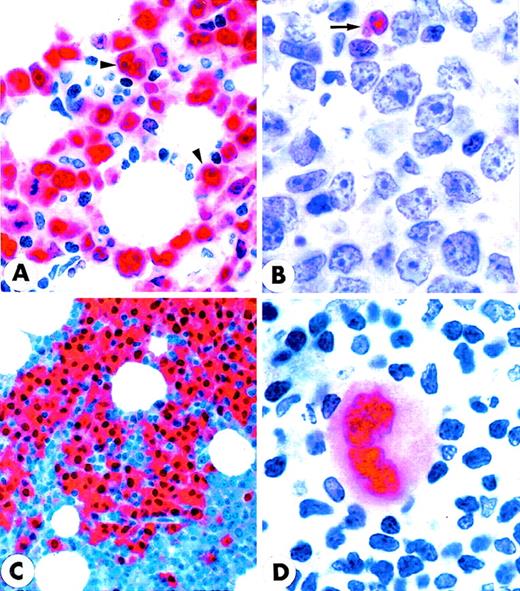
Double-stained GC of normal tonsil (paraffin sections).
(A) Double staining for MUM1 (brown) and CD23 (red) shows that GC MUM1+ cells (arrowhead) are located in the light zone in close association with CD23+ follicular dendritic reticulum cells (arrow). Asterisk indicates negative centroblasts in the dark zone (× 250; hematoxylin counterstain). (B) The intimate contact of brown MUM1+ cells (arrowhead) with red CD23+follicular dendritic cells (arrow) is shown at higher magnification (× 1000). Double arrowheads indicate a MUM1−/CD23− GC cell. (C) Expression of MUM1 (red) and Bcl-6 (green) are mutually exclusive within the GC of tonsil (double immunofluorescence; × 800). (D) Expression for brown nuclear Bcl-6 (arrowhead) and blue nuclear MUM1 (arrow) in GC B-cells appears to be mutually exclusive (× 1000; no counterstain). (E) Expression for brown nuclear MUM1 protein (arrowhead) and blue Ki67 proliferation antigen (arrow) appears to be mutually exclusive, with the exception of rare cells (double arrowheads) that double stain for the 2 antigens (× 1000; no counterstain). (F) The MUM1 protein (brown) and the CD68 antigen (blue) are clearly expressed in different cell types. The arrow points to a CD68+ tingible body macrophage, whereas the arrowhead indicates a MUM1+ GC cell (× 1000; no counterstain). (A-F) Biotin-avidin peroxidase/APAAP.
Double-stained GC of normal tonsil (paraffin sections).
(A) Double staining for MUM1 (brown) and CD23 (red) shows that GC MUM1+ cells (arrowhead) are located in the light zone in close association with CD23+ follicular dendritic reticulum cells (arrow). Asterisk indicates negative centroblasts in the dark zone (× 250; hematoxylin counterstain). (B) The intimate contact of brown MUM1+ cells (arrowhead) with red CD23+follicular dendritic cells (arrow) is shown at higher magnification (× 1000). Double arrowheads indicate a MUM1−/CD23− GC cell. (C) Expression of MUM1 (red) and Bcl-6 (green) are mutually exclusive within the GC of tonsil (double immunofluorescence; × 800). (D) Expression for brown nuclear Bcl-6 (arrowhead) and blue nuclear MUM1 (arrow) in GC B-cells appears to be mutually exclusive (× 1000; no counterstain). (E) Expression for brown nuclear MUM1 protein (arrowhead) and blue Ki67 proliferation antigen (arrow) appears to be mutually exclusive, with the exception of rare cells (double arrowheads) that double stain for the 2 antigens (× 1000; no counterstain). (F) The MUM1 protein (brown) and the CD68 antigen (blue) are clearly expressed in different cell types. The arrow points to a CD68+ tingible body macrophage, whereas the arrowhead indicates a MUM1+ GC cell (× 1000; no counterstain). (A-F) Biotin-avidin peroxidase/APAAP.
Notably, the totality of the MUM1+ GC cells failed to express the Bcl-6 protein (Figures 3C, 3D) that was in turn detectable in most centroblasts and centrocytes. Expression of the Ki67 proliferation antigen and MUM1 within the GC was also mutually exclusive (Figure 3E), and only scattered MUM1+ cells appeared to express a positivity for Ki67 usually associated to nucleoli (Figure 3E). Tingible body macrophages within the GC were consistently CD68+/MUM1− (Figure3F).
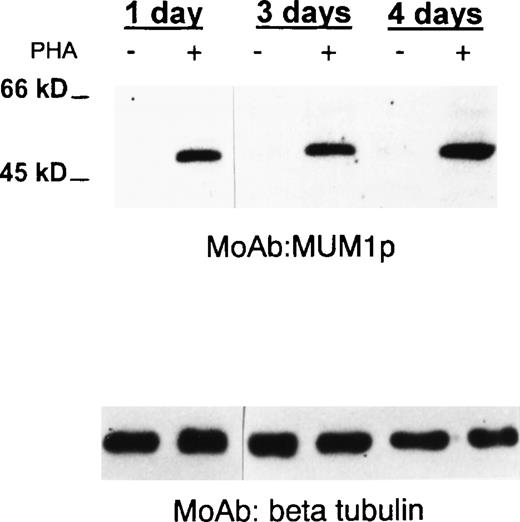
Most T cells in the interfollicular area of normal tonsil and reactive lymph nodes were negative for MUM1, but some of them (1%-5%) strongly expressed the protein (not shown). Similar findings (not shown) were observed in the GCs, whereas the MUM1+/CD3+elements represented only a minority (less than 5%) of all MUM1+ cells in the light zone. Most large CD30+cells in the tonsil coexpressed the MUM1 protein (Figures 4A and 4B). As expected, the CD30+/MUM1+ cells were mostly located in the area adjacent to the follicle mantle and appeared to be proliferating (Ki67+) (Figure4C). No MUM1 expression was detected in peripheral blood T lymphocytes under basal conditions, but the molecule was strongly induced after PHA stimulation, as demonstrated by Western blotting (Figure 5) and immunocytochemistry (not shown). Taken together, these findings strongly suggest that the MUM1 protein is expressed in activated T cells.
Double stainings in the interfollicular area of normal tonsil.
(A,C) Paraffin sections. (B) Frozen section. (A) A large cell coexpressing surface CD30 (blue) and nuclear MUM1 (brown) is indicated by the arrow; a MUM1+/CD30− small cell is also observed (arrowhead) (× 1000; no counterstain). (B) A large cell coexpressing surface CD30 (red) and nuclear MUM1 (brown) is indicated by the arrow (× 1000; hematoxylin counterstain). (C) Many large cells in the area adjacent to the follicle mantle (most likely CD30+ cells) coexpress the proliferation antigen Ki67 (blue labeling of nucleoli) and the brown nuclear MUM1 protein (double arrowheads). Arrowhead indicates a Ki67+/MUM1− cell. Arrow indicates a Ki67−/MUM1+ cell (× 800 ; no counterstain).
Double stainings in the interfollicular area of normal tonsil.
(A,C) Paraffin sections. (B) Frozen section. (A) A large cell coexpressing surface CD30 (blue) and nuclear MUM1 (brown) is indicated by the arrow; a MUM1+/CD30− small cell is also observed (arrowhead) (× 1000; no counterstain). (B) A large cell coexpressing surface CD30 (red) and nuclear MUM1 (brown) is indicated by the arrow (× 1000; hematoxylin counterstain). (C) Many large cells in the area adjacent to the follicle mantle (most likely CD30+ cells) coexpress the proliferation antigen Ki67 (blue labeling of nucleoli) and the brown nuclear MUM1 protein (double arrowheads). Arrowhead indicates a Ki67+/MUM1− cell. Arrow indicates a Ki67−/MUM1+ cell (× 800 ; no counterstain).
MUM1 expression in normal activated T cells.
A 50-kd band of the expected molecular size of the MUM1 protein is seen in lanes (+) corresponding to PHA-stimulated peripheral blood T cells (at days 1, 3, 4), whereas no band is observed in lanes (−) corresponding to T cells under basal conditions (Western blotting with the MUM1p monoclonal antibody). β Tubulin levels are shown below to control for the amounts of the loaded protein.
MUM1 expression in normal activated T cells.
A 50-kd band of the expected molecular size of the MUM1 protein is seen in lanes (+) corresponding to PHA-stimulated peripheral blood T cells (at days 1, 3, 4), whereas no band is observed in lanes (−) corresponding to T cells under basal conditions (Western blotting with the MUM1p monoclonal antibody). β Tubulin levels are shown below to control for the amounts of the loaded protein.
MUM1 expression appeared to be lymphoid-restricted because other cell types, such as follicular dendritic cells, macrophages, interdigitating reticulum cells, myeloid and erythroid precursors, megakaryocytes, and endothelial and epithelial cells, were consistently negative for the protein.
Single-cell polymerase chain reaction studies
Frozen sections of nonneoplastic tonsil were immunostained for MUM1. Single MUM1+ cells were isolated by micromanipulation from different GCs, mantle zones, and cryptic epithelia and studied by PCR. Results are summarized in Table 2 and show rearranged Ig genes in the MUM1+ cells of all regions, which confirms their B-cell nature. Sequence analysis of the rearrangements revealed a functional coding region and somatic mutations within the V segment. In terms of number of somatic mutations, the MUM1+ cells can be considered as GC or post-GC cells. This is also valid for the MUM1+ cells picked from the mantle zone, thus supporting the view that they did not represent mantle cells but GC-derived cells migrating through the mantle zone. In contrast, the MUM1−cells isolated from the mantle zone were devoid of somatic mutations and thus belonged to the unmutated mantle cell pool.
MUM1 expression in lymphohematopoietic neoplasms
The immunostaining results on paraffin sections from 150 cases of human lymphoma are summarized in Table 3. The MUM1 protein was usually absent in tumor cells of B-cell lymphoblastic leukemia. Most mantle cell–derived lymphomas were MUM1−, but approximately 30% showed weak to moderate nuclear positivity for the protein. Two of 3 nodal marginal zone lymphomas contained 30% to 50% MUM1-positive cells (usually showing plasmacytoid morphology). The tumor cells of follicular lymphomas (grades 1 and 2) were usually MUM1−, and only a small percentage (less than 20%) of MUM1+ cells (possibly representing normal residual GC cells) was observed within neoplastic follicles. In B-cell chronic lymphocytic leukemia, small neoplastic lymphocytes were usually MUM1− or showed only faint MUM1 expression, whereas prolymphocytes and paraimmunoblasts in pseudofollicles (proliferation centers) showed moderate MUM1 positivity.
MUM1 protein expression in paraffin sections of human lymphomas
| Type . | Cases (N = 150) . | MUM1 Expression by Tumor Cells (%) . |
|---|---|---|
| B-cell derived | ||
| Acute lymphoblastic leukemia | 5 | 0/5 |
| Chronic lymphocytic leukemia | 15 | 10/153-150 |
| Hairy cell leukemia | 6 | 0/6 |
| Mantle cell lymphoma | 6 | 2/63-151 (30%-50%) |
| Marginal zone lymphoma | 3 | 2/33-152 (30%-50%) |
| FCC lymphoma | 15 | 1/153-153 |
| DLCL-B | 30 | 23/30 (30%-100%) |
| Burkitt lymphoma | 3 | 0/3 |
| Myeloma | 15 | 15/15 (100%) |
| T-cell derived | ||
| Acute lymphoblastic leukemia | 5 | 0/5 |
| PTCL-NOS | 3 | 1/3 (30%) |
| PTCL-lymphoepit. | 2 | 1/2 (10%) |
| PTCL-AILD | 5 | 5/5 (10%-20%) |
| PTCL-intestinal | 3 | 1/3 (100%) |
| ATL | 3 | 3/3 (10%-40%) |
| Mycosis fungoides | 5 | 2/5 (40%) |
| Anaplastic large cell lymphoma | 10 | 10/10 (100%) |
| Hodgkin's disease | ||
| Lymphocyte predominance | 2 | 2/2 (100%) |
| Nodular sclerosis | 9 | 9/9 (100%) |
| Mixed cellularity | 5 | 5/5 (100%) |
| Type . | Cases (N = 150) . | MUM1 Expression by Tumor Cells (%) . |
|---|---|---|
| B-cell derived | ||
| Acute lymphoblastic leukemia | 5 | 0/5 |
| Chronic lymphocytic leukemia | 15 | 10/153-150 |
| Hairy cell leukemia | 6 | 0/6 |
| Mantle cell lymphoma | 6 | 2/63-151 (30%-50%) |
| Marginal zone lymphoma | 3 | 2/33-152 (30%-50%) |
| FCC lymphoma | 15 | 1/153-153 |
| DLCL-B | 30 | 23/30 (30%-100%) |
| Burkitt lymphoma | 3 | 0/3 |
| Myeloma | 15 | 15/15 (100%) |
| T-cell derived | ||
| Acute lymphoblastic leukemia | 5 | 0/5 |
| PTCL-NOS | 3 | 1/3 (30%) |
| PTCL-lymphoepit. | 2 | 1/2 (10%) |
| PTCL-AILD | 5 | 5/5 (10%-20%) |
| PTCL-intestinal | 3 | 1/3 (100%) |
| ATL | 3 | 3/3 (10%-40%) |
| Mycosis fungoides | 5 | 2/5 (40%) |
| Anaplastic large cell lymphoma | 10 | 10/10 (100%) |
| Hodgkin's disease | ||
| Lymphocyte predominance | 2 | 2/2 (100%) |
| Nodular sclerosis | 9 | 9/9 (100%) |
| Mixed cellularity | 5 | 5/5 (100%) |
FCC, follicle center lymphoma; DLCL-B, diffuse large B-cell lymphoma; PTCL, peripheral T-cell lymphoma; NOS, not otherwise specified; Lymphoepit, lymphoepithelioid type; AILD, angioimmunoblastic lymphadenopathy type; intestinal, enteropathy-type intestinal T-cell lymphoma; ATL, adult T-cell lymphoma/leukemia. The numbers in parentheses indicate the percentage of MUM1+ tumor cells.
Moderate positivity of prolymphocytes and paraimmunoblasts in pseudofollicles; weak nuclear staining is observed in the small neoplastic cells.
Usually weak nuclear staining.
Nuclear positivity is seen especially in cells with plasmacytoid morphology.
Strong positivity of tumor cells in FCC lymphoma, grade III.
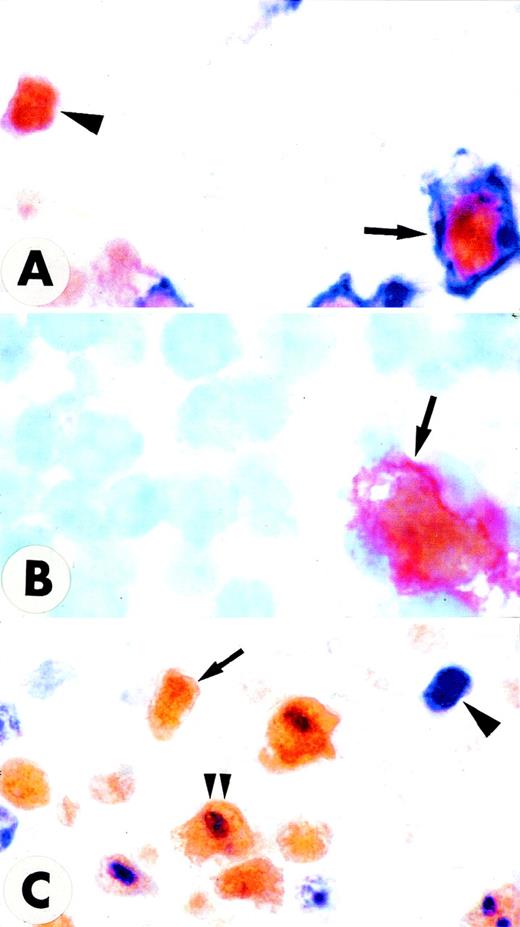
Among B-cell lymphomas, the strongest expression of MUM1 was observed in lymphoplasmacytoid lymphoma/immunocytoma and in multiple myeloma (Figure 6C). The MUM1 protein was also strongly expressed in approximately 75% of diffuse large B-cell lymphomas (range of positive tumor cells, 30%-100%) (Figure 6A), but it was absent in approximately 25% (Figure 6B). Double staining for MUM1/Bcl-6 showed that about 50% of MUM1+ diffuse, large B-cell lymphomas coexpressed the MUM1 and Bcl-6 proteins (data not shown). Notably, tumor cells in all cases of Hodgkin's disease were consistently and strongly MUM1+ (Figure 6D). In conclusion, the MUM1 protein appeared to be predominantly and strongly expressed in lymphoid neoplasms thought to be derived from late-stage B cells. In contrast to normal B cells, in which Bcl-6 and MUM1 expression were mutually exclusive, coexpression of the 2 proteins was commonly detected in diffuse, large B-cell lymphomas.
MUM1 expression in lymphomas and myeloma.
(A) Diffuse, large B-cell lymphoma showing nuclear and cytoplasmic positivity for the MUM1 protein. Arrowheads point to large MUM1+ tumor cells with prominent nucleoli (lymph node paraffin section; × 800). (B) Diffuse, large B-cell lymphoma showing no expression of the MUM1 protein. The arrow points to a normal residual MUM1+ cell (lymph node paraffin section; × 1000). (C) Multiple myeloma showing strong nuclear and cytoplasmic positivity of tumor cells for the MUM1 protein (paraffin section from bone marrow trephine biopsy; × 250). (D) Strong nuclear and weak cytoplasmic expression of the MUM1 protein in a Reed–Sternberg cell of Hodgkin's disease, nodular sclerosing type (lymph node paraffin section; × 1000). (A-D) Immunostaining with the MUM1p monoclonal antibody; APAAP procedure; hematoxylin counterstain.
MUM1 expression in lymphomas and myeloma.
(A) Diffuse, large B-cell lymphoma showing nuclear and cytoplasmic positivity for the MUM1 protein. Arrowheads point to large MUM1+ tumor cells with prominent nucleoli (lymph node paraffin section; × 800). (B) Diffuse, large B-cell lymphoma showing no expression of the MUM1 protein. The arrow points to a normal residual MUM1+ cell (lymph node paraffin section; × 1000). (C) Multiple myeloma showing strong nuclear and cytoplasmic positivity of tumor cells for the MUM1 protein (paraffin section from bone marrow trephine biopsy; × 250). (D) Strong nuclear and weak cytoplasmic expression of the MUM1 protein in a Reed–Sternberg cell of Hodgkin's disease, nodular sclerosing type (lymph node paraffin section; × 1000). (A-D) Immunostaining with the MUM1p monoclonal antibody; APAAP procedure; hematoxylin counterstain.
In keeping with the detection of MUM1 in normal activated T cells, we also found strong expression of the protein in CD30+anaplastic large-cell lymphomas, both ALK− and ALK+23 (not shown), that were thought to derive from activated T cells. In other peripheral T-cell lymphomas (Table 3), MUM1 expression usually paralleled that of CD30.
Discussion
In this article, we describe the characteristics of a new murine monoclonal antibody (MUM1p) specifically directed against the human MUM1 protein. The epitope recognized by MUM1p is fixative resistant, and the antibody is suitable for immunohistochemical detection of the MUM1 protein on routinely fixed, paraffin-embedded samples and Western blotting.
In cultured cells and primary tissues, MUM1+ cells strongly expressed the protein in the nucleus. This finding is expected because MUM1 is a member of the IRF family that acts as a transcription factor.1,6 Nuclear positivity was diffuse and microgranular and did not associate with specific nuclear organelles, such as nuclear bodies or nucleoli. In addition to the strong nuclear labeling, weak-to-moderate positivity was also observed in the cytoplasm of MUM1-expressing cells. It is unlikely that this was caused by a fixation/embedding-related artifactual diffusion of the protein from the nuclear to the cytoplasmic compartment because, in addition to paraffin sections, it was also observed in cytospin preparations of the IM9 myeloma cells and in tonsil frozen sections. Cross-reactivity of the MUM1p antibody with a cytoplasmic protein other than MUM1 also appears unlikely because it was observed with antibodies (monoclonal and polyclonal) directed against different epitopes of the MUM1 protein; all indicated a diffuse/microgranular cytoplasmic pattern similar to that observed in the nucleus. Moreover, cytoplasmic expression of MUM1 was observed at variable degrees in various normal and neoplastic cell types, including some GC B cells, plasma cells, and activated T cells, tumor cells in multiple myeloma, diffuse large B-cell lymphomas, T/null CD30+ anaplastic large-cell lymphoma, and Hodgkin's disease. Finally, no additional bands other than that typical (50 kd) of MUM1 were detected on Western blotting from the lysates of different cell types. The finding of MUM1 expression in the cytoplasm was in contrast to previous observations7 that nuclear, but not cytoplasmic, extracts of mouse lymph node cells expressed by Western blotting the 50-kd band typical of IRF4 (the murine homologue of MUM1). These conflicting findings may result from 1 or more of the following: lower sensitivity of Western blot analysis than that of APAAP immunocytochemistry for detecting small amounts of the protein; higher affinity of the MUM1p monoclonal antibody used in this study than that of the polyclonal antimurine IRF4 used by other investigators7; and different subcellular distribution of MUM1/IRF4 in humans and in mice. Colocalization and immunoelectron microscope studies should provide additional insights concerning the topographic distribution of the MUM1 protein in the nucleus and in the cytoplasm. In keeping with previous data in knockout mice,7 immunohistologic studies of various human tissues with MUM1p clearly proved that the expression of MUM1/IRF4 is lymphoid-restricted. Recently, it has been reported that IRF4 can be expressed in murine macrophages.25 Perhaps these two studies have conflicting results because in mice the MUM1/IRF4 protein has a different distribution than it has in humans or because Marecki et al25 used a lower specificity polyclonal anti-IRF4 antibody.
In normal lymphohematopoietic tissues, the MUM1 protein was mainly expressed in B cells, but it was mapped to B-cell compartments different from those occupied by other transcription factors involved in B-cell development. For example, Bcl-6 is strongly expressed in GC B cells (most centroblasts and centrocytes),12,26,27 and the Pax-5 protein28 has been predominantly found in lymphocytes of the mantle zone.29 The most striking characteristic of the MUM1p monoclonal antibody in tissue sections of lymphohematopoietic tissues was its strong reactivity with the nucleus and cytoplasm of mature plasma cells, whereas other B-cell types—eg, most GC B cells and IgD+/IgM+ virgin B lymphocytes of the follicle mantle—usually failed to express the protein. This finding strongly suggests that the MUM1 protein may play a key role in the terminal phases of B-cell differentiation toward the plasma cell, and it is in keeping with the finding that IRF4(−/−) mice show an absence of plasma cells associated with a dramatic reduction in serum immunoglobulins.7
In addition to plasma cells, a small percentage of GC B cells strongly expressed MUM1. Interestingly, these MUM1+ cells were mainly located in the light zone of the GC, but the highly proliferating, follicle-colonizing B blasts (centroblasts) of the dark zone failed to express the protein. Thus, it is unlikely that the MUM1 protein is involved in the process of clonal expansion and somatic hypermutation of the IgV-region genes known to occur in the dark zone of the GC,30,31 though such involvement is conceivable for the Bcl-6 protein that is strongly expressed and is a target for somatic hypermutation in the centroblasts.32-34 In the normal GC, the dividing centroblasts of the dark zone give rise to nondividing centrocytes that upregulate their immunoglobulin receptors and migrate to the light zone, where they interact with an extensive network of follicular dendritic cells (harboring the antigen on their surfaces in the form of immunocomplexes) and with T cells.30,31 The centrocytes that are not selected by follicular dendritic cell–held antigens are believed to die through apoptosis, whereas the centrocytes showing high affinity for the antigen on follicular dendritic cells are presumed to be positively selected and can follow 1 of 2 main pathways (memory B cell or immunoblastic/plasma cell differentiation), depending on the costimulatory signals they receive from the follicular dendritic cells and T cells.30,31 Our immunohistologic studies clearly identified a population of B cells in the light zone that was in intimate contact with follicular dendritic cells and expressed strongly the MUM1 protein. These MUM1+ cells often displayed enlarged plasma cell–like cytoplasm but showed, in contrast to mature plasma cells, markedly irregular nuclei (resembling those of centrocyte-type GC cells). These findings suggest that the expression of MUM1 protein was initiated within the light zone of the GC and that these MUM1+ B cells may represent antigen-selected surviving centrocytes and their progeny committed to differentiate further into plasma cells. This explanation is consistent with the observation that the MUM1+ GC B cells contain mutated Ig gene rearrangements with functional coding sequences and do not express Bcl-6, whose down-regulation has been associated with differentiation toward plasma cells.27 35
Although predominantly associated with late-stage B-cell differentiation, expression of the MUM1 protein did not appear to be B-cell specific. This was supported by the findings that a small percentage of T cells (1%-5%) in the GC and in the interfollicular areas of normal tonsil and reactive lymph nodes double stained for MUM1 and CD3, that most normal CD30+ cells in the interfollicular area did express MUM1, and that the expression of the MUM1 protein could be induced by the stimulation of peripheral blood T cells with PHA. These immunohistologic findings clearly indicated that MUM1 is expressed in activated T cells, and they are in keeping with the experimental evidence that knockout mice for the IRF4 gene (the murine homologue of MUM1) are unable to generate cytotoxic or antitumor responses.7 Moreover, the MUM1 gene shows complete homology to the ICSAT gene that was independently cloned from an HTLV1-positive adult T-cell leukemia cell line.6 Although the function of the MUM1 protein in T cells is unknown, it is of interest to note that other transcription factors known to play a fundamental role in B-cell development (Bcl-6, BOB1/OBF1, OCT2) have also been found in activated T cells.27 36-39
Among B-cell lymphomas, the strongest and most consistent expression of MUM1 was observed in lymphoplasmacytoid lymphoma/immunocytoma and multiple myeloma, and this finding was in keeping with the strong expression of the MUM1 protein detectable in normal and reactive plasma cells.
We also found that approximately 75% of diffuse large B-cell lymphomas expressed strongly the MUM1 protein. Unlike observations in normal GC B cells, in which Bcl-6 and MUM1 appear to be mutually exclusive, many tumor cells in approximately 50% of MUM1+ diffuse large B-cell lymphomas coexpressed the MUM1 and Bcl-6 proteins. These findings suggested that at least a proportion of diffuse large B-cell lymphomas may be derived from MUM1+ GC B cells with deregulated Bcl-6. In contrast, MUM1− diffuse, large B-cell lymphomas may be related to the GC B cells that do not express the protein.
Notably, MUM1 was consistently and strongly expressed in Hodgkin and Reed-Sternberg cells of classic Hodgkin's disease (nodular sclerosing and mixed cellularity). This finding is in keeping with the current concept that the tumor cells of classic Hodgkin's disease represent a clonal expansion of neoplastic B cells, probably related to some differentiation stage of GC B cells.40 41 Finally, we found that the MUM1 protein was expressed in lymphomas thought to be derived from activated T cells (eg, CD30+ anaplastic large-cell lymphomas).
In conclusion, this article describes a new monoclonal antibody, MUM1p, suitable for detecting the MUM1 protein on routine bioptic samples and Western blot analysis, and it provides novel data about MUM1 expression in normal and neoplastic lymphohematopoietic tissues. Based on these findings, it may be expected that the MUM1p monoclonal antibody will be a valuable tool for research and possibly diagnosis.
Acknowledgments
We thank A. Foerster and D. Jahnke for their skillful technical assistance in subcloning and sequencing of isolated MUM+cells. We also thank Mrs Claudia Tibidò for her excellent secretarial assistance.
Supported by the Associazione Italiana per la Ricerca sul Cancro. A.P. and C.S. were supported by the Federazione Italiana per la Ricerca sul Cancro.
Reprints:Brunangelo Falini, Istituto di Ematologia, Policlinico, Monteluce, 06122 Perugia, Italy; e-mail:faliniem@unipg.it
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal