The ABO blood group is clinically the most important blood group system. Elucidation of the molecular basis of the ABO polymorphism allows genotype determination without family studies. Described here is a new method based on the simultaneous amplification by polymerase chain reaction (PCR) of 3 fragments from exon 6, and 5′ and 3′ ends of exon 7 of the ABO gene, followed by single-strand conformation polymorphism (SSCP) analysis. This multiplex PCR-SSCP protocol allows the well-established base changes at 9 nucleotide positions 261, 297, 467, 526, 646, 657, 681, 1059, and 1096 to be assayed simultaneously so that 7 common alleles (A1, A1v, A2, B, O1, O1v, and O2) can be distinguished in a single-tube single-lane format. Each allele was characterized by a set of 3 haplotype-specific SSCP patterns. Chinese (n = 125) and white European (n = 98) samples were analyzed, and their genotypes were found consistent with the serologic phenotypes or could be deduced unambiguously. Fifteen samples (2 Chinese and 13 white European) were each found carrying at least 1 rare allele. Most of these alleles were new and some might be generated by intragenic recombination. This technique is the simplest, quickest, and most informative method reported to date and also readily identifies new alleles.
The ABO blood group system was discovered by Karl Landsteiner at the beginning of the 20th century. However, it was only in 1990 that the nucleotide (nt) sequence of the ABO gene was determined,1,2 and in 1995 that its genomic organization was elucidated.3 The ABO locus spans over 18 kilobases (kb) and consists of 7 exons, which range in size from 26 to 688 base pairs (bp), with most of the coding sequence lying in exon 7.3
The 2 major alleles, A and B, differ in 8 positions at nt 297, 526, 657, 703, 796, 803, 930, and 1096 (Table 1), and 4 of these base substitutions (nt 526, 703, 796, and 803) result in amino acid substitutions (residues 176, 235, 266, and 268).2,4The amino acid substitutions at the last 2 positions are critical in determining the specificity of theglycosytransferase.5There are 2 A1 alleles, A1 and A1variant (or A1v); they differ at nt 467 with the resulting substitution of leucine in A1v for proline in A1.2 Both are common, though with greatly different frequencies, in the few populations so far studied.6-9 The A2 allele is characterized by a single-base substitution at nt 467 (as in the A1v allele) and a single-base deletion at nt 1059.10 This deletion generates a frameshift and results in an additional domain at the carboxyl terminal of the mature protein, and is thought to be critical in determining the activity and substrate specificity of the transferase.
Sequence variations in common ABO alleles (base changes shown with reference to the A1 allele)
| Exon . | 3 . | 4 . | 5 . | 6 . | 7 . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR Fragment* . | . | . | . | . | F1 . | F2 . | . | . | . | . | . | . | F3 . | |||||||
| Nucleotide . | 106 . | 188 . | 189 . | 220 . | 261 . | 297 . | 467 . | 526 . | 646 . | 657 . | 681 . | 703 . | 771 . | 796 . | 802 . | 803 . | 829 . | 930 . | 10591061 . | 1096 . |
| A1 | G | G | C | C | G | A | C | C | T | C | G | G | C | C | G | G | G | G | C | G |
| A1v | T | |||||||||||||||||||
| A2 | T | — | ||||||||||||||||||
| B | G | G | T | A | A | C | A | A | ||||||||||||
| O1 | — | |||||||||||||||||||
| O1v | T | A | T | T | — | G | A | A | T | A | ||||||||||
| O2 | G | G | A | A | ||||||||||||||||
| Exon . | 3 . | 4 . | 5 . | 6 . | 7 . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR Fragment* . | . | . | . | . | F1 . | F2 . | . | . | . | . | . | . | F3 . | |||||||
| Nucleotide . | 106 . | 188 . | 189 . | 220 . | 261 . | 297 . | 467 . | 526 . | 646 . | 657 . | 681 . | 703 . | 771 . | 796 . | 802 . | 803 . | 829 . | 930 . | 10591061 . | 1096 . |
| A1 | G | G | C | C | G | A | C | C | T | C | G | G | C | C | G | G | G | G | C | G |
| A1v | T | |||||||||||||||||||
| A2 | T | — | ||||||||||||||||||
| B | G | G | T | A | A | C | A | A | ||||||||||||
| O1 | — | |||||||||||||||||||
| O1v | T | A | T | T | — | G | A | A | T | A | ||||||||||
| O2 | G | G | A | A | ||||||||||||||||
F1, F2, and F3 are the 3 PCR fragments used to assay the known single nucleotide polymorphisms. Note that deletions are denoted by dashes.
There are 3 common O alleles. The O1 allele differs from the A1 allele by a single-base deletion at nt 261 (Table1), which shifts the reading frame of the coding sequence and leads to the translation an enzymatically inactive protein.2 The O1 variant (or O1v) allele has not only the single-base deletion at nt 261 but also another 9 single-base substitutions when compared with the A1allele.2,11 The O2 allele does not possess the nt 261 deletion found in O1 and O1v alleles, and differs from the A1 allele in 4 nt substitutions, 297, 526, 802, and 1096, resulting in 2 amino acid substitutions.4,12 These 2 amino acid substitutions were found to abolish the activity of the transferase expressed in vitro. Both O1 and O1v are very common6-9,11,13-15, whereas O2 is less common.4,5,13,14 16-19
The molecular basis of many other rare alleles has also been delineated and was reviewed recently.20 This indicates the presence of extensive polymorphism in the coding sequence of the ABO locus. This is further substantiated by the identification of many more new rare alleles by single-strand conformation polymorphism (SSCP) analysis of 4 fragments separately amplified using polymerase chain reaction (PCR) from the last 2 exons of the gene, which account for 78% of the coding sequences.8,21,22 SSCP analysis detects single nucleotide polymorphisms (SNPs) and small insertions/deletions on the basis of the differential electrophoretic mobility of single-stranded DNA fragments with different sequences.23This report describes the development and use of an ABO genotyping method based on multiplex PCR-SSCP analysis that can discriminate the above-mentioned 7 common ABO alleles (A1, A1v, A2, B, O1, O1v, and O2) in a single-tube single-lane format. This technique is the simplest, quickest, most cost-effective, and informative ABO genotyping method reported to date. Another advantage is its capability of identifying new alleles very easily.
Materials and methods
Samples
Chinese donors' samples (n = 125) had previously been typed serologically and genotyped using denaturing gradient gel electrophoresis (DGGE) of PCR products amplified from exon 6,24 which was based on the protocol developed by Johnson and Hopkinson.25 DNA extraction was also carried out as previously reported.24 DNA samples from white Europeans (London area, n = 98) of unknown ABO blood groups were gifts from Dr David Whitehouse. Genotyped DNA samples carrying O2 or A2 alleles were given by Drs C. Gassner,18 M. J. Hessner,19 R. L. Sparrow,13 and R. Steffensen.16
Representative DNA samples carrying different combinations of the previously mentioned 7 common ABO alleles were also typed at 8 polymorphic sites of the gene by means of restriction digestion of appropriate PCR amplified fragments, according to reported protocols with slight modifications. These include the following: nt 188/189 (MvnI) using mo-41/mo-42 primer pair;11 nt 261 (KpnI) using ABOe6F1/ABOi6R1 primer pair (Table 2); and nt 467 (AluI and HpaII), nt 526 (BssHII and NarI), nt 646 (NdeII), nt 703 (AluI and HpaII), and nt 771 (DdeI) all using ABO-5S/ABO-4AS primer pair.11 26 All restriction enzymes (Boehringer Mannheim GmbH, Germany) were used according to the manufacturer's instructions. These DNA samples were used as controls for optimizing the electrophoretic conditions for SSCP analysis.
PCR fragments and primers used in this study
| Fragment (size) . | Forward and Reverse Primers . | Region Bound by the Primer Pair (excluding primer annealing regions) . | |
|---|---|---|---|
| Name . | Sequence (5′ → 3′)* . | ||
| F1 (192 bp) | ABOe6F1 | TGC AGT AGG AAG GAT GTG CTC GTG | nt 259 → 374/IVS6 +1 → +27 |
| ABOi6R1 | CAG TCA CTC GAC ACT ACC TGG GTC T | ||
| F2 (279 bp) | ABOe7F2 | CCG TGT CCA CTA CTA TGT CTT CAC C | nt 460 → 693 |
| ABOe7R2 | CCG TAG AAG CTG GGG TGC AG | ||
| F3 (165 bp) | ABOe7F3 | CCT GAG GAA GCT GAG GTT CAC TGC | nt 1029 → 1144 |
| ABOi7R4 | ACA ACA GGA CGG ACA AAG GAA ACA G | ||
| Fragment (size) . | Forward and Reverse Primers . | Region Bound by the Primer Pair (excluding primer annealing regions) . | |
|---|---|---|---|
| Name . | Sequence (5′ → 3′)* . | ||
| F1 (192 bp) | ABOe6F1 | TGC AGT AGG AAG GAT GTG CTC GTG | nt 259 → 374/IVS6 +1 → +27 |
| ABOi6R1 | CAG TCA CTC GAC ACT ACC TGG GTC T | ||
| F2 (279 bp) | ABOe7F2 | CCG TGT CCA CTA CTA TGT CTT CAC C | nt 460 → 693 |
| ABOe7R2 | CCG TAG AAG CTG GGG TGC AG | ||
| F3 (165 bp) | ABOe7F3 | CCT GAG GAA GCT GAG GTT CAC TGC | nt 1029 → 1144 |
| ABOi7R4 | ACA ACA GGA CGG ACA AAG GAA ACA G | ||
Underlined bases indicate mismatches with the A1 cDNA sequence.
PCR amplification
Six primers (Table 2) were designed using the software Oligo (version 4.0; National Bioscience Inc, Plymouth, MA) so that the self-complementarity of individual primers and primer dimer formation between any pair of primers was negligible. To achieve this, 1 or 2 mismatches with the genomic sequence were introduced in some primers. Multiplex PCR was carried out using a thermal cycler (CT1; Perkin Elmer, Foster City, CA) in a reaction volume of 25 μL containing 1 × reaction buffer (50 mmol/L KCl,10 mmol/L Tris-HCl, pH 9.0 at 25°C, and 0.1% Triton X-100) 1.5 mmol/L MgCl2, 0.2 mmol/L each of dNTP, 0.4 μmol/L each of the 6 primers, 50 to 100 ng of genomic DNA, and 0.5 U of Taq DNA polymerase (in Storage Buffer A; Promega, Madison, WI). To set up the PCR, all components, except Taq DNA polymerase, were included in each reaction and 40 μL of mineral oil was overlaid to prevent evaporation. The enzyme was added during the initial denaturation phase of 95°C/5 min (hot start). Amplification was carried out for 35 cycles of 95°C/20 s and 70°C/60 s, and followed by a final extension of 10 minutes at 72°C.
SSCP analysis
A mixture was prepared for each sample by mixing 2.5 μL PCR product, 4 μL SSCP loading solution (95% formamide, 20 mmol/L Na2EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF) and water made up to 8 μL. The mixture was denatured at 95°C for 7 minutes and cooled immediately in ice water before being loaded into the gel with a Hamilton syringe. Electrophoresis was carried out at 400 V for 2 hours 45 minutes using Hoefer SE600 tanks (Pharmacia Biotech, San Francisco, CA) and 9%T/1%C polyacrylamide gels measuring 160 × 140 × 0.75 mm. Buffer in the gel and the tank was 1 × TBE (89 mmol/L Tris, 89 mmol/L boric acid, 2.5 mmol/L Na2EDTA, pH 8.3). The buffer temperature was maintained at 25°C by connecting the heat exchanger of the tank to an external thermostatically controlled circulator and by continuously mixing the buffer throughout the run.
The bands were visualized by silver staining. The gel was fixed in 2 changes of 10% ethanol and 0.5% acetic acid, each 2 minutes. This was followed by incubation in 2 changes, each 7 minutes, of freshly prepared 0.1% silver nitrate solution with continuous shaking. The silver nitrate solution was then decanted and the silver stain developed for 10 to 15 minutes in a solution of 1.5% NaOH, 0.01% sodium borohydride, and 0.4% (v/v) formaldehyde. After being rinsed in 3 changes of water, the gel was dried at 83°C for 45 minutes for permanent record.
Direct sequencing of PCR products
For samples to be sequenced, a 2051-bp fragment spanning exon 6, intron 6, and exon 7 was amplified using the primer pair ABOe6F1/ABOi7R4 (Table 2) as described previously with the following modifications: 0.5 μmol/L of each primer, and amplification carried out for 35 cycles of 95°C/1 min and 68°C/3 min. The PCR products were electrophoresed in agarose gel and purified using QIAquick Gel Extraction Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions. The purified PCR products were sequenced using ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit and the sequenced samples electrophoresed using ABI PRISM 310 Genetic Analyzer (both from Perkin Elmer) according to the manufacturer's protocol with the following modifications. The cycle sequencing was instead carried out for 30 cycles of 96°C/30 s and 60°C/4 minutes 15 seconds in a final reaction volume of 10 μL (half the recommended volume). Three primers were used to sequence to exons 6 and 7: ABOi6R1, ABO-5S (see Samples) and ABOi7R5 (5′ TGC AGG CAG CCC TCC CAG AG 3′).
Results
SSCP banding patterns
Fragment 1 (F1) was amplified from exon 6 and its immediate flanking regions, fragment 2 (F2) from the 5′ end of exon 7, and fragment 3 (F3) from the 3′ end of exon 7 and the 3′ untranslated region (Table 1). Thus, this strategy assayed the known SNPs at nt 261 and 297; at nt 467, 526, 646, 657, and 681; and at nt 1059 to 1061 and 1096, respectively. The numbers of standard SSCP banding patterns were 4 for F1 (1a to 1d corresponding to alleles O1, O1v, A1/A1v/A2, and B/O2), 5 for F2 (2a to 2e corresponding to A1v/A2, A1/O1, O2, B, and O1v), and 3 for F3 (3a to 3c corresponding to B/O2, A1/A1v/O1/O1v, and A2) (Figure 1A and B, and Table1). In other words, the observed haplotypes of the known SNPs within every amplified fragment each produced a distinctive SSCP pattern.
Multiplex PCR-SSCP analysis for the determination of the ABO genotypes.
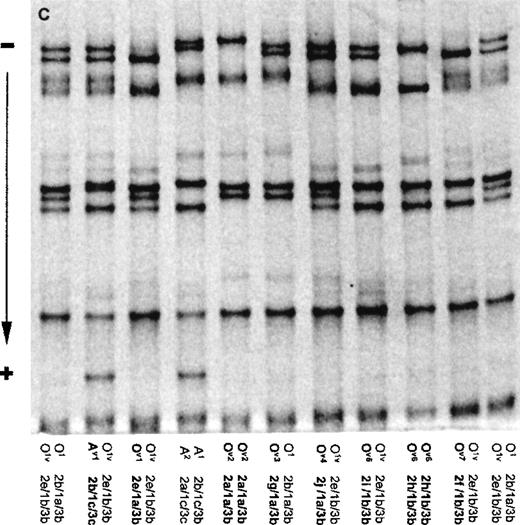
(A) The SSCP banding patterns for homozygotes (for O1, O1v, A1, A1v, and B) and heterozygotes (for less common alleles A2 and O2). (B) Schematic drawing showing the standard SSCP banding patterns for the 7 common ABO alleles, together with their designations. Note that the homozygous patterns for A2 and O2 are deduced from the heterozygous patterns. For alleles B and O2, the weakly stained fastest-moving band of the F1 fragment overlaps with the strongly stained slowest-moving band of the F3 fragment marked by asterisk (*). (C) The SSCP banding patterns for representative samples containing rare alleles, together with some common phenotypes (O1O1v and A1A2) for comparison. The rare alleles are designated as Av1, and Ov1 to Ov7.
Multiplex PCR-SSCP analysis for the determination of the ABO genotypes.
(A) The SSCP banding patterns for homozygotes (for O1, O1v, A1, A1v, and B) and heterozygotes (for less common alleles A2 and O2). (B) Schematic drawing showing the standard SSCP banding patterns for the 7 common ABO alleles, together with their designations. Note that the homozygous patterns for A2 and O2 are deduced from the heterozygous patterns. For alleles B and O2, the weakly stained fastest-moving band of the F1 fragment overlaps with the strongly stained slowest-moving band of the F3 fragment marked by asterisk (*). (C) The SSCP banding patterns for representative samples containing rare alleles, together with some common phenotypes (O1O1v and A1A2) for comparison. The rare alleles are designated as Av1, and Ov1 to Ov7.
The 7 common ABO alleles were each characterized by a set of 3 haplotype-specific SSCP patterns produced by the amplified fragments (Figure 1A and B). In particular, SSCP pattern 1a was exclusively observed in allele O1, whereas patterns 1b and 2e were unique to allele O1v. Pattern 3c was only associated with allele A2. Alleles A1 and A1v were identical for fragments F1 and F3, but differed in F2 (pattern 2b vs 2a). Alleles A1v and A2 were identical for F1 and F2, but different for F3 (pattern 3b vs 3c). Note that pattern 1c was common and unique to the A alleles. Although alleles B and O2 had the same patterns for F1 and F3 (1d and 3a, unique to themselves), they could be distinguished by their own characteristic patterns in F2 (2d vs 2c). It should also be noted that patterns 2b and 2c only differed in their fastest moving, weakly stained bands. Homozygotes gave SSCP patterns as shown in Figure 1A and B, whereas heterozygotes were characterized by SSCP patterns that were the combinations of 2 different allele specific patterns.
ABO genotyping of Chinese and white European samples
The distribution of ABO genotypes determined by the multiplex PCR-SSCP analysis is shown in Table 3 and the allele frequencies calculated by the method of gene counting are shown in Table 4. The genotypes of the Chinese samples could be determined unambiguously and were consistent with the serologic phenotypes. These genotypes also correlated with those determined by the DGGE method,24 except for 2 cases, given the following differences between the 2 methods. The DGGE method analyzed PCR products (250 bp) amplified from exon 6 and its immediate flanking regions, and thus could not distinguish between alleles A1 and A1v, which differ at nt 467 (exon 7, Table 1). The DGGE method was also unable to differentiate between alleles B and O2, which have the same sequence in exon 6 (Table 1), though allele O2 was not found in the Chinese samples in this study. Incidentally, the O1v allele was found to be identical to the “O2” allele defined by DGGE. Two group O samples, genotyped as O1O1vby the DGGE method, were each found to be heterozygous for O1v and a different rare O allele (Ov1 and Ov4 as shown in Table 5); the genotypes are written as O1vOv in Table 3. These 2 rare O alleles could indeed be typed as the O1allele when only the F1 fragment was considered (pattern 1a), but they did not have the expected pattern 2b for F2 (see below). Note that Ov is being used here as a generic notation for any “new” O allele other than O1, O1v, and O2.
Distribution of ABO genotypes in Chinese and white Europeans
| Blood Group3-150 . | Genotype3-151 . | Chinese . | white European . | ||
|---|---|---|---|---|---|
| n . | % . | n . | % . | ||
| A1 | A1 A1 | — | — | 1 | 1.02 |
| A1 A1v | 1 | 0.80 | — | — | |
| A1 A2 | — | — | 4 | 4.08 | |
| A1 O1 | — | — | 14 | 14.29 | |
| A1 O1v | — | — | 3 | 3.06 | |
| A1 O2 | — | — | — | — | |
| A1v A1v | 3 | 2.40 | 1 | 1.02 | |
| A1v A2 | — | — | — | — | |
| A1v O1 | 23 | 18.40 | — | — | |
| A1v O1v | 7 | 5.60 | — | — | |
| A1v O2 | — | — | — | — | |
| A2 | A2 A2 | — | — | — | — |
| A2 O1 | — | — | 5 | 5.10 | |
| A2 O1v | — | — | 1 | 1.02 | |
| A2 O2 | — | — | — | — | |
| A2 Ov | — | — | 1 | 1.02 | |
| B | B B | 1 | 0.80 | 2 | 2.04 |
| B O1 | 18 | 14.40 | 9 | 9.18 | |
| B O1v | 12 | 9.60 | 4 | 4.08 | |
| B O2 | — | — | 1 | 1.02 | |
| B Ov | — | — | 2 | 2.04 | |
| A1 B | A1 B | 2 | 1.60 | 4 | 4.08 |
| A1v B | 8 | 6.04 | 1 | 1.02 | |
| A2 B | A2 B | — | — | — | — |
| O | O1 O1 | 15 | 12.00 | 15 | 15.31 |
| O1 O1v | 25 | 20.00 | 17 | 17.35 | |
| O1 O2 | — | — | 1 | 1.02 | |
| O1 Ov | — | — | 4 | 4.08 | |
| O1v O1v | 8 | 6.40 | 2 | 2.04 | |
| O1v O2 | — | — | — | — | |
| O1v Ov | 2 | 1.60 | 2 | 2.04 | |
| O2 O2 | — | — | — | — | |
| Ov Ov | — | — | 3 | 3.06 | |
| A | O1v Av1 | — | — | 1 | 1.02 |
| Total | 125 | 100 | 98 | 100 | |
| Blood Group3-150 . | Genotype3-151 . | Chinese . | white European . | ||
|---|---|---|---|---|---|
| n . | % . | n . | % . | ||
| A1 | A1 A1 | — | — | 1 | 1.02 |
| A1 A1v | 1 | 0.80 | — | — | |
| A1 A2 | — | — | 4 | 4.08 | |
| A1 O1 | — | — | 14 | 14.29 | |
| A1 O1v | — | — | 3 | 3.06 | |
| A1 O2 | — | — | — | — | |
| A1v A1v | 3 | 2.40 | 1 | 1.02 | |
| A1v A2 | — | — | — | — | |
| A1v O1 | 23 | 18.40 | — | — | |
| A1v O1v | 7 | 5.60 | — | — | |
| A1v O2 | — | — | — | — | |
| A2 | A2 A2 | — | — | — | — |
| A2 O1 | — | — | 5 | 5.10 | |
| A2 O1v | — | — | 1 | 1.02 | |
| A2 O2 | — | — | — | — | |
| A2 Ov | — | — | 1 | 1.02 | |
| B | B B | 1 | 0.80 | 2 | 2.04 |
| B O1 | 18 | 14.40 | 9 | 9.18 | |
| B O1v | 12 | 9.60 | 4 | 4.08 | |
| B O2 | — | — | 1 | 1.02 | |
| B Ov | — | — | 2 | 2.04 | |
| A1 B | A1 B | 2 | 1.60 | 4 | 4.08 |
| A1v B | 8 | 6.04 | 1 | 1.02 | |
| A2 B | A2 B | — | — | — | — |
| O | O1 O1 | 15 | 12.00 | 15 | 15.31 |
| O1 O1v | 25 | 20.00 | 17 | 17.35 | |
| O1 O2 | — | — | 1 | 1.02 | |
| O1 Ov | — | — | 4 | 4.08 | |
| O1v O1v | 8 | 6.40 | 2 | 2.04 | |
| O1v O2 | — | — | — | — | |
| O1v Ov | 2 | 1.60 | 2 | 2.04 | |
| O2 O2 | — | — | — | — | |
| Ov Ov | — | — | 3 | 3.06 | |
| A | O1v Av1 | — | — | 1 | 1.02 |
| Total | 125 | 100 | 98 | 100 | |
For white Europeans, the blood groups (or phenotypes) are deduced based on their genotypes. The phenotype for O1vAv is group A, but whether it is group A1 or A2 cannot be determined reliably.
Av1 represents the single new rare A allele and Ov new rare O alleles. For the sake of simplicity, all the different rare O alleles are grouped under the name Ov. Thus, Ov Ov does not necessarily represent a homozygote.
ABO allele frequencies in Chinese and white Europeans
| Allele4-150 . | Chinese . | white European . |
|---|---|---|
| A1 | 0.0120 | 0.1378 |
| A1v | 0.1800 | 0.0153 |
| A2 | — | 0.0561 |
| Av1 | — | 0.0051 |
| B | 0.1680 | 0.1276 |
| O1 | 0.3840 | 0.4082 |
| O1v | 0.2480 | 0.1633 |
| O2 | — | 0.0102 |
| Ov | 0.0080 | 0.0765 |
| Allele4-150 . | Chinese . | white European . |
|---|---|---|
| A1 | 0.0120 | 0.1378 |
| A1v | 0.1800 | 0.0153 |
| A2 | — | 0.0561 |
| Av1 | — | 0.0051 |
| B | 0.1680 | 0.1276 |
| O1 | 0.3840 | 0.4082 |
| O1v | 0.2480 | 0.1633 |
| O2 | — | 0.0102 |
| Ov | 0.0080 | 0.0765 |
Av1 represents the single new rare A allele and Ov new rare O alleles. For the sake of simplicity, all the different rare O alleles are grouped under the name Ov.
Sequence variations in rare ABO alleles identified in this study (base changes shown with reference to the A1allele)
| Allele5-151 . | N5-152 . | SSCP Patterns . | Exon 6 . | Exon 7 . | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F15-150 . | F25-150 . | . | . | . | . | . | . | . | F35-150 . | ||||||||||||
| nt 261 . | 297 . | 318 . | 467 . | 526 . | 542 . | 579 . | 595 . | 646 . | 657 . | 681 . | 703 . | 771 . | 796 . | 802 . | 803 . | 829 . | 930 . | 1059- 1061 . | |||
| A1 | 2b/1c/3b | G | A | C | C | C | G | T | C | T | C | G | G | C | C | G | G | G | G | C | |
| Av1 | 1 | 2b/1c/3c | — | ||||||||||||||||||
| Ov1 | 1 | 2e/1a/3b | — | A | A | T | A | ||||||||||||||
| Ov2 | 5 | 2a/1a/3b | — | T | T | ||||||||||||||||
| Ov3 | 1 | 2g/1a/3b | — | T | |||||||||||||||||
| Ov4 | 1 | 2j/1a/3b | — | C | |||||||||||||||||
| Ov5 | 1 | 2i/1b/3b | — | G | A | A | A | T | A | ||||||||||||
| Ov6 | 5 | 2h/1b/3b | — | G | T | A | A | T | A | ||||||||||||
| Ov7 | 3 | 2f/1b/3b | — | G | A | T | A | ||||||||||||||
| Allele5-151 . | N5-152 . | SSCP Patterns . | Exon 6 . | Exon 7 . | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F15-150 . | F25-150 . | . | . | . | . | . | . | . | F35-150 . | ||||||||||||
| nt 261 . | 297 . | 318 . | 467 . | 526 . | 542 . | 579 . | 595 . | 646 . | 657 . | 681 . | 703 . | 771 . | 796 . | 802 . | 803 . | 829 . | 930 . | 1059- 1061 . | |||
| A1 | 2b/1c/3b | G | A | C | C | C | G | T | C | T | C | G | G | C | C | G | G | G | G | C | |
| Av1 | 1 | 2b/1c/3c | — | ||||||||||||||||||
| Ov1 | 1 | 2e/1a/3b | — | A | A | T | A | ||||||||||||||
| Ov2 | 5 | 2a/1a/3b | — | T | T | ||||||||||||||||
| Ov3 | 1 | 2g/1a/3b | — | T | |||||||||||||||||
| Ov4 | 1 | 2j/1a/3b | — | C | |||||||||||||||||
| Ov5 | 1 | 2i/1b/3b | — | G | A | A | A | T | A | ||||||||||||
| Ov6 | 5 | 2h/1b/3b | — | G | T | A | A | T | A | ||||||||||||
| Ov7 | 3 | 2f/1b/3b | — | G | A | T | A | ||||||||||||||
F1, F2, and F3 are the 3 PCR fragments used to assay the known single nucleotide polymorphisms. Nucleotide (nt) positions shown in boldface (nt 318, 542, and 595) are new mutation sites not previously reported. Nt 1096 in F3 is not shown here because it is within the region to which the sequencing primer ABOi7R5 anneals. Note that deletions are denoted by dashes.
Ov1 and Ov4 were found in Chinese and also previously reported in Japanese,8 whereas the rest were found in white Europeans and were new alleles.
N refers to the number of chromosomes found for each rare allele.
Eighty-five white European samples were found either heterozygous or homozygous for the 7 common ABO alleles and their blood groups could be deduced unambiguously (Table 3). The rest (n = 13) carried at least 1 new allele (1 A2Ov, 2 BOv, 4 O1Ov, 2 O1vOv, 3 OvOv, and 1 O1vAv; Table 3).
As the white European samples had not been serologically typed for the ABO blood group, representative white European samples carrying the various common alleles were sequenced for exons 6 and 7. These common alleles were found to carry the expected base changes as shown in Table1. These results were also consistent with those of restriction analysis performed for DNA samples used as controls for optimizing electrophoretic conditions for SSCP analysis (see above).
Description of new alleles
New alleles were identified by subtracting the SSCP patterns of the common allele, if present, from those observed in the heterozygotes. This study identified a total of 15 samples (2 Chinese and 13 white European) carrying at least 1 “new” allele (Table 3). Although the white European samples were not typed serologically, the new alleles could still be identified as being A or O alleles. These 15 samples were sequenced for exons 6 and 7. (The nucleotide sequences for new alleles identified in this study have been submitted to GenBank [accession numbers: AF182745 to AF182756].) The SSCP banding patterns for each distinct new allele are shown in Figure 1C, and the corresponding base changes underlying these alleles in Table 5.
An SSCP pattern of 2b/1c/3c identified 1 new allele as an A allele (Av1) because of its 1c pattern in F1. Apparently, this Av1 allele seemed to be a hybrid A1-A2 allele and resembled the classical A2 allele in carrying the single C deletion at nt 1059 to 1061, but not the base change at nt 467 (Table 5). It has been shown that the decreased A transferase activity of the A2 allele is due to the single C deletion.10 Thus, it is very tempting to suggest that the O1vAv individual would be typed as group A2.
All other new alleles had patterns 1a/3b or 1b/3b for F1 and F3 (Figure1C and Table 5). F1 producing patterns 1a or 1b had the single-base deletion at nt 261 (Figure 1B and Table 1), which would result in frameshift and thus produce a truncated and enzymatically inactive protein product. Thus, new alleles with F1 patterns 1a or 1b were predicted to be O alleles. Their F2 SSCP patterns were of 2 types.
In one type, the F2 pattern was either 2a or 2e, but not in combination with the expected F1/F3 patterns. One chromosome carried the Ov1 allele, which was characterized by the 2e/1a/3b pattern and appeared to be a hybrid O1-O1v allele. Five chromosomes carried the Ov2 allele defined by the 2a/1a/3b pattern. Although appearing to be a hybrid O1-A1v allele, the Ov2 allele was in fact shown by sequencing to carry in its “1a” F1 fragment a C318T transition in addition to the single G deletion at nt 261.
In another type, the F2 pattern was completely novel (2f to 2j) and different from the 5 standard patterns (2a to 2e). In other words, these F2 fragments harbored base changes other than those shown in Table 1. They were in combination with F1/F3 patterns 1a/3b or 1b/3b. Both Ov3 and Ov4, each found on 1 single chromosome only, were O1-like alleles with the patterns 2g/1a/3b and 2j/1a/3b, respectively. They have base changes in addition to the single C deletion at nt 261. Interestingly, the C657T transition of allele Ov3 was identical to that found in the B allele at the same position. The other 3 new alleles (Ov5, Ov6, and Ov7) were O1v-like with F2 patterns 2i, 2h, and 2f, respectively, in combination with 1b/3b. Ov5 and Ov6 each had an additional mutation on an O1v-specific haplotype background, whereas Ov7 is the same as O1v, except that the base at nt 681 was G instead of A (ie, A1-like rather than O1v-like).
In summary, 1 Av1 and 7 distinct Ov alleles (Ov1-Ov7) were identified in a total of 18 chromosomes (Table 5). Av1 and Ov1 appeared to be hybrid alleles. The rest were characterized by additional base changes on an O1 or O1v background with Ov2, Ov3, and Ov4 being O1-like and Ov5, Ov6, and Ov7, O1v-like. Nucleotides 318, 542, and 595 were new mutation sites not previously described. It is also interesting to note that Ov1 and Ov4 were found in Chinese in this study and corresponded to alleles *O202 and *O102, respectively, reported in a Japanese study.8 The others were found in white Europeans and were genuinely new alleles.
Discussion
Molecular cloning of the ABO gene and elucidation of the molecular basis of its various alleles allow the direct determination of the ABO genotypes without family studies. One group of methods relied on the restriction analysis of SNPs in PCR-amplified products, with or without multiplexing the PCRs.2,4,16,26-30 This approach culminated in the development of a single-tube single-lane genotyping method based on restriction analysis of duplex PCR products and claimed to discriminate 5 alleles (A1, A2, B, O1, and O2).4 However, this particular method and another30 solely relied on the SNP at nt 467 to differentiate A1 and A2, and thus could not distinguish between A1v and A2 (Table1).
Another approach uses allele-specific amplification, again with or without multiplexing the PCRs.14,18,19,31 In particular, one report described an elegant use of this approach to distinguish 6 alleles (A1, A2, B, O1, O1v, and O2) by mixing 10 primers in a single reaction to produce PCR products of different lengths and targeting 5 SNP sites.14 Still another approach was the combined use of allele-specific amplification and restriction analysis.6,7 9
The fourth approach uses mutation screening methods that detect both known and unknown SNPs in the assayed PCR products (eg, denaturing gradient gel electrophoresis24,25 and SSCP analysis).8,29 It is very exciting to note that 13 different alleles (common and rare) could be identified by SSCP analysis of 4 separate PCR products amplified from exons 6 and 7.8 The current study made use of this advantage and further improved the method by multiplexing the PCRs and the subsequent SSCP analysis.
To distinguish the 7 common alleles (Table 1), the minimum requirement is to assay SNPs at nt 261, 297, 467, and 1059 plus any one of the SNPs differentiating between B and O2 alleles (nt 657, 703, 796, 802, 803, and 930). As a result, 3 PCR products were amplified in a single tube from exon 6, 5′ and 3′ ends of exon 7, and analyzed with SSCP in a single lane to assay a total of 9 known SNP sites (Table 1). The redundancy in the known SNPs being assayed increased the accuracy and reliability of the genotyping method. The fact that any other unknown SNPs within the regions bounded by the primer pairs (Table 2) will most likely be detected underlies the capability of this muliplex PCR-SSCP approach to identify new alleles. It is expected that this method can also detect some known rare alleles with the base changes within the regions bounded by the 3 primer pairs (Table 2), eg, Ax and B3.20 This technique is the simplest, quickest, most cost-effective method reported to date to be able to discriminate 7 common ABO alleles in a single-tube single-lane format, and at the same time readily identifies new alleles (Table 5).
It should be noted that about half of exon 7 (nt 694-1028) and the coding sequences preceding exon 6 are not covered by the PCR amplification strategy described here. These regions contain many known SNP sites2,8,20,21 33 and are also likely to harbor new mutation sites yet to be identified. Thus, sequence variation in these regions, known or unknown, will not be detected by the current multiplex PCR-SSCP analysis. However, given the physical closeness of the amplified fragments in exon 7 and the sensitivity of the SSCP technique, this method has already made full use of the multiplexing and the SSCP strategies. Its success and reproducibility hinge on the precise control of the buffer temperature during electrophoresis of the large polyacrylamide gel. In addition, silver staining requires the use of freshly prepared reagents.
For the reported Chinese population, the frequencies of the major ABO alleles (A, B, and O) obtained in this study were similar to those obtained for a larger sample of random Chinese donors (n = 315).24 However, for the reported white European population from London, the frequency of the A allele (Table 4) was lower than previously reported32 (0.21 vs 0.28), whereas the B allele was much higher (0.13 vs 0.06). This indicates that the collected white European samples might not be truly random samples. It is also possible that some might come from offspring of white Europeans and other ethnic groups living in London.
The A1v allele was much more frequent than the A1 allele in the Chinese population (Table 4). This is in parallel with the findings in the Japanese and the Korean populations,7,9 and may probably be true for the Orientals in general. On the contrary, the reverse was true for the white Europeans: The A1 was much more common than the A1v allele (Table 4), a finding consistent with that from another white European population (German).6 The O2 allele was not found in Chinese (Table 4), neither was it found in Japanese7,14 and Koreans,9suggesting that this allele may be extremely rare or even does not exist in the Orientals. Interestingly, this allele was not found in Amerindians either.17 Although not very common, this allele was found in white Europeans at a frequency of 0.0102 (Table 4), comparable to those reported for other white European populations (0.017 to 0.037).4,6,14 16-19
Even with a medium sample size, the study found 15 samples (2 Chinese and 13 white European) each carrying at least 1 “new” allele. This gave an overall new allele frequency of 0.0080 in Chinese and 0.0867 in white Europeans (Table 4). Thus, new alleles were found more frequently in white Europeans than in Chinese. With the exception of one new A allele, all other new alleles were predicted to be O alleles on the basis of the presence of the single base deletion at nt 261. In addition, most of the base changes leading to these new alleles were found to be located in the F2 fragment amplified from the 5′ end of exon 7, which is downstream of the single-base deletion at nt 261. This should not be too surprising since the frameshift produced by this single-base deletion would release the downstream sequences from selection pressure, if any, which are thus vulnerable to more mutations without further altering the phenotype (a truncated product). Note that Ov1 and Ov4 found in Chinese in this study had previously been reported in Japanese,8 whereas the rest were genuinely new alleles and were found in white Europeans.
Most rare alleles identified in this study were characterized by additional base changes on an O1 or O1vbackground. On the other hand, alleles Av1 and Ov1 each had one of the standard SSCP patterns in their respective fragments F1 to F3, but in unexpected combinations: 2b/1c/3c and 2e/1a/3b (Figure 1C and Table 5). These unexpected combinations and the underlying nucleotide sequence data suggest that these 2 alleles might be hybrid alleles (A1-A2 and O1-O1v, respectively) probably generated by intragenic recombination between common alleles. As the ABO alleles were increasingly being analyzed at the DNA level, more and more hybrid alleles were reported in the literature and suggested to be produced by recombination in intron 6 or exon 7.21,33-35 Chi and chi-like sequences have been considered to be associated with recombination36 and are found in the 3′ end of intron 6.33 In light of these findings, it has been proposed that the ABO gene might contain a recombination hot spot around intron 6 and exon 7.33,35 A lot of SNPs were identified in intron 6 (about 1 kb in size) and their haplotypes found to be specific for the common ABO alleles.33 35 Thus, complete sequencing of the coding region and intron 6 will characterize these newly identified alleles and will shed light on the possible generation of these hybrid alleles by intragenic recombination.
Acknowledgments
I would like to thank Drs C. Gassner (Central Institute for Blood Transfusion and Immunological Department, General Hospital and University Clinics, Innsbruck, Austria), M. J. Hessner (The Blood Center of Southeastern Wisconsin, Milwaukee, WI), R. L. Sparrow (Red Cross, Blood Bank Victoria, Southbank, Victoria, Australia), and R. Steffensen (Regional Centre for Blood Transfusion and Clinical Immunology, Aalborg Hospital, Aalborg, Denmark) for their provision of DNA samples carrying O2 or A2 alleles. Thanks also go to Dr David Whitehouse (MRC Human Biochemical Genetics Unit, Galton Laboratory, University College, London, UK) for his white European DNA samples. Last but not least, I would also like to thank Dr M. C. Chow (Department of Applied Biology and Chemical Technology, Hong Kong Polytechnic University, HK) for allowing me to use the ABI PRISM 310 Genetic Analyzer.
This study was supported by a departmental research grant (G-S062), Department of Nursing and Health Sciences, Hong Kong Polytechnic University.
Reprints:S. P. Yip, Department of Nursing and Health Sciences, the Hong Kong Polytechnic University, Hung Hom, Kowloon, Hong Kong SAR, China; e-mail: hsspyip@polyu.edu.hk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal