Survivin is a member of the inhibitor of apoptosis protein (IAP) family that is believed to play a role in oncogenesis. To elucidate further its physiologic role(s), we have characterized the murinesurvivin gene and complementary DNA (cDNA). The structural organization of the survivin gene, located on chromosome 11E2, is similar to that of its human counterpart, both containing 4 exons. Surprisingly, 3 full-length murine survivin cDNA clones were isolated, predicting the existence of 3 distinct survivin proteins. The longest open reading frame, derived from all 4 exons, predicts a 140-amino acid residue protein, survivin140, similar to human survivin, which contains a single IAP repeat and a COOH-terminal coiled-coil domain that links its function to the cell cycle. A second cDNA, which retains intron 3, predicts the existence of a 121-amino acid protein, survivin121 that lacks the coiled-coil domain. Removal of exon 2-derived sequences by alternative pre-messenger RNA (mRNA) splicing results in a third 40-amino acid residue protein, survivin40, lacking the IAP repeat and coiled-coil structure. Predictably, only recombinant survivin140 and survivin121 inhibited caspase-3 activity. All 3 mRNA species were variably expressed during development from 7.5 days postcoitum. Of the adult tissues surveyed, thymus and testis accumulated high levels of survivin140 mRNA, whereas survivin121-specific transcripts were detected in all tissues, while those representing survivin40 were absent. Human counterparts to the 3 survivin mRNA transcripts were identified in a study of human cells and tissues. The presence of distinct isoforms of survivin that are expressed differentially suggests that survivin plays a complex role in regulating apoptosis.
A delicate balance between cell proliferation and cell death is required by multicellular organisms to maintain cell and tissue homeostasis and thereby prevent the development of a variety of pathologic outcomes, including hematologic malignancies, hematopoietic dysfunction, cancer and lymphoproliferative diseases, autoimmune and degenerative disorders, impaired wound repair, and developmental abnormalities.1,2 Apoptosis is the major physiologic means by which cell death is achieved. Although the cellular mechanisms responsible for mediating and regulating “programmed cell death” are not yet fully understood, it is clear that the process is complex and tightly regulated. Current evidence supports a model in which the induction of apoptotic cell death requires the activation of a series of cysteine proteases, the caspases. After their release and activation in response to a variety of stimuli, caspases lead to the destruction of the cell in an apparently stepwise process, by disrupting the nuclear membrane and chromatin structure, reorganizing the cytoskeleton, shutting down DNA replication and repair, destroying DNA, inducing phagocytosis, and finally disintegrating the cell into apoptotic bodies.3 The complexity of this system is underscored by recent data indicating that there also exist caspase-independent pathways of apoptosis.4
Physiologic mechanisms to regulate apoptosis, and thereby prevent uncontrolled cell destruction, are highly conserved through evolution. The prototypical members of the inhibitor of apoptosis (IAP) family were first identified in baculoviruses as proteins that could inhibit the apoptotic response of cultured insect cells to viral infections.5,6 Since that time, several additional members of this family have been identified. The human XIAP, cIAP-1, and cIAP-2 proteins each contain 3 amino-terminal tandem repeats of the so-called baculovirus repeat (BIR) domain, and a carboxy-terminal RING zinc finger domain. The neuronal apoptosis inhibitor protein (NAIP), however, contains no RING finger, whereas the “BIR repeat containing ubiquitin-conjugating enzyme” (BRUCE) contains only a single BIR repeat and lacks a RING finger domain.7 Although exhaustive structure-function studies remain to be performed, it appears that the BIR domain in these proteins is critical for antiapoptotic function, although each individual BIR repeat may vary in specific activity.8 Furthermore, the mechanism(s) by which IAPs interfere with apoptosis remains unclear. Although XIAP, cIAP-1, and cIAP-2 are able to inhibit caspase-3 and capase-7, NAIP is not, consistent with the notion that additional caspase-independent pathways of apoptosis may exist.9 10
Survivin is a recently described member of the IAP family.11 It resembles BRUCE in that it contains only a single BIR repeat and lacks a carboxy-terminal RING finger domain. In addition, both survivin and BRUCE contain an insertion of 3 amino acid residues within the BIR domain, which is absent in other IAP family members.7 The gene encoding human survivin was first identified by screening a human genomic DNA library with a complementary DNA (cDNA) putatively encoding the coagulation factor Xa receptor, effector cell protease receptor-1 (EPR-1).12,13Because the coding regions of human survivin demonstrate a high degree of sequence similarity with the corresponding regions ofEPR-1 in the antisense orientation, it was suggested that the 2 genes may be related evolutionarily11 and that the regulation of the 2 genes may somehow be linked.14 The functions of EPR-1 and survivin are not clearly related, however. EPR-1 is reported to be expressed widely in adult tissues and is believed to mediate the proinflammatory or mitogenic properties of factor Xa.15-18 In contrast, highest levels of steady-statesurvivin messenger RNA (mRNA) expression are found in solid tumors and during fetal development, with much lower levels found in most adult tissues. Recent studies reveal that the expression of human survivin is cell-cycle regulated and that its antiapoptotic function is mediated both by its BIR domain and by the interaction of its C-terminal coiled-coil domain with microtubules of the mitotic spindle.14,19,20 The cDNA encoding the murine counterpart of human survivin has recently been reported.21
To elucidate the role(s) of survivin in physiologic and pathologic conditions, before and after development, we characterized the murinesurvivin gene and cDNA. We report that there exist 3 distinct survivin cDNAs that are expressed differentially during development and in adults, and that encode functionally distinct proteins. In addition, we show that 3 distinct forms of survivin are also found in human tissues and that these human variants are expressed differentially as well.
Materials and methods
Reagents
DNA restriction enzymes were obtained from Boehringer Mannheim Canada (Dorval, Quebec) or New England Biolabs (Mississauga, Ontario), and radioisotopes were from ICN Biomedicals (Mississauga, Ontario). Murine 14.5 days postcoitum (dpc) embryo λ phage cDNA and 129SV genomic DNA libraries were purchased from Clontech (Palo Alto, CA) and from Stratagene Cloning Systems (San Diego, CA), respectively. The murine PAC library was provided by Genome Systems (South Bend, IN). Recombinant human caspase-3 (catalogue no. 235 417) and the caspase-3 substrate colorimetric kit (catalogue no. 235 400) were purchased from EuroBiochem (Bierges, Belgium). The caspase-3 has a specific activity of approximately 1 U/mg protein, defined by the amount of enzyme that cleaves 1.0 μmol of the substrate Ac-DEVD-pNA per minute at 25°C. A panel of cDNAs derived from normal human tissues was obtained from Clontech—Multiple Tissue cDNA Panel 1, catalogue no. K1420-1.
Identification of cDNA and genomic DNA for murine survivin
Based on the published human EPR-1 cDNA sequence, a murine expressed sequence tag (EST mc61a11.r1) was identified by BLAST search. Identification of the cDNA encoding humansurvivin12,13 revealed that this EST actually corresponded to the murine homologue of survivin. Indeed, the predicted amino acid sequence corresponding to this EST exhibited 86% identity with the reported human survivin sequence, between amino acid residues 1 and 128. EST mc61a11.r1 was subsequently purchased from Genome Systems, the DNA sequence was confirmed, and the insert was radiolabeled with α-32P-dCTP using the random primer synthesis method22 for use as a probe to screen both the murine embryo cDNA and genomic DNA libraries.
The λ phage cDNA library was prepared for plaque hybridization. Filters were hybridized in Quik-Hyb (Stratagene) for 1 hour at 65°C with the α-32P-dCTP-labeled EST probe and finally washed at 55°C with 0.5 × standard sodium citrate (SSC), 0.1% sodium dodecyl sulfate (SDS). After 4 rounds of plaque purification, the cDNA inserts were isolated and subcloned into pBS KS(+) (Stratagene). The genomic DNA library was screened using the same α-32P-dCTP-labeled probe, and a single clone, SRV10′, was isolated, containing the entire coding region of murine survivin. The ∼13 kb insert was purified from the λ phage and inserted into the Not1 site of pBS KS(+). A murine PAC library (Genome Systems) was also screened using the entire 13 kb SRV10′ insert as a probe. Three independent PAC clones were identified and were shown by Southern blotting with gene-specific radiolabeled oligonucleotide probes to contain the entire SRV10′ sequence.
Fluorescent in situ hybridization
Fluorescent in situ hybridization (FISH) analysis was performed following established procedures, by seeDNA Biotech Inc. (Toronto, Ontario) using the murine PAC survivin genomic clone as a probe.23,24 Lymphocytes were isolated from mouse spleen and cultured at 37°C in RPMI 1640 medium supplemented with 15% fetal calf serum, 3 μg/mL concanavalin A, 10 μg/mL lipopolysaccharide and 5 × 10−5 mol/L mercaptoethanol. After 44 hours, the cultured lymphocytes were treated with 0.18 mg/mL BrdU for an additional 14 hours. The synchronized cells were washed and recultured for 4 hours in α-minimum essential medium (MEM) with thymidine 2.5 μg/mL. Chromosome slides were made as reported.23 24 For FISH detection, the slides were heated at 55°C for 1 hour, treated with RNAse A, and denatured in 70% formamide in 2 × SSC for 2 minutes at 70°C, followed by dehydration with ethanol. The biotinylated probe was denatured at 75°C for 5 minutes in 50% formamide and 10% dextran sulfate, and hybridized overnight on the denatured slides. FISH signals and the DAPI banding patterns were recorded separately and photographed. The FISH signals were subsequently mapped to specific chromosomal bands by the superimposition of FISH and DAPI images.
RNA isolation, Northern analysis, and reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was isolated from confluent cell monolayers or from tissues by the method of Chomczynski and Sacchi.25 For adult tissue samples, adult mice (10-12 weeks) were killed and tissues were dissected and placed into liquid nitrogen. For embryonic samples, C57Bl/6 mice were mated, conception was assessed by the presence of a coital plug (the morning of coital plug being scored as 0.5 days postcoitum [dpc]), pregnant females were killed at various developmental time points, and embryos were carefully removed by dissection. Total RNA was extracted from adult and embryonic samples by homogenization of the tissues, and cDNA was synthesized by reverse transcription using murine leukemia virus (M-MLV) reverse transcriptase and a cDNA synthesis kit (Bethesda Research Laboratories, Burlington, Ontario). First-strand synthesis was primed using random hexanucleotides, and PCR was performed as reported,26 with the annealing temperature adjusted to 55°C. Pairs of oligonucleotide primers for PCR were synthesized such that they flanked the alternatively spliced mRNA forms. Primer srv86 (sense 5′-TCGCCACCTTCAAGAACTGGCCCTTCCTGGA), when paired with primer srvas311 (antisense 5′-GTTTCAAGAATTCACTGACGGTTAGTTCTT) was expected to generate PCR amplicons of 225 bp or 144 bp, depending on the mRNA splice form. Primer srv86, when paired with srvas6380 (antisense 5′-GGCTTCTGACAATGCTTG), was expected to generate a PCR amplicon of 332 bp. Resultant RT-PCR products were subsequently examined by agarose gel electrophoresis and by Southern blotting, using a radiolabeled oligonucleotide probe, srv295 (sense 5′-TGGCTGCGCCTTCCTCACTGT) corresponding to the sequence found within both amplicons. The amount of cDNA synthesized from each sample was calibrated according to the relative expression of hypoxanthine phosphoribosyl transferase (HPRT), as determined by agarose gel electrophoresis of RT-PCR products generated using the specific oligonucleotide primers, HPRT sense (5′–CACGGACTAGAACACCTGC) and HPRT antisense, (5′–GCTGGTGAAAAGGACCTCT), yielding a 221 bp product.27
Nested RT-PCR of RNA derived from human cell lines or tissue was used to examine expression of human survivin transcripts.11 For the same purpose, cDNAs derived from the mRNA of several normal human tissues were purchased from Clontech . To determine whether a cDNA lacking exon 2 was present, primers in exons 1 and 3 were paired. Thus, primer hsrv2841 (sense 5′-CAGCCCTTTCTCAAGGAC) was first paired with hsrv5175 (antisense 5′-GGAAAGCGCAACCGGACGAAT), and 2% of the PCR product was subjected to a further 30 cycles of PCR using the primer pair hsrv2861 (sense 5′-CCGCATCTCTACATTCAAGAAC) and hsrv5159 (antisense 5′-CGAATGCTTTTTATGTTCCTC), yielding an expected product of either 193 bp or 83 bp, the latter expected if exon 2 is excised by alternative mRNA splicing. Nested RT-PCR was also used to determine whether the homologue of murine survivin121 is present in human-derived tissue or cells. Primer hsrv2841 was paired first with primer hsrv5451 (antisense 5′-GAGGCAGGAGAATCACTTG), and 2% of the resultant amplicon was reamplified using primers hsrv2861 and hsrv5431 (antisense 5′-GAGAGGCAGAGGTTGCAGTG), yielding products of either 480 bp (retention of intron 3) or 370 bp (excision of exon 2). Resultant RT-PCR products were examined by agarose gel electrophoresis and by Southern blotting using a radiolabeled oligonucleotide probe, hsrv2888 (sense 5′-CTTCTTGGAGGGCTGCGCCTGCAC) corresponding to sequence in exon 1, which is found within both amplicons. For further verification, DNA sequencing of gel-purified amplicons was performed.
Southern hybridization
The RT-PCR products were separated by electrophoresis on a 2% agarose gel. Oligonucleotide probes were end-labeled with γ32P-ATP. Following Southern transfer, Genescreen nylon filters (NEN Life Science Products, Brussels, Belgium) were hybridized with radiolabeled probe in Quik-Hyb for 1 hour, washed with 0.1% SDS, 0.5 × SSC at 55°C, and exposed to Kodak XAR-5 film at −70°C.
Expression of recombinant murine survivin
cDNAs corresponding to the 3 forms of murine survivin were subcloned in-frame into the expression vector pSecTagA (Invitrogen), to produce survivin fusion proteins bearing an N-terminal Ig κ-chain leader sequence and a C-terminal myc epitope/polyhistidine tag. The resultant plasmid vectors—surv140/psec, surv121/psec, and surv40/psec—were then expressed stably into COS-7 cells using zeocin selection, conditioned media were collected, and the myc/His6-tagged proteins were purified by nickel-column chromatography. Recombinant proteins were eluted with 250 mM imidazole, 0.3 mol/L NaCl in phosphate-buffered saline (PBS), and the presence of the expected forms of survivin was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblotting using murine monoclonal anti-myc antibodies, followed by detection with horseradish peroxidase (HRP)-conjugated goat–anti-mouse antibodies and visualization with the ECL Western blotting system (Pharmacia-Amersham, Rosendaal, Netherlands). Protein concentrations were determined using bovine serum albumin as a standard.
Lysates of transiently transfected cells were also evaluated for the expression of recombinant survivin variants. PBS-washed cell pellets were suspended in a buffer containing 25 mM Hepes pH 7.5, 5 mM MgCl2, 5 mM EDTA, 5 mM dithiotreitol, exposed to 3 freeze-thaw cycles, and centrifuged at 16 000g for 30 minutes. The lysates were then divided into aliquots and frozen at −20°C for subsequent analysis.
In vitro analysis of apoptosis
The effect of recombinant murine survivins on caspase-3 activity was evaluated by using a specific colorimetric assay kit (EuroBiochem; catalogue no. 235 417) according to the manufacturer's instructions. Briefly, purified recombinant myc/His-tagged survivin (20-50 nmoles/L) was added to 30 ng (3 × 10−5 units) of human recombinant caspase-3 (EuroBiochem) in assay buffer (0.1 mol/L NaCl, 50 mM Hepes, 10 mM DTT, 1 mM EDTA, 10% glycerol, 0.1% CHAPS [3-([3-Cholamidopropyl] dimethylammoniol)-1-propane-sulfonate], pH 7.4) in a final volume of 120 μL. The reaction was started by addition of 10 μL of the colorimetric caspase-3 substrate I and the rate of change in absorbance at 405 nm was measured using a kinetic enzyme-linked immunosorbent assay plate reader. Controls without either caspase-3 or substrate resulted in no change in absorbance during 30 minutes of observation. Experiments were performed in triplicate, repeated 3 times, and the means and standard errors were calculated.
Antibody production
Polyclonal antibodies against murine survivin were generated by Eurogentec (Seraing, Belgium) using a standard immunization protocol whereby rabbits were immunized with KLH-conjugated peptides NH2-ALPQIWQLYLKNYRI-COOH and NH2-CFKELEGWEPDDNPI-CONH2, representing amino acid residues 5 to 19 and 60 to 74, respectively, of the deduced murine amino acid sequence. Immunoglobulin was purified from immune sera by protein-A sepharose chromatography, and specific antisurvivin antibodies (Ig-srv .336) were subsequently obtained by affinity chromatography of the immunoglobulin fractions on EAH-sepharose 4B columns (Pharmacia-Amersham Biotech) to which the survivin peptides had been coupled.
Biotinylation of proteins, immunoprecipitation, and Western blotting
Embryonic samples, 12.5 dpc, were pooled and homogenized in PBS with 5 mM EDTA, 5 mM MgCl2. The clear cell lysate, obtained following centrifugation at 14 000g at 4°C for 30 minutes, was incubated with biotin (EZ-Link Sulfo-NHS-LC-LC-Biotin, Pierce, Polylab N.V., Antwerp) for 60 minutes at 4°C, after which the reaction was stopped by the addition of 0.1 mol/L Tris-HCl. Biotinlyated proteins were separated from free biotin by size fractionation on a PD-10 column (Pharmacia) with PBS as the running buffer. The resultant biotinylated protein solution was incubated in PBS with 0.5 mol/L NaCl with either 20 μg of Ig-srv336 or preimmune immunoglobulin for 2 hours, after which protein A-sepharose was added for a further 1 hour. The immunoprecipitated pellets were washed 3 times with PBS/0.5 mol/L NaCl and twice with PBS. Samples were boiled in nonreducing Laemmli buffer and separated on an 18% SDS-polyacrylamide gel. After transfer, the nitrocellulose filters were blocked for 2 hours in Blotto, incubated with HRP-conjugated streptavidin for 1 hour, and then washed. The Amersham ECL Western-blotting system, followed by exposure of the blots to Hyperfilm-ECL, was used to detect specific bands. Prestained standard markers were used to determine the approximate molecular weight.
Analysis of data
Alignments of DNA and amino acid sequences were performed with the aid of MacVector software (Oxford Molecular Ltd, Oxford, UK).
Results
Identification of the murine survivin gene and cDNAs
A murine survivin cDNA probe was identified by searching the dbest database for a murine EST—clone mc61a11.r1—that matched closely the cDNA encoding human EPR-1.11 By FASTA analysis,28 EST mc61a11.r1 exhibited 85% and 79% sequence similarity to the corresponding portions of human survivin andEPR-1, respectively. Using this EST as a probe, 1 genomic and 9 cDNA clones (ranging in size from 800 bp to 2.1 kb) were identified by λ phage library screening. Each of the cDNA clones was sequenced in its entirety on both strands, while approximately 11 kb of the ∼13 kb genomic clone was also sequenced. A murine genomic PAC DNA library was subsequently screened with the purified survivin genomic DNA probe, resulting in the identification of 3 independent, overlapping PAC clones. A biotin-labeled PAC DNA clone containing the entiresurvivin coding region (see below) was then used for FISH to localize the survivin gene to chromosome 11E2 (Figure1). Restriction mapping and partial sequencing of DNA fragments from the PAC clones confirmed that the λ phage genomic clone and the PAC clones were overlapping and represented a single gene (data not shown).
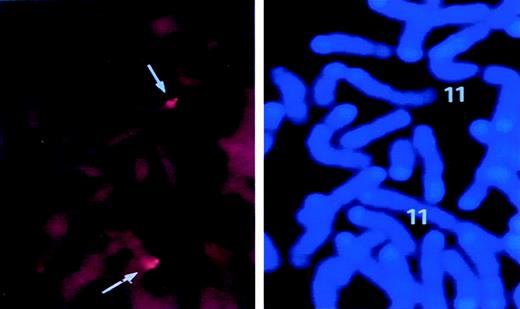
FISH analysis for chromosomal localization of murinesurvivin locus.
Lymphocytes were isolated from mouse spleen and cultured as detailed in “Materials and Methods.” A biotin-labeled PAC DNA clone containing the entire survivin gene was hybridized with metaphase lymphocyte chromosomes. FISH signals and the DAPI banding pattern were recorded separately, and the assignment of FISH mapping data with chromosomal bands was achieved by superimposing FISH signals with DAPI-banded chromosomes. In the left panel, dots represent the specific FISH signals detected on chromosome 11. The right panel shows the same mitotic figure, stained with DAPI to identify mouse chromosome 11.
FISH analysis for chromosomal localization of murinesurvivin locus.
Lymphocytes were isolated from mouse spleen and cultured as detailed in “Materials and Methods.” A biotin-labeled PAC DNA clone containing the entire survivin gene was hybridized with metaphase lymphocyte chromosomes. FISH signals and the DAPI banding pattern were recorded separately, and the assignment of FISH mapping data with chromosomal bands was achieved by superimposing FISH signals with DAPI-banded chromosomes. In the left panel, dots represent the specific FISH signals detected on chromosome 11. The right panel shows the same mitotic figure, stained with DAPI to identify mouse chromosome 11.
Four murine survivin cDNAs (clones 5.25, 92.16, 41.1, and 42.5) corresponded very closely to their full-length human counterparts. The largest of these, clone 5.25, comprised 48 bp of 5′ untranslated sequence (UTR) upstream of an ATG translation initiation codon, a 420 bp open reading frame (ORF), and 872 bp of 3′ UTR following an in-frame TAA stop codon, and predicted a 140 amino acid protein—survivin140— of molecular weight 16 286 Da, with 84% sequence similarity to human survivin (Figure2). Comparison of the DNA sequence of this full-length cDNA clone with that of the murine survivin λ phage and PAC genomic clones revealed that the structural organization of the murine survivin locus was very similar to that of its human counterpart (Figures 3 and4). The murine survivin gene comprises 4 exons. Exon 1, containing the ATG translational start site (position 2865), is preceded by a putative TATA-less promoter and a GC-rich region extending approximately 250 nucleotides in the 5′ direction. Exons 1 to 4 (111, 110, 118, and 81 bp, respectively) are separated by 3 introns 292, 2797, and 2448 nucleotides in length. Exon 4 contains an in-frame TAA stop codon at nucleotide position 8822.
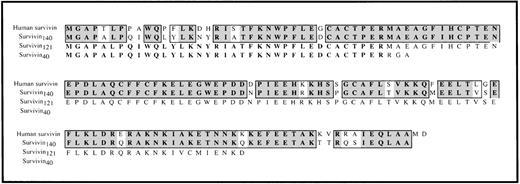
Comparison of human survivin and murine survivin.
The predicted amino acid sequences of murine survivin140(derived from cDNA clones 5.25, 92.16, 41.1, and 42.5), survivin121 (derived from cDNA clones 7HB and 7L .15), and survivin40 (derived from cDNA clones 5.20, 91.1, and 5.24) are aligned with the amino acid sequence of human survivin.
Comparison of human survivin and murine survivin.
The predicted amino acid sequences of murine survivin140(derived from cDNA clones 5.25, 92.16, 41.1, and 42.5), survivin121 (derived from cDNA clones 7HB and 7L .15), and survivin40 (derived from cDNA clones 5.20, 91.1, and 5.24) are aligned with the amino acid sequence of human survivin.
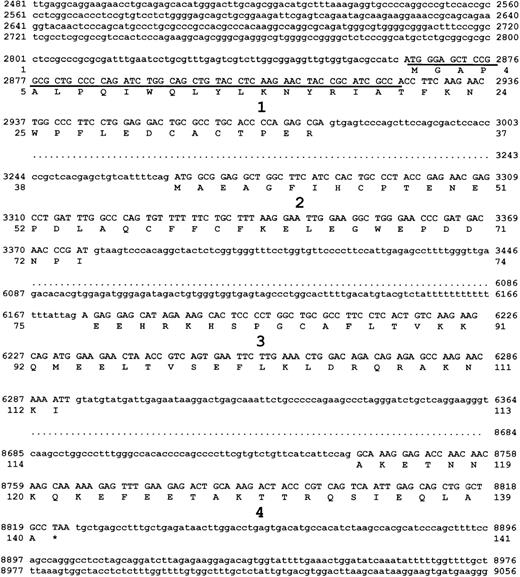
Sequence of the murine survivin gene.
The λ phage genomic DNA clone containing all 4 exons of murinesurvivin was sequenced on both strands. The 4 exons, with flanking intron sequence, are shown. Each exon is numbered, and the DNA encoding exons 1 to 4 are in uppercase letters. The predicted amino acid sequence of survivin140, derived from exons 1 to 4 is also shown. Survivin121 is predicted to be derived from retention of intron 3, whereas survivin40 is derived from pre-mRNA splice removal of exon 2. Underlined DNA sequence represents the only region of murine survivin cDNA without homology to putative cDNA of human EPR-1.
Sequence of the murine survivin gene.
The λ phage genomic DNA clone containing all 4 exons of murinesurvivin was sequenced on both strands. The 4 exons, with flanking intron sequence, are shown. Each exon is numbered, and the DNA encoding exons 1 to 4 are in uppercase letters. The predicted amino acid sequence of survivin140, derived from exons 1 to 4 is also shown. Survivin121 is predicted to be derived from retention of intron 3, whereas survivin40 is derived from pre-mRNA splice removal of exon 2. Underlined DNA sequence represents the only region of murine survivin cDNA without homology to putative cDNA of human EPR-1.
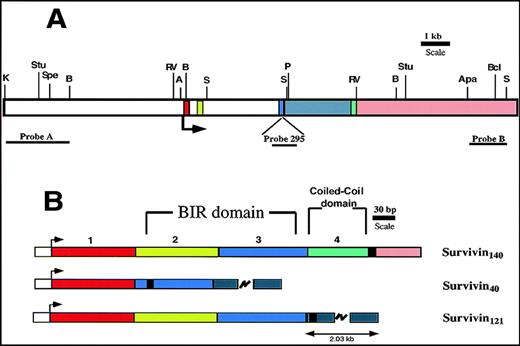
Murine survivin gene and cDNAs.
Exons 1 to 4 are denoted in red, light green, blue, and green, respectively. Intron 3 is gray, and the 3′ flanking region is pink. (A) Restriction map of murine survivin gene. The arrow indicates the ATG translational start site. Restriction enzyme sites are shown: K (Kpn1); Stu (Stu1); Spe (Spe1); B (BglII); RV (EcoRV); A (Asc1); S (Sac1); P (Pml1); Apa (Apa1); Bc (Bcl1). Location of DNA probes used for Southern blots are shown. (B) Exon organization of the 3 distinct cDNAs encoding the predicted survivin proteins. The arrow indicates the start of translation. The heavy black vertical line represents the end of the coding region. Survivin140 is derived from all 4 exons. Survivin40 is generated by removal of exon 2-derived sequences, with a resultant in-frame stop codon, and a truncated protein. Survivin121 is derived from exons 1 to 3, and a short part of intron 3, the latter being presumably retained during pre-mRNA processing.
Murine survivin gene and cDNAs.
Exons 1 to 4 are denoted in red, light green, blue, and green, respectively. Intron 3 is gray, and the 3′ flanking region is pink. (A) Restriction map of murine survivin gene. The arrow indicates the ATG translational start site. Restriction enzyme sites are shown: K (Kpn1); Stu (Stu1); Spe (Spe1); B (BglII); RV (EcoRV); A (Asc1); S (Sac1); P (Pml1); Apa (Apa1); Bc (Bcl1). Location of DNA probes used for Southern blots are shown. (B) Exon organization of the 3 distinct cDNAs encoding the predicted survivin proteins. The arrow indicates the start of translation. The heavy black vertical line represents the end of the coding region. Survivin140 is derived from all 4 exons. Survivin40 is generated by removal of exon 2-derived sequences, with a resultant in-frame stop codon, and a truncated protein. Survivin121 is derived from exons 1 to 3, and a short part of intron 3, the latter being presumably retained during pre-mRNA processing.
Although 9 independent survivin cDNA clones were isolated, only 4 corresponded to a full-length form comprising exons 1 to 4. The remaining 5 clones corresponded to 2 variant cDNAs that appeared to have arisen by alternative pre-mRNA splicing (see Figures 2 and 4). Clones 5.20, 91.1, and 5.24 corresponded to a shorter transcript that lacked exon 2-derived sequences, and therefore contained an ORF of only 120 nucleotides followed by a 3′ UTR, predicting a truncated 40 amino acid residue protein—survivin40—with a molecular weight of 4667 Da (see Figure 4). In contrast, clones 7HB and 7L .15 contained exons 1 to 3 and retained intron 3 as well. Acquisition of a new in-frame stop codon within intron 3 results in an ORF of 363 nt, predicting a 121 amino acid protein (survivin121) with a molecular weight of 14 172 Da (see Figure 4).
Alignment of survivin protein sequences with those of other BIR-containing proteins
The amino acid sequences of the 3 variant murine survivin proteins were aligned (see Figure 2) and compared with those of other BIR-containing IAP protein members (not shown). The amino-terminal 113 amino acid residues of survivin121 are identical to those of survivin140. The remaining carboxy-terminal portions of these 2 variants are distinct. Amino acids 15 to 87 comprise the cysteine/histidine-containing zinc-binding BIR domain of survivin140 (and survivin121). This region exhibits 30% to 40% sequence identity with each of the BIR-domains of murine XIAP, NAIP, and BRUCE. In common with human survivin, NAIP, and BRUCE, murine survivin does not contain a carboxy-terminal RING finger. Survivin40 shares sequence with only the amino-terminal 30% of the BIR domains of survivin140 and survivin121 and does not contain a region rich in cysteine or histidine residues, suggesting that it likely does not have direct antiapoptotic function.
Tissue distribution and developmental expression of murine survivin mRNAs
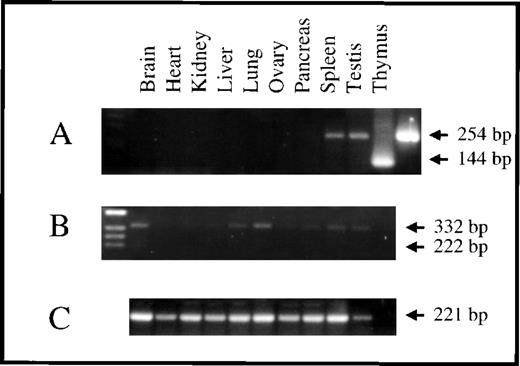
Using mRNA variant-specific RT-PCR, we determined the relative distribution of the 3 survivin transcripts in a variety of murine tissues, including brain, heart, kidney, liver, lung, ovary, pancreas, spleen, testis, and thymus. As illustrated in Figure5, survivin140-specific mRNA was detectable only in thymus and testis. In contrast, survivin121-specific transcripts were detectable in all tissues, although the signal was strongest in brain and ovary.Survivin40 mRNA, on the other hand, was not detectable in any of these tissues.
Tissue distribution of murine survivin mRNAs.
RT-PCR was performed on mRNA derived from adult tissues using primers that flanked the specific survivin mRNAs. (A) Accumulation of mRNA for survivin140 was detected in thymus and testis (primer pair srv86 and srvas311 results in amplicon of 254 bp). Positive controls using cDNA clones 5.20 and 5.25 (see Figure 2) as target DNA, are in 2 lanes at the right side, demonstrating prominent 144 bp and 254 bp bands, respectively. (B) Survivin121 mRNA was detected in all tissues (primer pair srv86 and srvas6380 results in amplicon of 332 bp). Survivin40 mRNA using primer pairs srv86 and srvas311, or srv86 and srvas6380 (expecting amplicons of 144 bp or 222 bp, respectively) was not detectable. (C) HPRT transcripts were present in all tissues (221 bp). The right lane in panels B and C are negative controls without target DNA.
Tissue distribution of murine survivin mRNAs.
RT-PCR was performed on mRNA derived from adult tissues using primers that flanked the specific survivin mRNAs. (A) Accumulation of mRNA for survivin140 was detected in thymus and testis (primer pair srv86 and srvas311 results in amplicon of 254 bp). Positive controls using cDNA clones 5.20 and 5.25 (see Figure 2) as target DNA, are in 2 lanes at the right side, demonstrating prominent 144 bp and 254 bp bands, respectively. (B) Survivin121 mRNA was detected in all tissues (primer pair srv86 and srvas6380 results in amplicon of 332 bp). Survivin40 mRNA using primer pairs srv86 and srvas311, or srv86 and srvas6380 (expecting amplicons of 144 bp or 222 bp, respectively) was not detectable. (C) HPRT transcripts were present in all tissues (221 bp). The right lane in panels B and C are negative controls without target DNA.
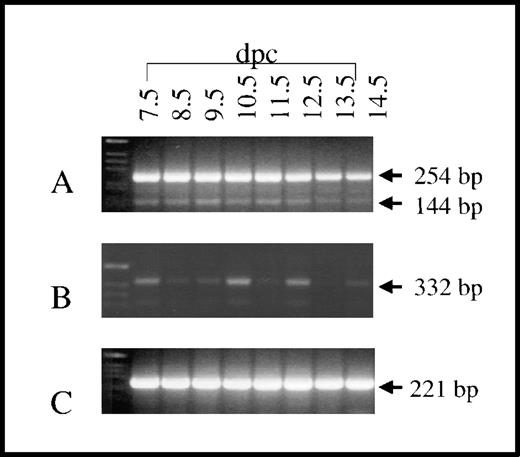
Previous studies have demonstrated that survivin is expressed in mouse embryos as early as 11.5 dpc.19 We examined the developmental expression of the 3 survivin mRNA variants by RT-PCR (Figure 6). In contrast to what we had observed in adult tissue, all 3 survivin mRNA species, including those corresponding to survivin40, were readily detectable throughout development from 7.5 dpc, the earliest time point examined, until 14.5 dpc. In all cases, the identity of the amplicons generated by RT-PCR was confirmed by Southern blotting using the radiolabeled internal oligonucleotide srv295. In addition, representative PCR products of each size were purified and subcloned, and their identity was verified by DNA sequencing.
Developmental expression of murine survivin mRNA.
RT-PCR was performed on mRNA derived from murine embryos at different developmental time points (7.5-14.5 dpc) using oligonucleotide primers as noted in Figure 5. (A and B) All 3 survivin mRNA species were detected, although survivin140 mRNA (254 bp) was the most prominent. (C) HPRT transcripts were present at all developmental time points (221 bp). These results are representative of experiments on 3 embryos at each time point.
Developmental expression of murine survivin mRNA.
RT-PCR was performed on mRNA derived from murine embryos at different developmental time points (7.5-14.5 dpc) using oligonucleotide primers as noted in Figure 5. (A and B) All 3 survivin mRNA species were detected, although survivin140 mRNA (254 bp) was the most prominent. (C) HPRT transcripts were present at all developmental time points (221 bp). These results are representative of experiments on 3 embryos at each time point.
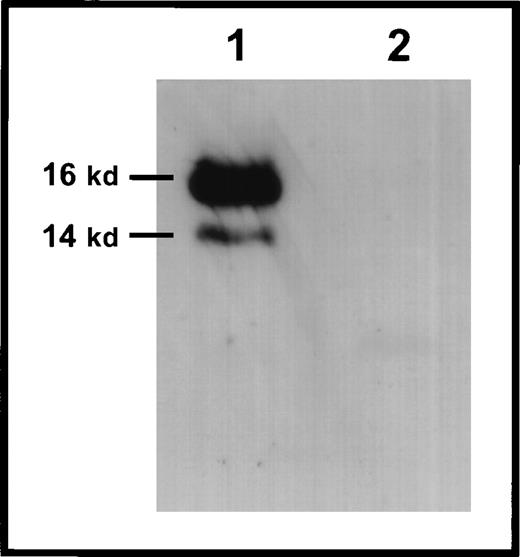
To confirm directly whether the different survivin transcripts detected in mouse embryos resulted in translation of distinct protein products, we isolated 12.5 dpc murine embryos and biotinylated the lysates for immunoprecipitation with an antisurvivin immunoglobulin that specifically identifies all 3 recombinant forms. We could detect bands representing survivin140 and survivin121, confirming that in embryos that are 12.5 dpc, at least 2 of the transcripts are translated into survivin proteins of the predicted sizes (Figure 7). The immunoprecipitation did not yield a band with a predicted molecular weight consistent with survivin40.
Immunoprecipitation of murine survivin from embryos.
Lysates of 12.5 dpc embyros were biotinylated and immunoprecipitated with anti-survivin antibodies (lane 1) or nonspecific antibodies (lane 2). Following separation by SDS-PAGE and transfer to nitrocellulose membranes, filters were incubated with streptavidin-HRP for detection with the ECL kit (Amersham), as detailed in “Materials and Methods.” Molecular weight markers are on the left. Bands representing survivin140 and survivin121 are visualized at approximately 16 kd and 14 kd, respectively.
Immunoprecipitation of murine survivin from embryos.
Lysates of 12.5 dpc embyros were biotinylated and immunoprecipitated with anti-survivin antibodies (lane 1) or nonspecific antibodies (lane 2). Following separation by SDS-PAGE and transfer to nitrocellulose membranes, filters were incubated with streptavidin-HRP for detection with the ECL kit (Amersham), as detailed in “Materials and Methods.” Molecular weight markers are on the left. Bands representing survivin140 and survivin121 are visualized at approximately 16 kd and 14 kd, respectively.
Interference of caspase-3 activity by recombinant murine survivin forms
To test directly the function of the variant murine survivin proteins, recombinant myc/His6-tagged survivin fusion proteins were created. The cDNAs encoding the survivin forms were subcloned into the expression vector pSecTagA, and the resultant vectors surv140/psec, surv121/psec, and surv40/psec cDNAs were each transfected stably into COS-7 cells. Western immunoblotting of cell lysates with anti-myc or antisurvivin antibodies (Ig-srv336) revealed single bands of appropriate molecular size for each of the survivin forms. Following purification of recombinant myc/His-tagged survivin variant forms from conditioned media, varying concentrations were tested in caspase-3 activity assays. In a representative set of experiments summarized in Table 1, both survivin140 and survivin121 suppressed exogenous caspase-3 activity with similar dose-response curves. At submaximal concentrations, survivin140 and survivin121 had an additive effect with respect to inhibiting caspase-3 activity. As predicted, survivin40 had no effect on caspase-3 activity, and furthermore, we did not observe any effect of survivin40 on the caspase-3 inhibiting function of survivin140 or survivin121.
Effect of survivin on caspase-3 activity
| . | Survivin140 (nmole/L) . | Survivin121 (nmole/L) . | Survivin40 (nmole/L) . | Caspase-3 Activity (ΔOD405/10 min) (n = 3) . |
|---|---|---|---|---|
| Caspase-3 (30 ng) | 0 | 0 | 0 | 0.023 ± 0.002 |
| 20 | 0 | 0 | 0.014 ± 0.003* | |
| 50 | 0 | 0 | 0* | |
| 0 | 20 | 0 | 0.016 ± 0.004* | |
| 0 | 50 | 0 | 0* | |
| 20 | 20 | 0 | 0* | |
| 0 | 0 | 50 | 0.021 ± 0.003 | |
| 20 | 20 | 50 | 0* | |
| Caspase-3 (0 ng) | 50 | 0 | 0 | 0 |
| . | Survivin140 (nmole/L) . | Survivin121 (nmole/L) . | Survivin40 (nmole/L) . | Caspase-3 Activity (ΔOD405/10 min) (n = 3) . |
|---|---|---|---|---|
| Caspase-3 (30 ng) | 0 | 0 | 0 | 0.023 ± 0.002 |
| 20 | 0 | 0 | 0.014 ± 0.003* | |
| 50 | 0 | 0 | 0* | |
| 0 | 20 | 0 | 0.016 ± 0.004* | |
| 0 | 50 | 0 | 0* | |
| 20 | 20 | 0 | 0* | |
| 0 | 0 | 50 | 0.021 ± 0.003 | |
| 20 | 20 | 50 | 0* | |
| Caspase-3 (0 ng) | 50 | 0 | 0 | 0 |
Purified recombinant myc/His6-tagged survivins were assessed for their effect on caspase-3 activity using a chromogenic assay as detailed in “Materials and Methods.” Varying concentrations of survivins were tested. Results show a representative experiment done in triplicate, with associated standard errors. Survivin140 and survivin121 similarly suppressed caspase-3 activity in a dose-dependent manner, whereas survivin40 had no effect. The asterisk denotes a significant difference in activity as compared with caspase-3 alone (first row) with P < .01.
Expression of survivin mRNA variants in human cells and tissues
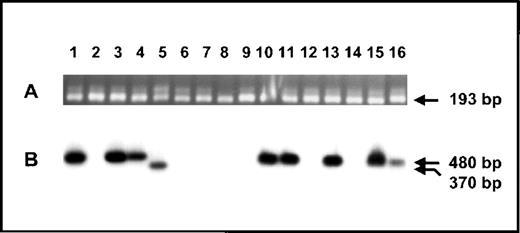
Using the published genomic DNA sequence of humansurvivin,11 we designed a series of oligonucleotides that were expected to identify similar humansurvivin cDNA variants by RT-PCR. Oligonucleotide primers hsrv2841, hsrv2861, and hsrv2888 lie within exon 1, hsrv5159 and hsrv5175 are within exon 3, whereas hsrv5431 and hsrv5451 lie within intron 3. As noted in Figure 8A, mRNA transcripts corresponding to full-length survivin were detected in all cell lines surveyed, including those derived from patients with leukemia, colon cancer, neuroblastoma, melanoma, hepatoma, neurofibrosarcoma, and carcinoma. Transcripts were also detected in mRNA from 4 patients with mucosa-associated lymphoid tissue-type (MALT) lymphomas, as well as in normal human tonsil, lymphoid tissue, kidney, pancreas, liver, brain, and lung (not shown). The identity of the transcripts was confirmed by Southern blotting with an internal oligonucleotide probe specific to survivin or EPR-1. As demonstrated in Figure 8B, we readily detected survivin transcripts corresponding to murine survivin121 in RNA from cell lines derived from 2 human leukemias, a germ cell tumor, a teratocarcinoma, melanoma, hepatoma, and carcinoma, but not in a colon cancer, neurofibrosarcoma, or in normal lymphoid tissue, tonsil, kidney, pancreas, brain, or lung. Weak signals corresponding to this transcript were also found in all 4 MALT lymphoma samples (not appreciated in Figure 8). Transcripts corresponding to the human counterpart of survivin40were detected in RNA derived from a neuroblastoma cell line and from 1 of the MALT lymphomas.
Expression of human survivin mRNA forms.
Prior to separation by agarose gel electrophoresis, nested RT-PCR was performed on mRNA derived from the following human cell lines and tissues: 1. K562 leukemia cell line; 2. Jurkat T-cell line; 3. germ-cell tumor derived cell line; 4. teratocarcinoma cell line; 5. neuroblastoma cell line; 6-9. malignant tumor from patients with MALT lymphoma; 10. AS19 melanoma cell line; 11. Del leukemia cell line; 12. SW620 colon cancer cell line; 13. Hep3B hepatoma cell line; 14. Ht1080 neurofibrosarcoma cell line; 15. A431 carcinoma cell line; 16. Raji leukemia cell line. (A) Ethidium bromide-stained agarose gel of nested RT-PCR products generated with oligonucleotide primers hsrv2861 and hsrv5431. Transcripts correspond to full-length murine survivin140. (B) Southern blot using32P-labeled hsrv2888 to detect specific nested RT-PCR products generated using oligonucleotide primers hsrv2861 and hsrv5159. Several cell lines express the specific transcript corresponding to murine survivin121, whereas only the neuroblastoma cell line expresses a transcript corresponding to murine survivin40.
Expression of human survivin mRNA forms.
Prior to separation by agarose gel electrophoresis, nested RT-PCR was performed on mRNA derived from the following human cell lines and tissues: 1. K562 leukemia cell line; 2. Jurkat T-cell line; 3. germ-cell tumor derived cell line; 4. teratocarcinoma cell line; 5. neuroblastoma cell line; 6-9. malignant tumor from patients with MALT lymphoma; 10. AS19 melanoma cell line; 11. Del leukemia cell line; 12. SW620 colon cancer cell line; 13. Hep3B hepatoma cell line; 14. Ht1080 neurofibrosarcoma cell line; 15. A431 carcinoma cell line; 16. Raji leukemia cell line. (A) Ethidium bromide-stained agarose gel of nested RT-PCR products generated with oligonucleotide primers hsrv2861 and hsrv5431. Transcripts correspond to full-length murine survivin140. (B) Southern blot using32P-labeled hsrv2888 to detect specific nested RT-PCR products generated using oligonucleotide primers hsrv2861 and hsrv5159. Several cell lines express the specific transcript corresponding to murine survivin121, whereas only the neuroblastoma cell line expresses a transcript corresponding to murine survivin40.
Discussion
The human survivin gene has been reported to be highly similar to the reverse complement of the EPR-1gene.11 Indeed, the human survivin gene was originally identified by screening a genomic library with a humanEPR-1 cDNA. Several lines of evidence indicate that the murine gene and cDNAs that we have isolated correspond to the murine homologue of survivin, rather than of EPR-1. First, the humanEPR-1 gene comprises 5 exons, with the mature protein being encoded by an ORF of 1014 nucleotides. In contrast, the human and murine survivin genes contain 4 exons, and the mature proteins are encoded by ORFs of 426 and 420 nucleotides, respectively. Second, both the sizes and nucleotide sequences of the murine survivinexons are similar to those of their human counterpart. Third, although the reverse complement of the most 3′ end of human EPR-1exhibits sequence similarity to survivin, and the corresponding exons—although transcribed in opposite directions—demonstrate considerable overlap, despite extensive DNA sequencing of our murinesurvivin genomic clones, regions homologous to exon 1 or the 5′ part of exon 2 of EPR-1 (those regions ofEPR-1 which are not similar to the reverse complement of human survivin) could not be found. Finally, attempts to transcribe the opposite strand of the murine survivin gene by computer analysis did not reveal additional potential intron-exon structures that could encode a protein similar to EPR-1. We are confident, therefore, that we have isolated the murine homologue ofsurvivin, rather than of EPR-1. Furthermore, in view of recent claims that survivin and EPR-1 may be members of a closely linked family of genes,14 we tried to determine whether the murine genome also contains a family ofsurvivin/EPR-1-related genes (data not shown). Southern blot analyses of murine ES cell genomic DNA and a murine survivinPAC genomic DNA clone did not suggest the presence of more than a single gene, nor did FISH analysis detect more than a single locus. Nonetheless, these are negative results and do not exclude definitively the possibility that a family of genes does exist.
In these studies, we report the isolation and characterization of 3 independent survivin mRNA species from a murine embryonic cDNA library. Examination of the corresponding cDNA and genomic DNA sequences indicated that each of these transcripts originates from the same gene. The cDNA encoding murine survivin140 is derived from all 4 exons. The corresponding deduced amino acid sequence predicts a protein with striking similarity to full-length survivin of human origin, containing a single amino-terminal BIR repeat of approximately 70 amino acids, and lacking a RING finger domain, yet containing a 40 residue coiled-coil carboxy-terminal domain. A second cDNA, derived from the same survivin gene, but lacking exon 2-derived sequence, predicts the existence of a truncated 40 amino acid protein—survivin40— that does not contain the BIR element believed to be critical for antiapoptotic function. Finally, a third cDNA, which retains intron 3, and thereby acquires a new in-frame stop codon, predicts the existence of an additional 121 amino acid protein—survivin121. Both survivin140 and survivin121 contain an intact BIR domain, and as expected, interfere with caspase-3 activity. In contrast to survivin140, survivin121 does not contain the tubulin-interacting carboxy-terminal coiled-coil region. Although we have not yet demonstrated that survivin140 and survivin121 differ in their ability to interact with tubulin-containing structures, we anticipate that in the absence of the coiled-coil domain, survivin121 activity will not be restricted spatially in a cell cycle-dependent manner.
Consistent with previous reports for humansurvivin,11 expression of murinesurvivin140, as detected by RT-PCR, is most prominent in proliferating adult tissues. In contrast, murinesurvivin121 mRNA transcripts appear to be more widely distributed, being found in most adult tissues surveyed, a finding in keeping with the observations of Kobayashi et al21 who also showed that murine survivin is present in several adult tissues and all T-cell and B-cell lines. We were unable to detect survivin40 mRNA transcripts in any of the adult murine tissues sampled. The failure of Ambrosini et al11 to detect humansurvivin121-specific RNA by Northern blot analysis likely indicates that the 3′ UTR survivin probe that they used lies beyond the region encompassed by this variant transcript. This notion is borne out by our studies in which each of the survivin mRNA forms is detectable by RT-PCR in a variety of human cells and tissues. Examination of the human survivingenomic DNA sequence11 predicts that the human counterparts for murine survivin121 and survivin40 would be 128 (human survivin128) and 40 (human survivin40) amino acid residues in length, respectively. Human survivin128, similar to its murine counterpart, would retain the BIR domain and lose its C-terminal coiled-coil structure. Human survivin40 would lack both a BIR domain and the coiled-coil structure. In several normal adult tissues, only transcripts corresponding to full-length human survivin were detected. In contrast, in addition to transcripts for full-length human survivin, most human tumors and tumor cell lines examined also expressed transcripts corresponding to survivin128, whereas a neuroblastoma cell line and 1 of the MALT lymphomas contained survivin40-specific transcripts as well. Based on our observation of an apparent change in the pattern of transcription of the variant survivin forms, it seems reasonable to assume that the differential expression of 1 or more of the functionally distinct survivin variants might affect malignant behavior of a tumor, and thereby alter prognosis. A recent report attempted to correlate the tumor-specific expression of survivin with prognosis in patients with neuroblastoma.20 Notably, this study detected survivin expression immunohistochemically using antibodies that would not be expected to distinguish among the 3 survivin forms. Our studies did not allow the roles of survivin variants in defining tumorigenicity to be assessed. We believe, however, that a more extensive evaluation, coupled with specific in situ localization of the survivin variants, will provide further insights into the role of the survivin variants in normal and abnormal cell growth and proliferation.
Based on the putative structure of human survivin, investigators hypothesized that its function would be similar to that of other members of the IAP family.11 The antiapoptotic function of IAP-class proteins is believed to be partially mediated via their BIR-domains. In Drosophila, several proteins, including the so-called death proteins, reaper and doom, are known to interact with IAP member proteins, thereby altering the binding of the BIR-containing protein to caspases, and diminishing their antiapoptotic function.10,29,30 Both human and murine full-length survivins have been shown to bind specifically to caspases 3 or 7 (or both) via the BIR-domain and to inhibit apoptosis in in vitro systems.21,31 Regulation of this interaction has been further elucidated by recent studies in which survivin expression in HeLa cells was shown to be cell-cycle dependent and largely restricted to the G2/M phase, a period during which the C-terminal coiled-coil structure of survivin binds to microtubules of the mitotic spindle, thereby enhancing interaction with target caspases.32 It has been hypothesized that the overexpression of survivin observed in most human cancer cell lines31 reflects, at least in part, escape from this apoptotic checkpoint during mitosis.32
These new insights into the mechanisms by which survivin may regulate the apoptotic pathway are particularly intriguing in light of our finding that there exist 3 variant forms of survivin, each containing distinct structural domains. Survivin140, similar to human survivin, contains both a BIR-domain and a coiled-coil carboxy-terminal region, and consequently would be expected to regulate apoptosis in a cell cycle-dependent fashion. In contrast, murine survivin121, and its human counterpart, human survivin128, lacks the coiled-coil structure, but contains a BIR-domain. We have further confirmed that both murine survivin140 and murine survivin121 can directly inhibit caspase-3 activity. Because survivin121 is likely unable to interact with microtubules, one would not expect it to regulate terminal caspases 3 and 7 in a localized manner that is similarly coupled to the cell cycle. Thus, murine survivin140 and survivin121, despite their similarity, may have quite different spectra of biologic activity. The complexity of this system is further underscored by the existence of survivin40, which lacks both a complete BIR domain and a coiled-coil region. Therefore, it is reasonable to speculate that the differential expression of these forms of survivin during the cell cycle might affect the balance between cell proliferation and programmed cell death.
We propose that the various forms of survivin differ in their ability to interact with other proteins involved in the regulation of apoptosis (or possibly to self-associate), and therefore, that the role of survivin in apoptosis regulation may vary in a manner that is defined by the spectrum of survivin proteins present, and consequently, by the combinatorial protein-protein interactions that are permitted. In a purified system, we were not able to detect any effect of survivin40 on the inhibition of caspase-3 activity by either survivin121 or survivin140. Despite these preliminary findings, it is still conceivable that through protein-protein interactions, survivin40, for example, might inhibit and thus regulate the antiapoptotic function attributed to survivin. It follows that alterations in expression of 1 or more of the survivin variants may be predicted to result in a variety of disease processes, either degenerative or proliferative. In that respect, mutations in the NAIP gene have been linked to motor neuron degeneration in one of the spinal muscular atrophies,33 while genetic deregulation of human IAP1 has been associated with the development of MALT lymphomas.34The wide tissue distribution and early developmental expression of the variant forms of survivin make it a likely candidate to be involved in several disease processes. Elucidation of the site-specific function(s) of these survivin variants may therefore provide further insights into the regulation of the apoptotic pathway in a variety of physiologic and pathologic conditions.
Supported in part by the Heart and Stroke Foundation of Ontario and the Medical Research Council of Canada.
Reprints:Edward M. Conway, Center for Transgene Technology and Gene Therapy KU Leuven Campus Gasthuisberg, O&N Herestraat 49, 9th floor, B-3000 Leuven, Belgium; e-mail: ed.conway@med.kuleuven.ac.be.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal