Toll-like receptors (TLRs) are a family of mammalian proteins homologous to Drosophila Toll. Human TLR2 was shown to mediate the responsiveness to lipopolysaccharide (LPS). On the other hand, gene mutations of mouse TLR4 (mTLR4) in LPS-hyporesponsive strains have suggested that mTLR4 is essential for LPS-signaling in mice, but the role of mTLR2 has not been explored. This report describes molecular cloning of the mTLR2 cDNA. Overexpression of mTLR2 and mouse CD14 conferred LPS-inducibility of c-Jun N-terminal kinase phosphorylation and nuclear factor-κB activation to COS7 cells, suggesting that mTLR2 is a signaling receptor for LPS. BothmTLR2 and mTLR4 genes were expressed in T cells. Treatment with anti-CD3ɛ, PMA plus ionomycin, or interleukin-2 (IL-2)/IL-15 increased mTLR2 but not mTLR4 messenger RNA (mRNA) in some T cell lines. Specific inhibitors of mitogen-activated extracellular signal-regulated kinase and fusion protein 38 (p38) kinase inhibited mTLR2 mRNA up-regulation by PMA plus ionomycin. This suggests that extracellular signal-regulated kinase and p38 kinase pathways were involved. Additionally, LPS treatment of EL-4 cell line decreasedIL-4 gene expression. Our results indicate that both mTLR2 and mTLR4 are involved in LPS signaling, but their expressions are regulated differently in T cells, and that LPS may directly affect T-cell functions by binding to TLRs.
Gram-negative bacteria represent a major group of pathogens causing serious infection, especially among the elderly and immunocompromised humans. A glycolipid known as endotoxin or lipopolysaccharide (LPS) is the principal bacterial constituent recognized by the innate immune system. LPS is a complex glycolipid composed of a hydrophilic polysaccharide portion and a hydrophobic domain known as lipid A.1 The conserved lipid A has been identified as responsible for LPS-induced biological effects.1 LPS stimulates host cells such as monocytes, macrophages, and B cells through the activation of transcription factors and protein kinases including NF-κB,2,3AP-1,3 extracellular signal-regulated kinases (ERKs),4,5 c-Jun N-terminal kinases (JNKs),6,7and fusion protein 38 (p38) kinases.8,9 Although there are some lines of evidence that a significant fraction of T cells respond to LPS,10 11 its direct effects on T cells have not been fully elucidated.
CD14 is involved in mediating LPS responses by binding LPS with high affinity.12 This binding requires a serum factor, LPS binding protein (LBP),13,14 which is a plasma lipid transfer protein that transfers LPS from the bacterial membrane to a binding site on CD14.15 In cells lacking CD14, the soluble form of CD14 (sCD14) in serum functionally replaces membrane-bound CD14.16 17 However, since CD14 is a glycosylphosphatidylinositol-anchored (GPI-anchored) protein, the existence of a signaling component was presumed in the LPS receptor complex.
Toll, first identified as a protein-controlling dorsoventrad pattern formation in the early development ofDrosophila,18 was shown to participate in antimicrobial immune responses.19 Toll has been shown to be conserved in various species, and it encodes a transmembrane protein with an intracellular portion that is homologous to that of the interleukin-1 (IL-1) receptor family proteins.18 Recently, several mammalian Toll homologues have been identified.20In addition to their cytoplasmic portion, they share repeating leucine-rich motifs (LRRs) in their extracellular region. One of the human Toll homologues, Toll-like receptor 2 (TLR2), has been shown to be involved in LPS signaling by 2 groups.21,22Cells transfected with the human TLR2 (hTLR2) complementary DNA (cDNA) acquired the capability for LPS-mediated signaling. There are 2 strains in mice (C3H/HeJ and C57BL10/ScCr) that exhibit an impaired ability to respond to LPS. Two recent studies found that the TLR4 gene in the chromosomal region was responsible for this defect (lps).23 24 These studies also found a missense mutation in the cytoplasmic domain of TLR4 in C3H/HeJ, which strongly suggests that TLR4 is the dominant receptor for at least some types of LPS. On the other hand, the role of mouse TLR2 (mTLR2) in LPS signaling has not been explored.
In this paper we report the molecular cloning of mTLR2 cDNA. Exogenous expression of mTLR2 in cells mediates LPS-induced intracellular signals, which suggests that mTLR2 is also involved in LPS responsiveness in mice. Although both mTLR2 and mTLR4genes are expressed in T cells, the gene expression of mTLR2but not that of mTLR4 is significantly increased by stimuli such as TCR stimulation and cytokine treatment. This suggests that theexpressions of LPS receptors, mTLR2 and mTLR4, are differently regulated in T cells.
Materials and methods
Reagents and antibodies
We used the following for the study: recombinant mouse IL-2 and human IL-15 (Peprotech, Seattle, WA); PD98 059, a specific inhibitor of mitogen-activated ERK kinase (MEK), and SB208 530, a specific inhibitor of p38 kinase (Calbiochem, San Diego, CA); LPS fromEscherichia coli serotype B6: 026 and PMA (Sigma, St. Louis, MO); RPMI 1640 medium (Gibco BRL, Rockville, MD); and fetal calf serum (FCS) (Sigma).
Antibodies included antimouse CD14 monoclonal antibody (mAb) (PharMingen, San Diego, CA); anti-Flag M2 mAb (Sigma Science); anti–phospho-JNK mAb (New England Biolabs, Beverly, MA); and anti-JNK1 polyclonal antibody (pAb) (Santa Cruz Biotechnology, Santa Cruz, CA), which recognizes JNK1, 2, and 3 isoforms.
Cell lines
All cell lines were grown in tissue culture flasks at 37°C in 5% carbon dioxide/95% air and passaged every 2 or 3 days to maintain logarithmic growth. Mouse T cell lines, EL-4 and S49.1 (American Type Culture Collection (ATCC), Rockville, MD), were maintained in RPMI 1640 medium with 10% FCS. A mouse T cell line, CTLL-2 (RIKEN Cell Bank, Tsukuba, Japan), was maintained in RPMI 1640 medium with 10% FCS and 10 ng/mL mouse IL-2. A Simian virus (SV)-40–transformed monkey kidney cell line, COS7 (ATCC), was maintained in Iscove's Modified Dulbecco's Medium (IMDM) supplemented with 10% FCS.
Enrichment of splenic T cells
T-cell enrichment was performed using nylon wool columns as previously described.25 Briefly, packed sterile nylon wool in a 10 mL syringe was equilibrated with complete medium. Then splenocytes suspended in complete medium were applied to the column. The cells were incubated for 1 hour at 37°C, and the nonadherent cells were obtained as a T-cell–enriched population by washing the column with 10 mL of complete medium. The obtained enriched cells that were more than 95% positive for CD3 by flow cytometry were used for the further study.
Isolation of the full-length cDNA clone encoding mTLR2
In an effort to clone mTLR2 cDNA, 2 primers (TGCTGGAGCCCATTGAGAGGA and GGACTTTATTGCAGTTCTCAG) were prepared based on the sequence information of the 2 mouse expressed sequence tag (est) clones (accession numbers AI020 960 and AA863 729) homologous to hTLR2 cDNA. A partial cDNA was prepared by reverse transcriptase–polymerase chain reaction (RT-PCR) using these primers and the total RNA prepared from a mouse macrophage cell line, J774.1. The synthesized 0.9-kilobase (kb) cDNA fragment was32P-labeled by random priming and used for screening a J774.1 cDNA library that was constructed in a cloning vector (Uni-Zap; Stratagene, La Jolla, CA) (gift of Dr H. Yagita, Juntendo University, Tokyo, Japan). Plaque hybridization was conducted as previously described.26 The inserts of the positive phage clones were excised (Rapid Excision Kit, Stratagene) according to the manufacturer's instructions to generate subclones in the plasmid (pBlueScript, Stratagene). DNA sequence analysis was performed on these plasmid clones using a DNA sequencer and a cycle sequencing kit (model 373A sequencer, Thermo Sequence; PE Biosystems, Forest City, CA).
Expression plasmids
The coding region of mouse CD14 was amplified by RT-PCR from total RNA isolated from J774.1 and inserted into a mammalian expression vector, pcDNA3.1(+) (Invitrogen, Carlsbad, CA). For the Flag-tagged mTLR2 expression plasmid, the coding region of mTLR2 was amplified by PCR from the isolated mTLR2 cDNA and cloned into the AscI site of the expression plasmid pEFBOS-Flag,27 which encodes a C-terminal Flag epitope.
Transient transfection
COS7 cells were plated onto 60-mm plates at 1 × 106 cells/plate on the day before transfection. Combinations of expression plasmid DNAs (6 μg total/plate) were transfected (Lipofectamine, Gibco BRL) according to the manufacturer's instructions. Cells were harvested 48 hours later with PBS and used for further analyses.
Flow cytometry
COS7 cells were incubated with anti-mCD14 mAb. After washes with staining buffer, goat antirat immunoglobulin G fluorescein isothiocyanate (IgG-FITC) (PharMingen) was added. Cells were analyzed by a fluorescence-activated cell sorter (FACSCalibur; Becton Dickinson, San Jose, CA).
Northern blot analysis
Total cellular RNA was extracted (TRIZOL reagent; Gibco BRL, Rockville, MD) according to the manufacturer's instructions. Messenger RNA (mRNA) was extracted (QuickPrep Micro mRNA Purification Kit; Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's instructions. We fractionated 10-μg aliquots of the total RNA or 5-μg aliquots of the total mRNA on a 1% agarose gel containing 20 mmol/L morpholinopropane sulfonic acid (MOPS), 5 mmol/L sodium acetate, 1 mmol/L EDTA (ethylenediaminetetraacetic acid, pH 7.0), and 6% (vol/vol) formaldehyde. The aliquots were then transferred to a nylon membrane. After ultraviolet-crosslinking (UV-crosslinking), membranes were soaked in prehybridization solution (6 × SSC [standard saline citrate], 5 × Denhardt's reagent, 0.5% SDS [sodium dodecyl sulfate], 100 mg/mL denatured salmon sperm DNA, and 50% formamide) for 3 hours at 42°C followed by incubation with a 32P-labeled probe in hybridization solution (6 × SSC, 0.5% SDS, 100 mg/mL denatured salmon sperm DNA, and 50% formamide) for 14 hours at 42°C. The membranes were washed twice in 2 × SSC and 0.1% SDS for 10 minutes at room temperature, washed twice in 0.1 × SSC and 0.1% SDS for 10 minutes at 50°C, and then exposed to film (Fuji RX-U films; Fuji Film, Tokyo, Japan).
Reverse transcriptase–polymerase chain reaction
Total cellular RNA was prepared (TRIZOL reagent, Gibco BRL). cDNA was synthesized from 2 μg of the total RNA by extension of random primers with 200 units (Superscript II, Gibco BRL). PCR of the cDNA was performed in a final volume of 50 μL containing 2.5 mmol/L magnesium dichloride (MgCl2), 2.5 units (AmpliTaq; Perkin-Elmer, Norwalk, CT), and 1 μmol/L specific primers (geneAmp 2400 PCR system, Perkin-Elmer). The synthesized PCR products were separated by electrophoresis on a 2% agarose gel and visualized by ethidium bromide staining. The primers were: mouse (m) β-actin sense, TGGAATCCTGTGGCATCCATGAAAC; β-actin antisense, TAAAACGCAGCTCAGTAACAGTCCG; mTLR2 sense, CAGCTTAAAGGGCGGGTCAGAG; mTLR2 antisense, TGGAGACGCCAGCTCTGGCTCA; mTLR4 sense, AGTGGGTCAAGGAACAGAAGCA; mTLR4 antisense, CTTTACCAGCTCATTTCTCACC; mIL-4 sense, CTAGCCTGAGTTCTCTTG; mIL-4 antisense, GAGTCAACATCTGCCTTCAC; mouse interferon γ (mIFNγ) sense, CTTCAAGATACAAGTGACCG; and mIFNγ antisense, TTCGGTAATGGACTTGCACA.
Isolation of S49.1 stable transfectants
S49.1 cells were transfected with an electroporator using 20 μg plasmid DNA and a setting of 800 millifarad (mF) and 300 V. Transfectants were selected with G418 (1 mg/mL). Resistant clones were screened for green fluorescent protein (GFP) fusion protein expression by flow cytometry. The expression of the right-sized protein was confirmed by Western blot analysis.
Extract preparation and immunoblotting
Cells were lysed in the following phospholipase C (PLC) lysis buffer at 108 cells/mL: 50 mmol/L HEPES (4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.0), 150 mmol/L sodium chloride (NaCl), 10% glycerol, 1% Triton X-100, 1.5 mmol/L MgCl2, 1 mmol/L EGTA, 100 mmol/L sodium fluorine (NaF), 10 mmol/L NaPPi, 1 mmol/L Na3VO4, 1 mmol/L phenylmethanesulfonyl fluoride, 10 mg/mL aprotinin, and 10 mg/mL leupeptin. The lysates were separated on SDS-polyacrylamide gels, then electrotransferred to polyvinylidene difluoride membranes (Immobilon; Millipore Corporation, Bedford, MA). The membranes were blocked for 2 hours in 2% bovine serum albumin (BSA) TBST and 20 mmol/L tris(hydroxymethyl aminomethane hydrochloride (Tris-HCl [pH 7.6], 0.15 mol/L sodium chloride, 0.1% Tween 20), incubated with primary antibodies in TBST for 1 hour, washed 3 times with TBST, and incubated for 1 hour with horseradish peroxidase–conjugated antimouse or antirabbit Ig (Amersham Pharmacia Biotech) diluted 1:10 000 in TBST. After 3 washes in TBST, the blot was developed with the enhanced chemiluminescence system (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Electrophoretic mobility shift assay
A κB oligonucleotide probe (Promega, Madison, WI) was phosphorylated with [32P]ATP using T4 polynucleotide kinase (Takara Biochemicals, Tokyo, Japan). Nuclear extracts were prepared as previously described.31 Specific binding of extract proteins to the κB probe was assessed by incubation for 30 minutes at room temperature in a solution containing 10 mmol/L HEPES (pH 7.9), 50 mmol/L potassium chloride (KCl), 0.2 mmol/L EDTA, 0.25 mmol/L DTT, 10% glycerol, 0.05% NP-40, and 0.5 μg poly (dI-dC) and then separated by electrophoresis in a 6% polyacrylamide gel.
Results
Isolation of the full-length cDNA clone encoding the mTLR2
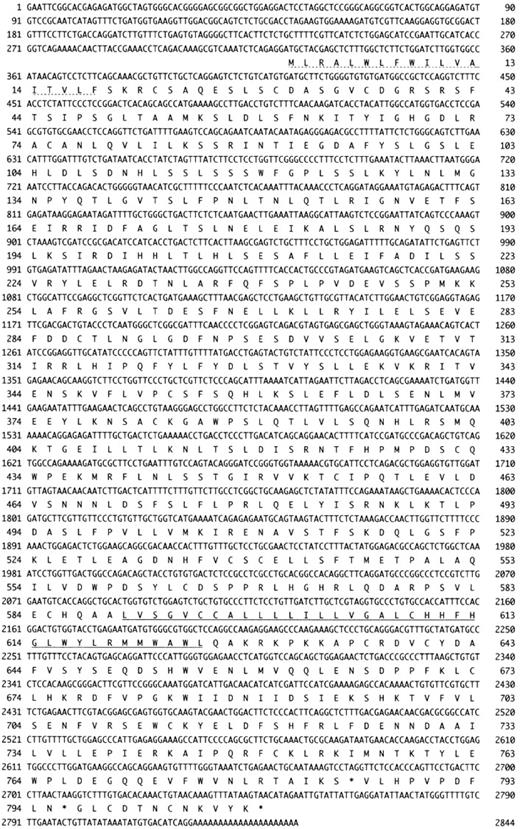
To clone mTLR2 cDNA, 2 primers were prepared based on the sequence information of the 2 mouse est clones (accession numbers AI020 960 and AA863 729) homologous to hTLR2 cDNA. A partial cDNA was prepared by RT-PCR using these primers, and the total RNA was prepared from a mouse macrophage cell line, J774.1, as a template. To isolate the complete mTLR2 cDNA, this 0.9-kb cDNA fragment was used as a probe for screening a J774.1 cDNA library. Altogether, we obtained 23 independent cDNA clones from 8 × 105 plaques, and DNA sequence analysis was performed on several of them. The longest cDNA clone was 3.0 kb and had an open reading frame of 784 codons (Figure1). This sequence contains an in-frame stop codon at 108 bp upstream of the first methionine codon. It coded for a signal peptide of 18 amino acid residues and a single transmembrane domain (amino acids 588-608).
Cloning of mTLR2 cDNA.
The amino acid sequences of mTLR2 (GenBank accession number AF216289) and its human homologue, hTLR2. The signal peptide is indicated by a dotted underline, and the putative transmembrane domain is underlined. Amino acid identity is shown between the 2 sequences. A “+” represents similarity between the corresponding pair of amino acid residues.
Cloning of mTLR2 cDNA.
The amino acid sequences of mTLR2 (GenBank accession number AF216289) and its human homologue, hTLR2. The signal peptide is indicated by a dotted underline, and the putative transmembrane domain is underlined. Amino acid identity is shown between the 2 sequences. A “+” represents similarity between the corresponding pair of amino acid residues.
The isolated cDNA clone was 70% identical and 83% homologous with hTLR220 at the amino acid level, which suggests that we had isolated mTLR2. Interestingly, the amino acid identity between hTLR2 and mTLR2 was higher in the cytoplasmic domain (86%) than in the extracellular domain (65%), although most of the leucines in the extracellular domain were conserved. The remarkable similarity of the cytoplasmic domain strongly indicated that the intracellular signaling mechanism of TLR2 should be conserved well between the 2 species.
In a recent publication, Heine et al32reported on their study to clone mTLR2 cDNA. Their reported nucleotide sequence of the 5′ untranslated region was different from ours, which may suggest the existence of an alternative splicing.
Mouse TLR2 mediates LPS signals in the presence of CD14
To determine if mTLR2 mediates LPS signals the same as hTLR2, we inserted the coding region of mTLR2 cDNA into a mammalian expression plasmid, pEFBOS, with a C-terminal Flag tag. The Flag-tagged mTLR2 was transiently expressed in a monkey kidney cell line, COS7, by itself or in combination with mouse CD14 (mCD14). The expression of the Flag-tagged mTLR2 and mCD14 was confirmed by Western blot analysis and flow cytometry, respectively (Figure 2A and B). Because LPS is known to induce JNK phosphorylation,6 7we monitored LPS signals using an antibody specific to phosphorylated forms of JNK. Phosphorylation of p46JNK could not be analyzed due to the comigrating nonspecific band recognized by this antibody; therefore we examined the LPS-induced phosphorylation of p54JNK. Transfection of either the control vector or the CD14 expression plasmid did not mediate LPS-induced p54JNK phosphorylation (Figure 2C, lanes 1, 2, 5, and 6), which indicates that the parental COS7 cells were hyporesponsive to LPS.
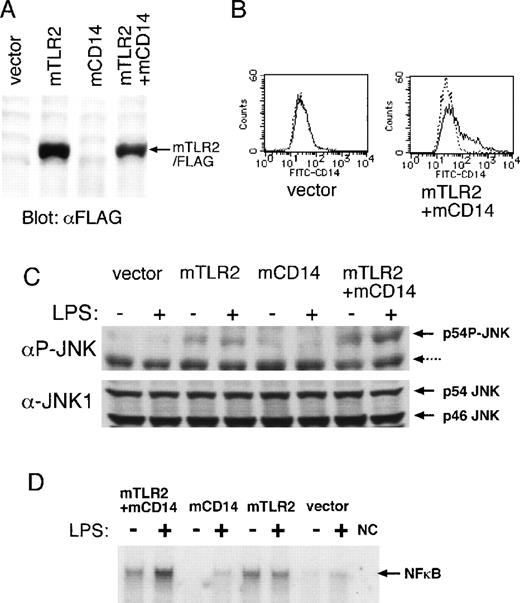
Effects of mTLR2 and mCD14 on LPS-induced JNK and NF-κB activation.
(A) Expression of the Flag-tagged mTLR2 in COS7 cells. COS7 cells were transiently transfected with the following: the vector alone, an expression plasmid of the Flag-tagged mTLR2, an expression plasmid of mCD14, or a combination of mTLR2/Flag and mCD14 expression plasmids. Flag-tagged mTLR2 expression was detected by immunoblotting with anti-Flag mAb. (B) Surface expression of mCD14 on COS7 cells. COS7 cells transiently transfected with expression plasmids as in (A) were stained with a control mAb (dotted line) or an anti-mCD14 mAb (solid line) followed by FITC-conjugated antirat IgG mAb. Flow cytometric analyses of COS7 cells transiently transfected with only vector and with Flag-tagged mTLR2 plus mCD14 are shown. A similar amount of mCD14 surface expression seen in double transfection was also observed in mCD14-transfected COS7 cells (data not shown). (C) LPS-induced JNK phosphorylation by mTLR2 and mCD14. Cytoplasmic cell extracts were prepared from COS7 cells transiently transfected as in (A), which were either left untreated or stimulated with 10 μg/mL LPS for 20 minutes. JNK phosphorylation was detected by Western blot analysis using an mAb specific for the phosphorylated form of JNK. Nonspecific 46-kd bands recognized by this mAb are indicated by a dotted arrow. The same filter was reblotted with an anti-JNK1 pAb. (D) LPS-mediated NF-κB DNA-binding activation is dependent on both mTLR2 and mCD14. Nuclear cell extracts were prepared from COS7 cells transiently transfected as in (A), which were either left untreated or stimulated with 10 μg/mL LPS for 20 minutes. EMSA was performed with a radio-labeled double-stranded probe containing a NF-κB binding site. NC represents negative control without nuclear extract.
Effects of mTLR2 and mCD14 on LPS-induced JNK and NF-κB activation.
(A) Expression of the Flag-tagged mTLR2 in COS7 cells. COS7 cells were transiently transfected with the following: the vector alone, an expression plasmid of the Flag-tagged mTLR2, an expression plasmid of mCD14, or a combination of mTLR2/Flag and mCD14 expression plasmids. Flag-tagged mTLR2 expression was detected by immunoblotting with anti-Flag mAb. (B) Surface expression of mCD14 on COS7 cells. COS7 cells transiently transfected with expression plasmids as in (A) were stained with a control mAb (dotted line) or an anti-mCD14 mAb (solid line) followed by FITC-conjugated antirat IgG mAb. Flow cytometric analyses of COS7 cells transiently transfected with only vector and with Flag-tagged mTLR2 plus mCD14 are shown. A similar amount of mCD14 surface expression seen in double transfection was also observed in mCD14-transfected COS7 cells (data not shown). (C) LPS-induced JNK phosphorylation by mTLR2 and mCD14. Cytoplasmic cell extracts were prepared from COS7 cells transiently transfected as in (A), which were either left untreated or stimulated with 10 μg/mL LPS for 20 minutes. JNK phosphorylation was detected by Western blot analysis using an mAb specific for the phosphorylated form of JNK. Nonspecific 46-kd bands recognized by this mAb are indicated by a dotted arrow. The same filter was reblotted with an anti-JNK1 pAb. (D) LPS-mediated NF-κB DNA-binding activation is dependent on both mTLR2 and mCD14. Nuclear cell extracts were prepared from COS7 cells transiently transfected as in (A), which were either left untreated or stimulated with 10 μg/mL LPS for 20 minutes. EMSA was performed with a radio-labeled double-stranded probe containing a NF-κB binding site. NC represents negative control without nuclear extract.
Overexpression of mTLR2 constitutively mediated slight phosphorylation of JNK, which was not strengthened further by LPS stimulation (Figure 2C, lanes 3 and 4). When mTLR2 was coexpressed with CD14, LPS significantly increased JNK phosphorylation (Figure 2C, lanes 7 and 8). Thus, mTLR2 mediates LPS responsiveness in a CD14-dependent fashion.
Using electrophoretic mobility shift assay (EMSA), we also examined NF-κB activation, which is a well-known downstream event of the LPS signal (Figure 2D). COS7 cells expressing mTLR2 constitutively showed slightly increased NF-κB DNA-binding activity. Coexpression of mCD14 with mTLR4 conferred LPS inducibility of NF-κB activation.
Tissue distribution of mTLR2 gene expression
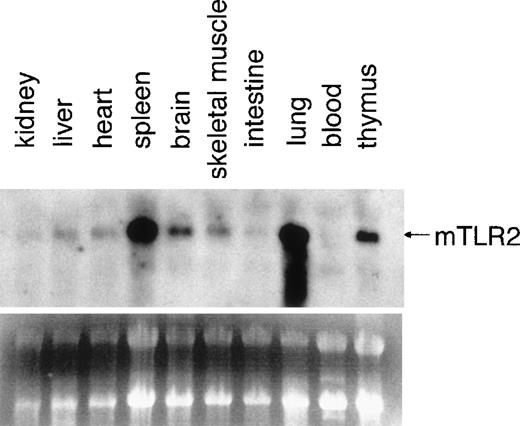
Northern blot analysis was performed on total RNA isolated from various mouse tissues (Figure3). The gene expression of mTLR2was detected in every tissue examined, and the highest expressions were observed in the spleen and the lung. Mouse TLR2 mRNA expression was also enriched in the brain and the thymus. We were particularly interested in mTLR2 expression in the thymus because the role of LPS signaling in T cells has not been fully established. We therefore investigated the regulation of mTLR2 gene expression in T cells.
Expression of mTLR2 in mouse tissues.
Total RNAs from indicated mouse tissues were resolved by formaldehyde gel electrophoresis, transferred to a nitrocellulose membrane, and hybridized with an mTLR2 cDNA probe. A picture of the ethidium bromide-stained gel is also shown.
Expression of mTLR2 in mouse tissues.
Total RNAs from indicated mouse tissues were resolved by formaldehyde gel electrophoresis, transferred to a nitrocellulose membrane, and hybridized with an mTLR2 cDNA probe. A picture of the ethidium bromide-stained gel is also shown.
Mouse TLR2 gene expression in T cells
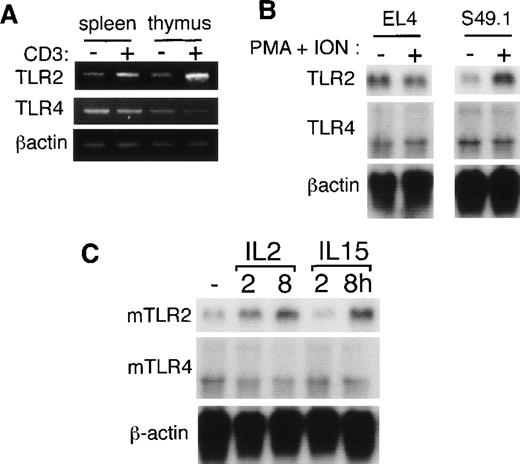
To elucidate the regulatory mechanisms for mTLR2 gene expression in T cells, purified mouse T cells from the thymus and the spleen were stimulated with immobilized anti-CD3ε mAb for 2 hours, and total RNA was isolated. Although the mTLR2 gene was constantly expressed in unstimulated T cells, as assessed by semiquantitative RT-PCR, triggering by anti-CD3ε mAb resulted in a substantial increase of the mTLR2 gene expression (Figure4A). In contrast, mTLR4 gene expression, which was also detected in unstimulated T cells, remained constant after anti-CD3ε treatment (Figure 4A).
Mouse TLR2 and TLR4 gene expressions in mouse T cells.
(A) Mouse T cells purified from thymuses and spleens were either unstimulated or stimulated with immobilized anti-CD3ε mAb for 2 hours. Total RNAs were extracted, and mTLR2 and mTLR4expressions were analyzed by semiquantitative RT-PCR. (B) S49.1 and EL4 cells were either unstimulated or stimulated with 100 ng/mL PMA plus 1 μg/mL ionomycin for 2 hours. We isolated 5 μg mRNA from the cells and analyzed them by Northern blot analysis using mTLR2 and mTLR4 cDNA probes. (C) CTLL-2 cells were starved overnight without factors and either left unstimulated or stimulated with 10 ng/mL mIL-2 or 10 ng/mL hIL-15 for the indicated time.
Mouse TLR2 and TLR4 gene expressions in mouse T cells.
(A) Mouse T cells purified from thymuses and spleens were either unstimulated or stimulated with immobilized anti-CD3ε mAb for 2 hours. Total RNAs were extracted, and mTLR2 and mTLR4expressions were analyzed by semiquantitative RT-PCR. (B) S49.1 and EL4 cells were either unstimulated or stimulated with 100 ng/mL PMA plus 1 μg/mL ionomycin for 2 hours. We isolated 5 μg mRNA from the cells and analyzed them by Northern blot analysis using mTLR2 and mTLR4 cDNA probes. (C) CTLL-2 cells were starved overnight without factors and either left unstimulated or stimulated with 10 ng/mL mIL-2 or 10 ng/mL hIL-15 for the indicated time.
To exclude the possibility that the positive RT-PCR products were due to a small number of contaminating macrophages or B cells, using Northern blot analysis, we tested TLR gene expressions in several mouse T cell lines (EL4, S49.1, and CTLL-2). Gene expression ofmTLR4 was easily detected in these 3 cell lines (Figure 4B and C). When compared with mTLR4, mTLR2 gene expression was relatively lower in S49.1 cells (Figure 4B). When this T cell line was stimulated with the combination of PMA and ionomycin, which mimics TCR stimulation, mTLR2 gene expression increased. On the other hand, mTLR4 expression remained constant after PMA plus ionomycin treatment in S49.1 cells (Figure 4B). Next, we examined the effect of cytokine stimulation on mTLR2 gene expression in CTLL-2 cells that are responsive to IL-2 and IL-15. The 2 cytokines share IL-2Rβ and γc for their signal transduction.33 Both IL-2 and IL-15 treatment significantly increased mTLR2 gene expression, whereas no remarkable change was detected in the mTLR4 mRNA level (Figure 4C).
ERK and p38 kinase pathways are involved in mTLR2 mRNA induction in mouse T cells
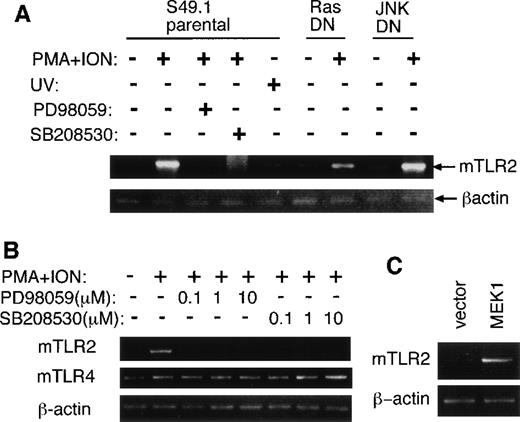
Both cytokine stimulation and anti-CD3ε mAb treatment of T cells are known to activate MAP kinase pathways including ERK, JNK, and p38 kinase.34 To investigate whether ERK and p38 kinase pathways are involved in mTLR2 mRNA up-regulation, we pretreated S49.1 cells with specific inhibitors of ERK (PD98 059) and p38 kinase (SB208 530) pathways followed by stimulation with PMA plus ionomycin. Pretreatment with each of these MAP kinase inhibitors abrogated the increase of mTLR2 mRNA at 10 μmol/L, as assessed by semiquantitative RT-PCR (Figure 5A). We next examined S49.1 cells that constitutively expressed a dominant negative form of JNK1 (JNKDN), which has 2 amino acid substitutions of the phosphorylation sites of JNK1. Although JNK activation by PMA plus ionomycin was markedly inhibited by the expression of JNKDN in this transfectant (data not shown), mRNA up-regulation of mTLR2 by PMA plus ionomycin treatment was comparable to that of parental S49.1 cells (Figure 5A). Additionally, UV treatment, which is a potent activator of JNK, did not induce significant increase of mTLR2 gene expression (Figure5A). Therefore, JNK activation may not be necessary for mTLR2 mRNA up-regulation by PMA plus ionomycin in S49.1 cells.
Involvement of MAP kinase pathways in mTLR2 induction in S49.1 cells.
(A) S49.1 cells were either untreated or treated with PMA plus ionomycin for 2 hours with or without the pretreatment of 10 μmol/L PD98 059 or SB208 530 for 30 minutes. S49.1 cells were also treated with UV irradiation (40 J/m2) and left for 1 hour at 37°C. In some experiments, S49.1 cells constitutively expressing a dominant negative form of JNK1 or H-Ras were treated with PMA plus ionomycin for 2 hours. Total RNAs were extracted, and the expression ofmTLR2, mTLR4, and β-actin was examined by semiquantitative RT-PCR. (B) S49.1 cells were pretreated with a series of concentrations of PD98 059 or SB208 530 for 30 minutes followed by treatment with PMA plus ionomycin for 2 hours. Total RNAs were extracted, and the expressions of mTLR2, mTLR4, andβ-actin were examined by semiquantitative RT-PCR. (C) Total RNAs were extracted from S49.1 cells transfected with pEGFP-C1 or pEGFP-MEK1A. Expression of mTLR2 and β-actin was examined by semiquantitative RT-PCR.
Involvement of MAP kinase pathways in mTLR2 induction in S49.1 cells.
(A) S49.1 cells were either untreated or treated with PMA plus ionomycin for 2 hours with or without the pretreatment of 10 μmol/L PD98 059 or SB208 530 for 30 minutes. S49.1 cells were also treated with UV irradiation (40 J/m2) and left for 1 hour at 37°C. In some experiments, S49.1 cells constitutively expressing a dominant negative form of JNK1 or H-Ras were treated with PMA plus ionomycin for 2 hours. Total RNAs were extracted, and the expression ofmTLR2, mTLR4, and β-actin was examined by semiquantitative RT-PCR. (B) S49.1 cells were pretreated with a series of concentrations of PD98 059 or SB208 530 for 30 minutes followed by treatment with PMA plus ionomycin for 2 hours. Total RNAs were extracted, and the expressions of mTLR2, mTLR4, andβ-actin were examined by semiquantitative RT-PCR. (C) Total RNAs were extracted from S49.1 cells transfected with pEGFP-C1 or pEGFP-MEK1A. Expression of mTLR2 and β-actin was examined by semiquantitative RT-PCR.
On the other hand, a dominant negative form of H-Ras (N17) only partially blocked mTLR2 mRNA increase (Figure 5A), which suggests that either the stimulation by PMA plus ionomycin works on downstream molecules of Ras or there is an alternative Ras-independent pathway for the induction of mTLR2 mRNA. Furthermore, when a series of concentrations of PD98 059 and SB208 530 was tested, both of them effectively inhibited mTLR2 mRNA up-regulation at concentrations as low as 0.1 μmol/L, which suggests that the inhibition by these chemicals was specific (Figure 5B).
Next, to test if constitutive activation of the ERK pathway is sufficient to induce mTLR2, S49.1 cells were transfected with an expression plasmid encoding a constitutively activated form of MEK1 (MEK1A), an upstream activator of ERK. The mRNA levels of mTLR2 in 3 independently isolated S49.1 cell lines overexpressing the active mutant of MEK1 were constitutively higher than that of the parental S49.1 cells. (A typical result is shown in Figure 5C.) Thus, mTLR2 induction by PMA plus ionomycin treatment is dependent on both ERK and p38 kinase activation, whereas continuous activation of the ERK pathway seems sufficient to induce mTLR2 gene expression in S49.1 cells.
LPS treatment directly affects the cytokine expression of the EL4 cell line
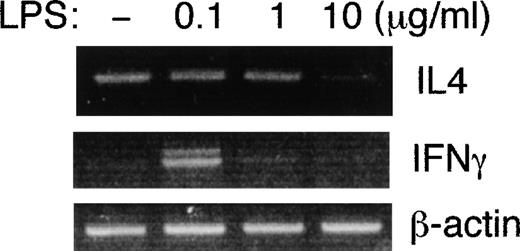
It has recently been reported that LPS and lipid A directly inhibit IL-4 production by mouse T helper cell 2 (Th2) clones.10 The EL4 cell line, which constitutively expresses both mTLR2 and mTLR4 (Figure 4B), is known to have a Th2-like phenotype and to produce IL-4.35 To investigate the direct effects of LPS on the cytokine production of EL4 cells, we treated EL4 cells with serial dilutions of LPS and analyzed IL-4 and IFNγ mRNA expression by semiquantitative RT-PCR (Figure6). The gene expression of IFNγwas weakly induced at 0.1 μg/mL LPS, but it was undetectable at higher concentrations of LPS. In contrast, IL-4 mRNA was constitutively detectable and decreased in the presence of 10 μg/mL of LPS. Thus, LPS directly affects the cytokine expression profile of the EL4 cell line.
LPS inhibits IL-4 gene expression in EL4 cells.
EL-4 cells were either untreated or treated with 0.1, 1, or 10 μg/mL of LPS for 6 hours. Total RNAs were extracted, and IL-4 andIFNγ gene expressions were examined by semiquantitative RT-PCR.
LPS inhibits IL-4 gene expression in EL4 cells.
EL-4 cells were either untreated or treated with 0.1, 1, or 10 μg/mL of LPS for 6 hours. Total RNAs were extracted, and IL-4 andIFNγ gene expressions were examined by semiquantitative RT-PCR.
Discussion
In this study, we describe the isolation and cloning of mTLR2, which mediates LPS-induced cellular signals similar to its human counterpart.21 22 We also show that mTLR2, as well as another putative LPS receptor, mTLR4, is expressed in mouse T cells, and that the gene expression of mTLR2 is differently regulated from that of mTLR4. In addition, LPS directly inhibited IL-4 expression in a Th2-type cell line, EL-4, which expresses both mTLR2 and mTLR4. LPS may thus directly affect T-cell functions through TLR-mediated signal pathways.
The extracellular segments of TLR proteins are distinctively composed of 24-28 amino acid leucine-rich repeats (LRRs).20 LRRs are also found in the Drosophila homologues, Toll and 18 Wheeler (18w).36,37 Although amino acid sequence homology between hTLR2 and mTLR2 is higher in the cytoplasmic domain, the LRR structure is conserved well, suggesting that it plays a critical role in their biological functions. CD14, which binds LPS with high affinity, also contains 10 copies of the LRR motif. However, LRRs of CD14 are not necessary for LPS binding, as shown by mapping studies.38,39 Human TLR2 has been reported to bind LPS by itself, but the Kd is low and may not be physiologically important.21 We have not tested the binding affinity of mTLR2 for LPS. However, it is unlikely that mTLR2 reacts to physiological concentrations of LPS by itself because COS7 cells expressing mTLR2 alone did not show a detectable response to 10 μg/mL LPS in either JNK or NF-κB activation (Figure 2).
It is noteworthy that the cytoplasmic domain is conserved well at the amino acid level between hTLR2 and mTLR2. TLRs 1-5 share a highly significant homology called the Toll-homology (TH) domain.20 The TH domain was first discovered as a resemblance of a cytoplasmic domain between Toll and IL-1 receptor type I (IL-1RI).18,40 It was later shown that the TH domain was also shared by numerous proteins including plant disease-resistance proteins,41 an intracellular myeloid differentiation marker (MyD88),42,43 ST2,43 IL-1Racp,44and IL-1RrP/IL-18R.45 The TH domain contains 10 conserved blocks consisting of 5 alternating β-strands and α-helices,20 and all the blocks are conserved in mTLR2 as well as in hTLR2 (Figure 1). Since both Toll and IL-1R mediate signaling by similar Rel-type transcription factors,18,40the TH domain presumably plays an important role in the activation of the NF-κB pathway. In assays using carboxy-terminal truncations of hTLR2, deletions of carboxy-terminal 13 or 15 amino acid markedly impaired the LPS-mediated NF-κB activation by hTLR2.21 22These deletions remove only the last block of α-helices, which suggests that the complete conformation of the TH domain is necessary for proper signaling. This finding is consistent with the fact that all 10 blocks of TH are conserved between mTLR2 and hTLR2 (Figure 1).
Our results have demonstrated that overexpression of mTLR2 in combination with mCD14 confers responsiveness to LPS stimulation in COS7 cells. Although hTLR2 was shown to mediate LPS signaling,21,22 the role of mTLR2 in LPS signaling remains questionable. Two recent studies examined a single autosomal locus (lps), which is responsible for the LPS hyporesponsiveness (lpsd) of 2 mouse strains (C3H/HeJ and C57BL10/ScCr).23,24 The mTLR4 gene was found in thelps locus and mutations in the mTLR4 gene in both strains were identified. This strongly suggests that a defectivemTLR4 is responsible for the LPS-hyporesponsiveness of these mice. More recently, mice lacking the mTLR4 gene have been generated, and these mice show LPS-hyporesponsiveness very similar to that in the 2 lpsd mouse strains.46 These findings are consistent with the idea that the mTLR4 gene is essential in LPS signaling.
Then what is the role of mTLR2 in LPS responsiveness? There are four possible explanations. First, it is possible that mTLR2 works as an alternative to mTLR4. Lpsd mice are hyporesponsive but not unresponsive to LPS. They showLPS-induced gene expressions in the presence of a larger dose of LPS,47 which suggests that there is some redundancy in LPS signaling. Our present data, which confirm that mTLR2 mediates LPS signals, may indicate that mTLR2 works to back up mTLR4. Another explanation is that mTLR2 and mTLR4 may respond to different types of LPS. Cells from C3H/HeJ mice are as sensitive as their normal counterparts to certain types of LPS (eg, Porphyromonas gingivalis LPS).48 There is divergence in LPS structure among Gram-negative bacteria, and it is presumable that mTLR4 responds to certain types of LPS better than mTLR2, while mTLR2 responds better to others. The third possibility is that mTLR2 and mTLR4 may cooperate to respond to LPS. It has not been shown how TLRs are involved in the formation of the LPS receptor complex, and it is possible that mTLR2 associates with mTLR4 to form a dimer or oligomer. Further studies, including a study of mice lacking themTLR2 gene, need to be done to fully understand the involvement of mTLR2 in the LPS receptor complex.
The fourth possibility is based on the inducibility of mTLR2expression (Figures 4 and 5). In our current study we have shown that in some T cell lines, the expression level of mTLR2 is very low, but it significantly increases after stimulation with anti-CD3ε mAb, PMA plus ionomycin, and IL-2/IL-15. However, no change is detected in mTLR4 expression after such treatments (Figure 4). Also,mTLR2 is significantly up-regulated in macrophages by stimulation with LPS and IFNγ (T. Matsuguchi, unpublished data, June 1999). Interestingly, the sensitivity oflpsd mice is restored by treatment with IFNγ or BCG,49-52 and the expression level ofmTLR2 in immune cells is likely to be higher after these treatments in lpsd mice. All these findings may suggest that immune-component cells react to LPS mainly with TLR4 at first, and TLR2 is the second LPS receptor boosting the immune response by the host. This hypothesis explains well why mTLR4 is essential in LPS responsiveness.
It is of note that the coexpression of mCD14 with mTLR2was necessary for the proper activation of JNK and NF-κB by LPS in COS7 cells. In contrast, in 2 earlier reports using hTLR2, 293 cells transfected with only hTLR2 increased the transcription ofNF-κB–dependent reporter genes in response to LPS.21,22 The exact reason for this discrepancy is unknown. It is possible that mTLR2 and hTLR2 respond differently to LPS in the absence of CD14. It is also possible that the discrepancy is due to the difference of the cell lines used for the transfection. The parental COS7 cells that we used in our experiments did not show any CD14 surface expression (Figure 2B). The 293 cells tested in the earlier reports might contain a small amount of endogenous CD14, which worked with the exogenous TLR2. It has recently been reported that hTLR4 needs a small molecule called MD-2 for proper transduction of the LPS signal.53 Therefore, it is also possible that COS7 and 293 may in different ways express an unknown coactivator protein, similar to MD-2, which is necessary for proper TLR2 function.
We have also shown that both mTLR2 and mTLR4 mRNAs are expressed in mouse T cells and some T cell lines. Although we could not show the protein expressions due to the lack of appropriate mAbs, LPS directly inhibited IL-4 expression in a mouse Th2-type cell line, EL4, suggesting the surface expression of LPS receptors on this T cell line. In spite of great interest, the direct effects of LPS on T cells have not been fully understood. Because it is now known that both TLR2 and TLR4 are expressed on T cells, it is reasonable to presume that LPS can directly affect some of the T-cell functions. To support our findings, it has recently been reported that both LPS and lipid A decrease IL-4 production from purified mouse CD4+ T cells and Th2 clones, indicating that LPS directly modulates cytokine expressions by T cells.10 Additionally, an earlier report showed the activation and subsequent apoptosis of T cells in LPS-treated mice.11 Although T cells usually do not express CD14, a soluble form of CD14 (sCD14) in the serum has been shown to bind LPS and induce signals in cells lacking CD14.16,17 It has also recently been shown that CD11c/CD18, which is expressed on some T cells,54 may work as a component of the LPS receptor.55 56
In the present study, we have shown that a low concentration (0.1 μg/mL) of LPS induces weak expression of IFNγ mRNA in EL4 cells (Figure 6). The same result was repeatedly observed in different sets of experiments (data not shown). Recent reports have shown that both JNK1 and JNK2 are involved in the development of Th1 cells producing IFNγ.57 58 Our results have revealed that mTLR2 is a potent inducer of JNK phosphorylation (Figure 2C). Therefore, it is a reasonable to presume that LPS induces signal pathways promoting Th1 cell development. Interestingly, the up-regulation of IFNγ mRNA was not observed at higher concentrations of LPS (Figure 6). The reason for this phenomenon is not clear. It was not due to the increased cell death because LPS as high as 100 μg/mL did not affect the viability of EL4 cells (data not shown). LPS treatment of this cell line might activate different downstream signals in a concentration-dependent manner. The LPS-binding affinities for TLR2 and TLR4 have not been clearly shown, and it is interesting to presume that either TLR2 or TLR4 works preferably at the low concentration of LPS, thereby mediating better IFNγ expression. EL4 cells constitutively express both TLR2 and TLR4 (Figure 4B).
The expression level of mTLR2 but not that of mTLR4increases in T cells with the stimulation by immobilized anti-CD3ε mAb, PMA plus ionomycin, IL-2, and IL-15. All of these stimulators are known to activate both ERK and p38 kinase,59-64 and our data indicate that mTLR2 mRNA up-regulation by PMA plus ionomycin is dependent on both ERK and p38 kinase pathways (Figure 5). It is becoming evident that the binding of MAP kinases to transcription factors is a critical determinant of their specificity. ERK is specifically targeted to the ets domain transcription factors, Elk-1 and Lin-1, while p38 has recently been shown to be targeted to the MADS-box transcription factors, MEF2A and MEF2C.65Mouse TLR2 gene may be under the regulation of multiple transcription factors that are activated by different MAP kinases, ERK, and p38 kinase. Collaboration between ERK and p38 kinase has also been reported for the induction of cytokine and iNOS synthesis in macrophages.66 67 However, the overexpression of a constitutively active mutant of MEK1 increased mTLR2 mRNA (Figure 5C), which suggests that the continuous activation of the ERK pathway is sufficient to induce the mTLR2 gene expression.
Lastly, it is interesting that mTLR2 is induced by IL-15 in some T cells. Since LPS is a potent inducer of IL-15 production from macrophages,68 these findings may suggest that at least a significant fraction of T cells responds to LPS in a 2-step manner: first by using constitutively expressed mTLR4, then by using mTLR2, which is induced later by cytokines like IL-15. It is interesting to speculate that mTLR2 and mTLR4 induce different intracellular signals; as a result, the same T-cell population might respond differently to LPS during the course of Gram-negative bacterial infection.
In conclusion, the present study has shown that both mTLR2 and mTLR4 are able to mediate LPS signaling. The expression of these proteins in mouse T cells indicates that LPS may have direct effects on T-cell activity during Gram-negative bacterial infection.
Acknowledgments
We thank Ms K. Itano, Ms A. Kato, and Ms A. Nishikawa for their technical assistance. We also thank Dr H. Yagita for providing a mouse cDNA library.
Supported in part by grants from the Ono Pharmaceutical Company (Japan; T.M.); the Yokoyama Research Foundation for Clinical Pharmacology (Japan; Y.Y.); the Center of Excellence; and the Core Research for Evolutional Science and Technology (CREST) Project and by grant JSPS-RFTF97L00703 from the Ministry of Education, Science and Culture of the Japanese Government.
Reprints:T. Matsuguchi, Laboratory of Host Defense and Germfree Life, Research Institute for Disease Mechanism and Control, Nagoya University School of Medicine, 65 Tsurumai-cho, Showa-ku, Nagoya 466-8550, Japan; e-mail: tmatsugu@med.nagoya-u.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal