Understanding the regulation of genes controlling fibrinolysis and matrix homeostasis is essential for elucidating the basis of tissue repair. A recently described novel Krüppel-like factor, Zf9, is up-regulated in acute liver injury in activated hepatic stellate cells. Because Zf9 can be induced widely, its activity was examined in vascular endothelium, a key cell in vascular injury. Zf9 is induced as an immediate-early response gene in bovine aortic endothelial cells (BAECs) following treatment with serum or phorbol ester. Zf9 transcriptionally activates urokinase plasminogen activator (uPA). Recombinant Zf9-GST binds to wild-type but not mutated ‘GC-box’ motifs within the human uPA promoter (−63 to −32), with greatest affinity to the middle of 3 contiguous GC boxes. Transient transfection of Zf9 drives transactivation of a full-length uPA promoter- and GC box-construct, but not a uPA promoter-construct devoid of GC boxes. Transactivation of uPA by Zf9 is also supported in DrosophilaS2 cells. Most importantly, transiently transfected Zf9 up-regulates endogenous uPA messenger RNA and activity in BAECs, resulting in increased bioactive transforming growth factor-beta (TGF-β) via enhancement of proteolytic activation of the latent molecule. Furthermore, concomitant expression of Zf9 and uPA proteins was observed in arterial endothelial cells after balloon injury in rats, suggesting a potential role of Zf9 in uPA expression not only in vitro but also in vivo. These findings suggest a role of Zf9 in the injury response by enhancing uPA synthesis and subsequent activation of latent TGF-β.
Coordinated gene expression is a key component of the response to tissue injury. Classes of induced genes include extracellular matrix proteins,1-4 growth factors/cytokines such as transforming growth factor-β (TGF-β),1-6 and proteases such as urokinase plasminogen activator (uPA).1-9uPA was originally discovered as a fibrinolytic factor responsible for conversion of plasminogen to plasmin in dissolving vascular thrombosis.10,11 However, it also has a key role in tissue remodeling,7,9,12 metastasis,13,14apoptosis,14,15 and immunity14 either directly or through activation of other enzymes including matrix proteases9,13,16 and cytokines.5,17 uPA has been implicated in the activation of latent TGF-β. By activating plasminogen to plasmin, uPA can stimulate proteolytic activation of latent TGF-β in several tissues and cell types especially in pathogenic situations.15,17-25 TGF-β in turn mediates key components of the injury response. It down-regulates the inflammation, modulates growth inhibition and differentiation, and enhances extracellular matrix production in a variety of cell types.1-4 Vascular endothelial cells are a potent source of uPA, and given their critical position in tissue remodeling, are a key regulator of the injury response.6 Thus, control of uPA/plasmin and thereby TGF-β levels in endothelial cells is an important event in the response to tissue injury.
Circulating levels of plasminogen are fairly constant, so cellular plasmin levels are controlled primarily by plasminogen activator (PA) activity. Cellular PA activity is, in turn, controlled not only by regulation of uPA synthesis but also by levels of its receptor (uPAR)7,11 and inhibitor (PAI-1).10 uPAR is essential for localizing the PA activity on the cell surface leading to acceleration of plasminogen activation,7,11 whereas PAI-1 rapidly inhibits PA activity.10 Thus, the balance between uPA, uPAR, and PAI-1 levels is critical for regulating the level of net fibrinolytic activity.10,11 Regulation of uPA gene expression is therefore a key control point of cellular PA activity. It is known that uPA expression by vascular cells is up-regulated during the process of tissue repair in vivo.7,8,12 However, the molecular basis for injury-induced uPA gene expression is not completely understood. Several transcription factors have been reported to participate in this response. These include LFB3/HNF,26CRE-binding protein,7 Ets-1,27PEA3,28-30 UEF1-4,28-30 AP-1,28-31and NF-κ B32 demonstrated in the cultured cells and Egr-1 in injured rat aorta.33 Sp1, the initial Krüppel-like zinc finger protein,34,35 is also shown to be another important transcriptional activator of uPA in vitro through its interaction with 3 contiguous GC boxes immediately upstream of the TATA box in the uPA promoter.36 37
Zf9/COPEB/GBF (soon to be renamed KLF6) is a novel zinc finger transcription factor recently cloned from liver, placenta, and leukocyte complementary DNA (cDNA) libraries.38-41 The molecule belongs to the family of Krüppel-like transcription factors,42-52 all of which contain 3 contiguous C2H2 zinc fingers at the carboxyl-terminal domain, and recognize either GC box motifs or CACCC motifs in responsive promoters.39-52 A proximal role for Zf9 in response to tissue injury has been suggested by its rapid induction in activated hepatic stellate cells, the key fibrogenic cell type in liver injury and repair.38,39 Moreover, Zf9 transactivates key genes comprising the injury response, including collagen α1(I), TGF-β1, and types I and II TGF-β receptors in hepatic stellate cells.39 53
Given its widespread expression and interaction with GC box motifs,39-41 we have explored a potential role for Zf9 inuPA gene expression by endothelial cells. We have identified Zf9 messenger RNA (mRNA) and protein in cultured endothelial cells, which are induced by serum or phorbol esters. Zf9 stimulates uPA transcription and endogenous uPA activity in these cells, leading to proteolytic activation of latent TGF-β1. Furthermore, concomitant expression of Zf9 and uPA proteins was induced in arterial endothelial cells following carotid balloon injury in rats. The data suggest that Zf9 may have an important role in regulation of PA/plasmin activity and TGF-β1 activity in vascular endothelium.
Materials and methods
Materials
Phorbol myristate acetate (PMA) was purchased from Sigma (Chemical Co, St Louis, MO). Rabbit polyclonal antibodies against rat Zf9 and rat uPA were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and American Diagnostica (Greenwich, CT), respectively. Recombinant human TGF-β1 and its antibody, which neutralizes all subtypes of TGF-βs, were from R&D System (Minneapolis, MN) and Genzyme Diagnostics (Cambridge, MA), respectively.
Plasmids and cells
Zf9 mammalian (Zf9-pCIneo) and Drosophila(Zf9-pAC) expression plasmids were constructed as previously described.39 The uPA-luciferase expressing vector, pGL2-2350, containing the human uPA promoter (−2345 to 32), its deletion mutant, pGL2-GC, containing the first 65 bp and GC box and TATA box region (−2345 to −2280 and −91 to +32, respectively), and another deletion mutant, pGL2-ΔGC, which contains uPA promoter devoid of the GC box region (−71 to −28), were as described.36 37 Bovine aortic endothelial cells (BAECs) were isolated and grown in α minimal essential medium (αMEM; GIBCO BRL, Life Technologies, Rockville, MD) containing 10% fetal calf serum (FCS). Drosophila S2 cells were generous gifts from Dr H. Tanaka, National Institute of Sericultural and Entomological Science, Tsukuba, Japan and maintained in Shields and Sang M3 insect medium (Sigma) containing 10% FCS.
Northern blotting
Isolation of total RNA from BAECs and its Northern blot analyses were performed as described previously.54 RNA was separated through 1% agarose-formaldehyde gel electrophoresis and transferred onto the Biodyne nylon membranes (Pall Biosupport, New York, NY). Membranes were hybridized with 32P-labeled cDNA for either human Zf9,39 bovine uPA,37 bovine uPAR,54 or human PAI-1,54 and rehybridized with chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control.54 Signals were quantitated using a Fujix BAS 2000 Bioimaging analyzer (Fuji Photo-Film, Tokyo, Japan).
Western blotting
Western blotting was performed as described previously37using rabbit anti-Zf9 polyclonal antibody (final 1:10,000 dilution), and goat antirabbit IgG antibodies conjugated with peroxidase (Jackson ImmunoResearch Laboratories, West Grove, PA). The signals were detected with an Amersham-Pharmacia (Buckinghamshire, UK) ECL system. The blot was reprobed with antirabbit GAPDH (Chemicon Int., Temecula, CA).
Gel shift assay
Rat zf9 cDNA was cloned into pGEX-3T (Pharmacia Biotech, Sweden), and transformed in Escherichia coli BL21. Following induction by 2 mM isopropyl β-d-thiogalactoside for 4 hours, the cells were disrupted by sonication, and Zf9-glutathione S-transferase (GST) fusion protein was purified using glutathione-Sepharose (Amersham-Pharmacia) from cell homogenates. Purified GST protein (Santa Cruz) was used as a control. Oligonucleotides corresponding to −63 to −32 of the human uPA promoter (wild-type uPA GC box),37 which contains 3 GC boxes, as well as mutant oligos, in which mutations were made in 1 or all of 3 GC boxes, were synthesized. Sequences of these oligonucleotides are indicated in Figure 2A. Oligonucleotides were double-stranded and labeled with γ32-P adenosine triphosphate (DuPont, Wilmington, DE) by polynucleotide kinase (Boehringer Mannheim, Mannheim, Germany). For gel shift assays, 20 ng of Zf9-GST fusion protein was preincubated for 15 minutes at 4°C with or without unlabeled oligonucleotide, then 3.5 ng labeled oligonucleotide (10,000 μCi/mol) was added in the presence of 1 μg of dI-dC (Pharmacia) in 20 μL binding buffer (20 mM Hepes, pH 7.4, containing 1 mM MgCl2, 10 μM ZnSO4, 20 mM KCl, and 8% glycerol). The reaction mixture was incubated for 15 minutes at 4°C and separated on a 6% polyacrylamide gel. The gel was dried and exposed on films for a Fujix BAS 2000 Bioimaging analyzer (Fuji Photo-Film).
Transient transfection
The BAEC cultures were plated on 35-mm dishes at 80% confluency.Drosophila S2 cells were grown in 35-mm dishes (5 × 106 cells/dish). Transient transfection was performed using Lipofectamine Plus reagents (GIBCO, BRL, Life Technologies Inc, Rockville, MD) in 1 mL serum-free medium containing either Zf9-pCIneo or Zf9-pAC, with or without 1 μg pGL2-2350, along with pRL-CMV (Renillaluciferase, 100 ng/dish, Promega, Madison, WI) as an internal standard to normalize transfection efficiency. pCIneo or pAC was used as empty vector to adjust total amount of DNA transfected always to 2 μg per each sample. After a 4-hour incubation, 1 mL of medium containing 20% FCS was added to the cultures, and then further incubated for 48 hours. Thereafter, either luciferase activity or endogenous PA activity was determined in cell lysates, or total RNA was isolated and used for Northern blotting.
Luciferase assay
Cells were harvested by scraping (BAECs) or by centrifugation (S2 cells) and lysed into 120 μL of the commercial lysis buffer (Promega). Luciferase activity was determined in 10 μL of lysate using a commercial kit (Promega) and luminometer (Turner Designs Instrument, Sunnyvale, CA). Transfection efficiency was normalized by Renilla luciferase activity measured in the same cell lysate at the same time.
Assay of cellular PA activity and membrane plasmin activity
Cellular PA activity was measured using the chromogenic substrate S2403 as described previously and expressed as urokinase unit per milligram of protein in the sample.37 Protein concentration was measured by bicinchoninic acid (BCA) (Pierce, Rockford, IL) assay using bovine serum albumin (BSA) as the standard. Plasmin bound to the cell surface was recovered with tranexamic acid and assayed as described previously.54 This plasmin is originally derived from plasminogen present in the serum.
Zf9 and uPA expression in vivo
The distal half of the left common carotid artery of a 10-week-old Sprague-Dawley rat was denuded of endothelium by 3 passages of a 2-F balloon catheter inserted through the left external carotid artery as reported previously.55 Pentobarbital (intraperitoneal, 30 mg/kg) and 1% xylocaine (local) were used for anesthesia. Animals were killed with an overdose of pentobarbital, and the artery was perfusion fixed with 3% paraformaldehyde and excised. Paraffin-embedded serial sections were stained using Vectastain ABC elite kit (Vector Laboratories, Burlingame, CA) according to the manufacturer's instructions with antibodies against rat Zf9 (final 1:500 dilution), rat uPA (final 1:50 dilution), or nonimmune rabbit serum (final 1:50 dilution). Color development was made with diaminobenzidine tetrahydrochloride and nuclear counterstain with hematoxylin. Procedures were examined and authorized by the institutional committee.
TGF-β assays
Measurement of TGF-β was performed using either a bioassay (cellular PA assay with BAECs) or enzyme-linked immunosorbent assays (ELISAs) specific for either TGF-β1 or TGF-β2 (Promega). BAEC cultures were transfected with Zf9 expression vector and incubated for 36 hours. The cultures were then rinsed with phosphate-buffered saline (PBS) and further incubated with 1 mL of αMEM containing 0.1% BSA for an additional 12 hours to generate conditioned medium. Cellular PA assays for TGF-β were performed as described previously.56 Briefly, following an 8-hour incubation of test BAECs with each sample conditioned medium in the presence or absence of 20 μg/mL either anti-TGF-β antibody or nonimmune antibody, cell lysate was prepared and PA activity levels of each cell lysate were determined. The concentration of TGF-β in the conditioned medium was calculated from the decrease in PA levels by comparison with a standard curve made with recombinant human TGF-β1. The specificity of the assays was verified by controls using anti-TGF-β antibody. The amount of total (active plus latent) TGF-β was determined following conversion of all latent TGF-β to active TGF-β by acidification of the sample (pH 3, 1 hour at room temperature), followed by neutralization. The lowest limit of this bioassay is about 2 pg/mL. For ELISAs, conditioned medium was concentrated 10-fold on Centriprep concentrator (Amicon Inc, Beverly, MA) and assayed for either active TGF-β1 or TGF-β2 according to the manufacture's directions, because both ELISAs detect a minimum of 25 pg/mL TGF-β1 and 32 pg/mL TGF-β2, respectively. Final concentrations were calculated accounting for measured losses. The concentration of total TGF-βs was determined in unconcentrated medium immediately after acid activation of all latent TGF-βs.
Statistics
Significance was determined by the 2-tailed t test.
Results
Expression of Zf9 by BAECs
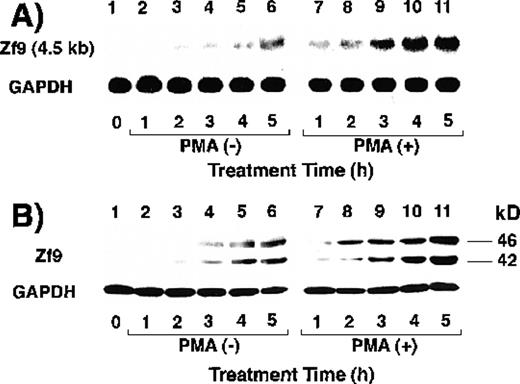
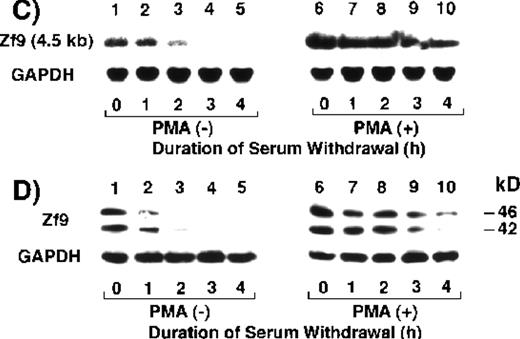
We first determined whether BAECs express Zf9. As seen in Figure1A, BAECs maintained in serum-free medium barely expressed a zf9 mRNA transcript of 4.5 kb (lane 1). Incubation of the cells with 10% FCS-containing medium inducedzf9 mRNA expression in a time-dependent manner after 2 hours (lanes 3-6). This induction was potentiated by inclusion of 10 ng/mL PMA (lanes 7-11). Similar induction of Zf9 was also observed in protein levels using Western blot (Figure 1B). PMA alone induced zf9mRNA and protein expression in the absence of serum (data not shown). When serum was removed after a 5-hour exposure in the absence of PMA, Zf9 expression disappeared within 3 to 4 hours (Figure 1C and D,lanes 1-5). On the other hand, the down-regulation of Zf9 by serum withdrawal in the presence of PMA is only partial (Figure 1C and D, lanes 6-10), because PMA can stimulate Zf9 production. These results indicate that Zf9 is a serum- and PMA-inducible factor expressed by BAECs. Furthermore, PMA may slow the degradation of Zf9. None of the following serum factors induced Zf9 when added individually to serum-free medium: platelet-derived growth factor, TGF-β, tumor necrosis factor-α, and interleukin-1β, or retinoic acid.
Expression of Zf9 by BAECs.
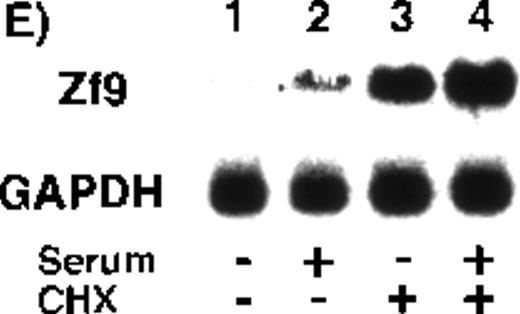
(A) and (B) Time course in induction by serum and PMA. After a 5-hour preincubation with serum-free medium confluent BAEC cultures in either 15-cm dishes or 6-cm dishes were incubated with 10% FCS-containing medium for the indicated times, in the absence and presence of 10 ng/mL PMA. Cell lysate was prepared from each 15-cm dish, total RNA was isolated, and 30 μg of each RNA was used for detection of zf9mRNA in Northern blotting (panel A). Cells on each 6-cm dish were scraped into 60 μL of sodium dodecyl sulfate (SDS) sample buffer, and rapidly passed 20 times through a 17-gauge needle to shear nucleic acids. A portion (30 μL) of each cell lysate was immediately subjected to SDS-polyacrylamide gel electrophoresis on 13% resolving gel without reducing conditions, and changes in Zf9 protein levels were analyzed by Western blotting as described in “Materials and Methods” (panel B). Lane 1, control; lanes 2-6, without PMA; lanes 7-11, with PMA. Each experiment was repeated 5 times with similar results and representative results are shown. (C) and (D) Disappearance of Zf9 expression by BAECs after depletion of serum. Following a 5-hour incubation with 10% serum ± 10 ng/mL PMA, BAECs were then incubated with serum-free medium ± 10 ng/mL PMA, and harvested at increasing intervals thereafter. The zf9 mRNA levels (panel C) and Zf9 protein levels (panel D) were analyzed by Northern blotting and Western blotting, respectively. Lanes 1-5, without PMA; lanes 6-10, with PMA. Each experiment was repeated 3 times with similar results and representative results are shown. (E) Superinduction of Zf9 mRNA in the presence of cycloheximide. After a preincubation in serum-free medium for 5 hours, confluent BAEC cultures were incubated for 3 hours with either fresh serum-free medium or 10% serum-containing medium in the absence or presence of 6 μg/mL cycloheximide (CHX). Cell lysates were prepared and the changes of zf9 mRNA levels were assessed by Northern blotting. Lane 1, no stimulus; lane 2, serum alone; lane 3, cycloheximide alone; lane 4, serum plus cycloheximide. The experiment was repeated 4 times with similar results and a representative result is shown.
Expression of Zf9 by BAECs.
(A) and (B) Time course in induction by serum and PMA. After a 5-hour preincubation with serum-free medium confluent BAEC cultures in either 15-cm dishes or 6-cm dishes were incubated with 10% FCS-containing medium for the indicated times, in the absence and presence of 10 ng/mL PMA. Cell lysate was prepared from each 15-cm dish, total RNA was isolated, and 30 μg of each RNA was used for detection of zf9mRNA in Northern blotting (panel A). Cells on each 6-cm dish were scraped into 60 μL of sodium dodecyl sulfate (SDS) sample buffer, and rapidly passed 20 times through a 17-gauge needle to shear nucleic acids. A portion (30 μL) of each cell lysate was immediately subjected to SDS-polyacrylamide gel electrophoresis on 13% resolving gel without reducing conditions, and changes in Zf9 protein levels were analyzed by Western blotting as described in “Materials and Methods” (panel B). Lane 1, control; lanes 2-6, without PMA; lanes 7-11, with PMA. Each experiment was repeated 5 times with similar results and representative results are shown. (C) and (D) Disappearance of Zf9 expression by BAECs after depletion of serum. Following a 5-hour incubation with 10% serum ± 10 ng/mL PMA, BAECs were then incubated with serum-free medium ± 10 ng/mL PMA, and harvested at increasing intervals thereafter. The zf9 mRNA levels (panel C) and Zf9 protein levels (panel D) were analyzed by Northern blotting and Western blotting, respectively. Lanes 1-5, without PMA; lanes 6-10, with PMA. Each experiment was repeated 3 times with similar results and representative results are shown. (E) Superinduction of Zf9 mRNA in the presence of cycloheximide. After a preincubation in serum-free medium for 5 hours, confluent BAEC cultures were incubated for 3 hours with either fresh serum-free medium or 10% serum-containing medium in the absence or presence of 6 μg/mL cycloheximide (CHX). Cell lysates were prepared and the changes of zf9 mRNA levels were assessed by Northern blotting. Lane 1, no stimulus; lane 2, serum alone; lane 3, cycloheximide alone; lane 4, serum plus cycloheximide. The experiment was repeated 4 times with similar results and a representative result is shown.
Next, the effect of the protein synthesis inhibitor, cycloheximide, onzf9 mRNA expression was assessed to determine if induction by serum comprised an immediate-early response. Addition of 6 μg/mL cycloheximide did not abrogate the induction of zf9 mRNA by serum, but rather resulted in prominent superinduction of the transcript (Figure 1E, lanes 3 and 4), confirming the immediate-early nature of zf9 induction in BAECs.
Binding of recombinant Zf9 to the uPA GC box
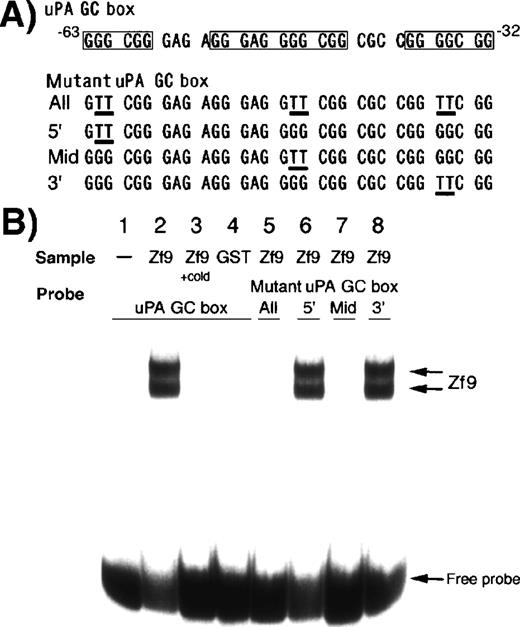
To determine if Zf9 interacts with the uPA promoter sequences, we examined binding of Zf9-GST to native and mutated GC boxes within the uPA promoter by gel shift assay (Figure2B). Zf9-GST fusion protein strongly bound to the wild-type uPA GC box (lane 2), whereas none was detected for GST alone (lane 4). Two shifted bands were detected. In experiments not shown, the higher molecular weight band corresponds to intact Zf9 based on Western blotting of the nondenaturing gel following transfer to nylon, and a lower molecular weight band represents a degradation product containing only the DNA binding domain.39Interaction of both intact and degraded Zf9 with labeled oligonucleotide was completely inhibited by a 20-fold excess of unlabeled oligonucleotide (lane 3). Addition of anti-Zf9 antibody abolished rather than supershifted the interaction between recombinant Zf9 and the oligonucleotide (data not shown), similar to what we reported previously using a consensus GC box oligonucleotide as a probe.39
Binding of Zf9-GST to GC boxes in the uPA promoter.
Binding of Zf9-GST fusion protein to the 32P-labeled uPA GC box oligonucleotide was determined by gel shift assay. Four different mutant oligonucleotides were tested, which contained mutations in one or all of the GC boxes as shown. Sequences of these oligonucleotides are indicated in panel A. Protein-DNA complexes were separated through a 6% polyacrylamide gel and visualized on an image analyzer (panel B). Lane 1, wild uPA GC box alone; lane 2, wild uPA GC box + Zf9-GST; lane 3, wild uPA GC box + Zf9-GST in the presence of a 20-fold excess of unlabeled (cold) oligonucleotide; lane 4, wild uPA GC box + GST; lanes 5-8, mutant uPA GC boxes + Zf9-GST. Lane 5, oligonucleotide with mutations in all 3 GC boxes (abbreviated as “All”); lane 6; mutant oligonucleotide in which 5′ site GC box was mutated (abbreviated as “5′”); lane 7, mutant oligonucleotide in which the middle GC box was mutated (abbreviated as “Mid”); lane 8; mutant oligonucleotide in which 3′ site GC box was mutated (abbreviated as “3′”). The experiment was repeated 3 times with similar results in all experiments; representative results are shown.
Binding of Zf9-GST to GC boxes in the uPA promoter.
Binding of Zf9-GST fusion protein to the 32P-labeled uPA GC box oligonucleotide was determined by gel shift assay. Four different mutant oligonucleotides were tested, which contained mutations in one or all of the GC boxes as shown. Sequences of these oligonucleotides are indicated in panel A. Protein-DNA complexes were separated through a 6% polyacrylamide gel and visualized on an image analyzer (panel B). Lane 1, wild uPA GC box alone; lane 2, wild uPA GC box + Zf9-GST; lane 3, wild uPA GC box + Zf9-GST in the presence of a 20-fold excess of unlabeled (cold) oligonucleotide; lane 4, wild uPA GC box + GST; lanes 5-8, mutant uPA GC boxes + Zf9-GST. Lane 5, oligonucleotide with mutations in all 3 GC boxes (abbreviated as “All”); lane 6; mutant oligonucleotide in which 5′ site GC box was mutated (abbreviated as “5′”); lane 7, mutant oligonucleotide in which the middle GC box was mutated (abbreviated as “Mid”); lane 8; mutant oligonucleotide in which 3′ site GC box was mutated (abbreviated as “3′”). The experiment was repeated 3 times with similar results in all experiments; representative results are shown.
We determined which GC boxes among 3 uPA promoter GC boxes are critical for Zf9 binding. Intact Zf9 did not bind to the oligonucleotides containing mutations in the middle GC box (lanes 5 and 7), but did bind to the mutants preserving the middle GC box (lanes 6 and 8), indicating that this middle GC box is the most critical for interaction between Zf9 and the uPA promoter.
Enhancement of PA activity by Zf9 via transactivation of uPA gene
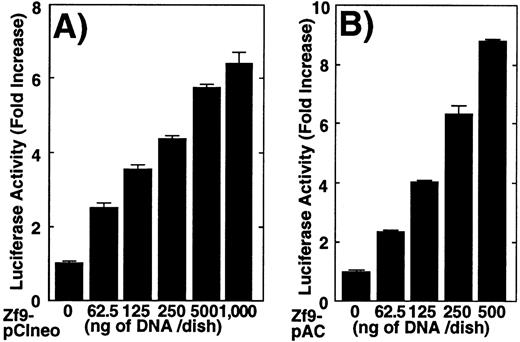
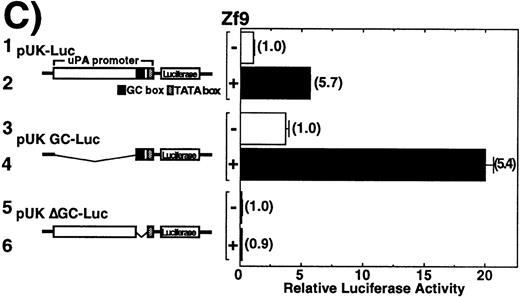
Transactivation of uPA by Zf9 was explored using transient transfection. Cotransfection of a uPA reporter (pGL2-2350) and the Zf9 expression vector (Zf9-pCIneo) in BAECs resulted in a concentration-dependent enhancement of luciferase activity (Figure3A). Relative luciferase activity increased maximally to 6-fold compared to the control cells transfected only with pCIneo empty vector. To confirm that transactivation of uPA by Zf9 was not dependent on coexpression of Sp1, the same analysis was performed in Drosophila S2 cells, which are devoid of endogenous Zf9 as well as Sp1-like activity.35,39 A similar effect was obtained and interestingly, the relative activity was greater in this cell type, probably due to low basal activity (Figure 3B). In Figure 3C we compared the effect of Zf9 expression on transactivation of wild-type uPA promoter (pUK-Luc) or its mutants, either containing only GC and TATA boxes (pUK GC-Luc) or deficient in GC boxes (pUK ΔGC-Luc). The basal activities ofpUK GC-Luc (sample 3) and pUK ΔGC-Luc (sample 5) were, respectively, 360% and 13% of that of the wild-typepUK-Luc (sample 1), suggesting that the GC box region is necessary for full transactivation of the uPA gene as reported previously.36 Zf9 transfection enhanced transactivation of both wild-type pUK-Luc and pUK GC-Luc to the same extent (5.4∼5.7-fold), but not of pUK ΔGC-Luc, indicating that transactivating activity of Zf9 requires GC boxes within uPA promoter.
Transactivation of the uPA promoter by Zf9 in BAECs andDrosophila S2 cells.
(A) and (B) BAEC and Drosophila S2 cell cultures grown on 35-mm dishes were cotransfected with a combination of the indicated amounts of Zf9 expression vector (Zf9-pCIneo or Zf9-pAC) and 1 μg of pGL2-2350, luciferase reporter gene fused with the uPA promoter, as described in “Materials and Methods.” After a 48-hour incubation, cell lysates were prepared, luciferase activity in each lysate was determined and expressed as fold increase. Panel A, BAECs; panel B, Drosophila S2 cells. Each value represents the average ± SD from triplicate determinations. Each experiment was repeated 3 times with similar results and representative results are shown. (C) Transactivation by Zf9 of the uPA promoter via GC box regions. BAEC cultures were cotransfected with a combination of 500 ng each of either pCIneo or Zf9-pCIneo plus 1 μg each of either pGL2-2350 (pUK-Luc), pGL2-GC (pUK GC-Luc), or pGL2-ΔGC (pUK ΔGC-Luc). Cell lysates were prepared, and luciferase activity of each lysate was determined. Data are expressed as relative luciferase activity compared to the activity of pUK-Luc cotransfected with pCIneo alone. The numbers in parentheses to the right of each bar indicate fold-induction calculated for each reporter. Samples 1 and 2, pUK-Luc; samples 3 and 4, pUK GC-Luc; samples 5 and 6, pUK ΔGC-Luc. Odd numbers, pCIneo; even numbers, Zf9-pCIneo. Each value represents the average ± SD from triplicate determinations. Experiment was repeated 3 times with similar results and representative results are shown.
Transactivation of the uPA promoter by Zf9 in BAECs andDrosophila S2 cells.
(A) and (B) BAEC and Drosophila S2 cell cultures grown on 35-mm dishes were cotransfected with a combination of the indicated amounts of Zf9 expression vector (Zf9-pCIneo or Zf9-pAC) and 1 μg of pGL2-2350, luciferase reporter gene fused with the uPA promoter, as described in “Materials and Methods.” After a 48-hour incubation, cell lysates were prepared, luciferase activity in each lysate was determined and expressed as fold increase. Panel A, BAECs; panel B, Drosophila S2 cells. Each value represents the average ± SD from triplicate determinations. Each experiment was repeated 3 times with similar results and representative results are shown. (C) Transactivation by Zf9 of the uPA promoter via GC box regions. BAEC cultures were cotransfected with a combination of 500 ng each of either pCIneo or Zf9-pCIneo plus 1 μg each of either pGL2-2350 (pUK-Luc), pGL2-GC (pUK GC-Luc), or pGL2-ΔGC (pUK ΔGC-Luc). Cell lysates were prepared, and luciferase activity of each lysate was determined. Data are expressed as relative luciferase activity compared to the activity of pUK-Luc cotransfected with pCIneo alone. The numbers in parentheses to the right of each bar indicate fold-induction calculated for each reporter. Samples 1 and 2, pUK-Luc; samples 3 and 4, pUK GC-Luc; samples 5 and 6, pUK ΔGC-Luc. Odd numbers, pCIneo; even numbers, Zf9-pCIneo. Each value represents the average ± SD from triplicate determinations. Experiment was repeated 3 times with similar results and representative results are shown.
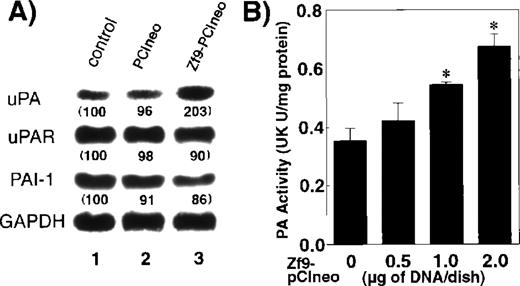
We next examined the effect of transiently transfected Zf9 on endogenous uPA expression and activity in BAEC cultures. uPA mRNA expression in BAEC cultures was up-regulated 200% as determined by densitometry normalized to GAPDH expression following transfection ofzf9 cDNA despite the typically low (< 20%) efficiency of transient transfection (Figure4A, upper bands). Because both uPAR and PAI-1 promoters also contain GC boxes,57 58 we examined the effect of Zf9 on these mRNAs. In contrast to uPA, mRNA expression of these genes was not affected by Zf9 (Figure 4A, second and third bands). The selective increase in uPA mRNA but not its inhibitor or receptor suggested that Zf9 might enhance net PA activity. Indeed, transient transfection with Zf9 increased endogenous cellular PA activity to 200% (Figure 4B). Furthermore, membrane-associated plasmin levels were increased from 9.8 ng/106 cells to 27.5 ng/106 cells after transfection with Zf9 for 48 hours. These results suggest that Zf9 promotes uPA transcription through GC box region and enhances surface plasmin levels of BAEC cultures.
Effect of Zf9 on endogenous PA activity in BAEC cultures.
(A) Effect on mRNA expression of uPA, uPAR, and PAI-1. BAEC cultures grown on 10-cm dishes were transfected with 8 μg ofZf9-pCIneo. As a control, cells were transfected with the same amount of pCIneo. Forty-eight hours after transfection, cell lysates were prepared, total RNA was extracted, and mRNA expression of uPA, uPAR, PAI-1, and GAPDH in each sample was assessed by Northern blotting. Radioactivity of each band was detected on an image analyzer, and relative intensity of each band was quantitated, normalized against the intensity of GAPDH, and expressed as arbitrary units in parentheses under each band. Each experiment was repeated 3 times with similar results in all, and representative results are shown. (B) Effect on BAEC PA activity levels. BAEC cultures grown on 35-mm dishes were transfected with the indicated amounts of Zf9-pCIneo. Forty-eight hours after transfection, cells were harvested in lysis buffer, and PA levels in each lysate were determined using chromogenic substrate, S-2403, and expressed as urokinase (UK) unit (U)/mg of protein in the sample. Each value represents the average ± SD from triplicate determinations. Points marked by an asterisk differ significantly (P < 0.05) from control cells transfected with 2 μg of pCIneo alone. Experiment was repeated 4 times with similar results in all, and representative results are shown.
Effect of Zf9 on endogenous PA activity in BAEC cultures.
(A) Effect on mRNA expression of uPA, uPAR, and PAI-1. BAEC cultures grown on 10-cm dishes were transfected with 8 μg ofZf9-pCIneo. As a control, cells were transfected with the same amount of pCIneo. Forty-eight hours after transfection, cell lysates were prepared, total RNA was extracted, and mRNA expression of uPA, uPAR, PAI-1, and GAPDH in each sample was assessed by Northern blotting. Radioactivity of each band was detected on an image analyzer, and relative intensity of each band was quantitated, normalized against the intensity of GAPDH, and expressed as arbitrary units in parentheses under each band. Each experiment was repeated 3 times with similar results in all, and representative results are shown. (B) Effect on BAEC PA activity levels. BAEC cultures grown on 35-mm dishes were transfected with the indicated amounts of Zf9-pCIneo. Forty-eight hours after transfection, cells were harvested in lysis buffer, and PA levels in each lysate were determined using chromogenic substrate, S-2403, and expressed as urokinase (UK) unit (U)/mg of protein in the sample. Each value represents the average ± SD from triplicate determinations. Points marked by an asterisk differ significantly (P < 0.05) from control cells transfected with 2 μg of pCIneo alone. Experiment was repeated 4 times with similar results in all, and representative results are shown.
Coordinated expression of Zf9 and uPA proteins by vascular endothelial cells of the carotid artery and aorta after balloon injury
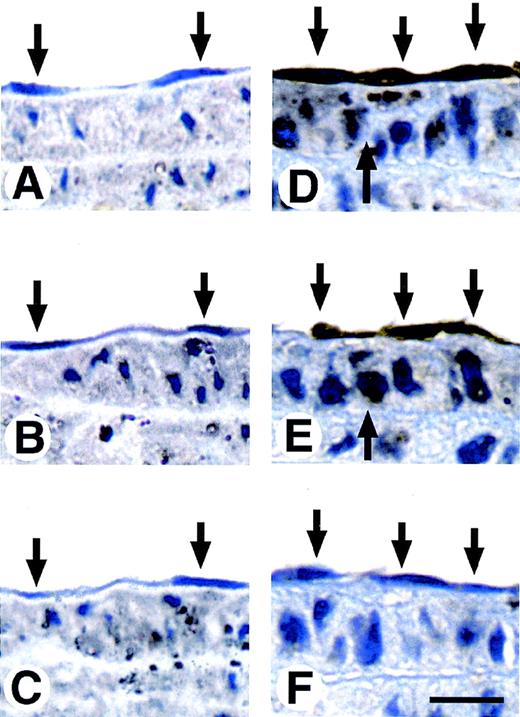
Vascular endothelial cells express a small amount or no uPA in normal arteries, but uPA expression dramatically increases after vascular injury.12 55 To study the role of Zf9 in response to vascular injury in vivo, we injured the distal half of the left carotid artery of rats with a balloon catheter and immunostained carotid artery with Zf9 and uPA antibodies at various time points after the injury. At 3 hours after the injury, neither Zf9 nor uPA immunoreactivity was detected in the vascular endothelial cells or in the smooth muscle cells (Figure 5, panels A and B, respectively). Within 2 days of injury, however, both Zf9 and uPA immunoreactivities were detected in the vascular endothelial cells (down arrows in Figure 5, panels D and E, respectively), whereas neither was detected in the endothelial cells of uninjured control rats. Weak immunoreactivity was also detected in the medial smooth muscle cells of the injured carotid artery (up arrows). Interestingly, Zf9 was induced not only in endothelial cells near the wound edge, but also in the adjacent proximal half of carotid artery, as well as in the endothelial cells of the aorta in the same animal. Nonimmune serum did not give any positive staining (Figure 5, panels C and F).
Concomitant expression of Zf9 and uPA in arterial endothelial cells after carotid balloon injury in rats.
The distal half of the left carotid artery of rats was injured with a balloon catheter. At 3 hours and 2 days after injury, the carotid artery was perfusion fixed with 3% paraformaldehyde, excised, fixed again with 3% paraformaldehyde, and paraffin embedded. Serial sections were stained with anti-Zf9 antibody (panels A and D), anti-uPA antibody (panels B and E) or nonimmune antibody (panels C and F) as described in the Materials and Methods. Panels A through F show the typical staining pattern of the adjacent proximal half of carotid artery at 3 hours (panels A-C) and 2 days (panels D-F) after injury. Down arrows indicate the endothelial cells. Up arrows indicate the smooth muscle cells. Scale bar, 50 μm.
Concomitant expression of Zf9 and uPA in arterial endothelial cells after carotid balloon injury in rats.
The distal half of the left carotid artery of rats was injured with a balloon catheter. At 3 hours and 2 days after injury, the carotid artery was perfusion fixed with 3% paraformaldehyde, excised, fixed again with 3% paraformaldehyde, and paraffin embedded. Serial sections were stained with anti-Zf9 antibody (panels A and D), anti-uPA antibody (panels B and E) or nonimmune antibody (panels C and F) as described in the Materials and Methods. Panels A through F show the typical staining pattern of the adjacent proximal half of carotid artery at 3 hours (panels A-C) and 2 days (panels D-F) after injury. Down arrows indicate the endothelial cells. Up arrows indicate the smooth muscle cells. Scale bar, 50 μm.
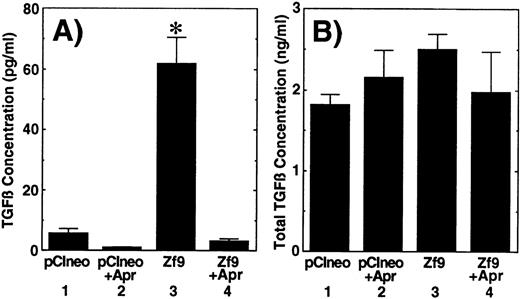
Induction of TGF-β by Zf9 via enhancement of PA activity
Finally, we examined whether Zf9 has biologic activity in this system via enhancement of PA/plasmin activity. We speculated that up-regulation of PA/plasmin by Zf9 might result in the activation of latent TGF-β as has been demonstrated in other systems.25,59,60 This hypothesis was tested by 2 different assays. First, as determined by bioassays, we observed that transient transfection of Zf9 cDNA induced the generation of active TGF-β in BAEC cultures during a subsequent 12-hour incubation with serum-free medium (Figure 6A, sample 3). This increase was eliminated by inclusion of a plasmin inhibitor, aprotinin, during the preparation of the conditioned medium (Figure 6A, sample 4), suggesting that TGF-β formation resulted from proteolytic activation of latent TGF-β.17-25,59 60 On the other hand, total TGF-β levels only moderately (35%) increased (Figure6B, sample 3), and there were no statistically significant differences between each sample. A similar result was also obtained with TGF-β1 and TGF-β2 ELISAs, although the basal levels of active TGF-βs (sum of TGF-β1 and TGF-β2 concentrations) were higher than those determined with the bioassay (Table 1). Transient transfection of Zf9 did not alter endogenous TGF-β1 and TGF-β2 mRNA levels in BAEC cultures (data not shown). These data indicate that Zf9 can induce TGF-β activity in BAECs and that the increased TGF-β in this system derived mainly from conversion of the latent to the active form of the cytokine, due at least in part to increased fibrinolytic activity.
Activation of latent TGF-β following transfection of Zf9.
BAEC cultures grown on 35-mm dishes were transfected with 1 μg of pCIneo or Zf9-pCIneo as described above. After a 36-hour incubation, medium was changed to serum-free αMEM containing 0.1% BSA, and cells were further incubated for 12 hours in the absence and presence of 50 μg/mL aprotinin (Apr). The concentration of active (panel A) and total (panel B) TGF-β in each conditioned medium was determined by the bioassays as described in “Materials and Methods.” Each value represents the average ± SD from triplicate determinations. Each experiment was repeated 3 times with similar results in all, and representative results are shown. Sample 1, pCIneo; sample 2, pCIneo with inclusion of aprotinin; sample 3,Zf9-pCIneo; sample 4, Zf9-pCIneo with inclusion of aprotinin. Points marked by an asterisk differ significantly (P < 0.05) from the control (sample 1).
Activation of latent TGF-β following transfection of Zf9.
BAEC cultures grown on 35-mm dishes were transfected with 1 μg of pCIneo or Zf9-pCIneo as described above. After a 36-hour incubation, medium was changed to serum-free αMEM containing 0.1% BSA, and cells were further incubated for 12 hours in the absence and presence of 50 μg/mL aprotinin (Apr). The concentration of active (panel A) and total (panel B) TGF-β in each conditioned medium was determined by the bioassays as described in “Materials and Methods.” Each value represents the average ± SD from triplicate determinations. Each experiment was repeated 3 times with similar results in all, and representative results are shown. Sample 1, pCIneo; sample 2, pCIneo with inclusion of aprotinin; sample 3,Zf9-pCIneo; sample 4, Zf9-pCIneo with inclusion of aprotinin. Points marked by an asterisk differ significantly (P < 0.05) from the control (sample 1).
Summary of the results of TGF-β1 and TGF-β2 ELISAs
| Vector . | Aprotinin (50 μg/mL) . | Active TGF-β (pg/mL) . | Total TGF-β (pg/mL) . | ||
|---|---|---|---|---|---|
| TGF-β1 . | TGF-β2 . | TGF-β1 . | TGF-β2 . | ||
| pClneo | − | 20.2 ± 3.3 | 18.1 ± 3.0 | 850 ± 64 | 1035 ± 65 |
| pClneo | + | 15.2 ± 2.2 | 14.7 ± 2.0 | 963 ± 47 | 851 ± 126 |
| Zf9-pClneo | − | 65.3 ± 7.2* | 48.6 ± 6.5* | 1,029 ± 47* | 1,118 ± 51 |
| Zf9-pClneo | + | 37.4 ± 1.1† | 29.8 ± 1.0† | 1,041 ± 79 | 1,035 ± 53 |
| Vector . | Aprotinin (50 μg/mL) . | Active TGF-β (pg/mL) . | Total TGF-β (pg/mL) . | ||
|---|---|---|---|---|---|
| TGF-β1 . | TGF-β2 . | TGF-β1 . | TGF-β2 . | ||
| pClneo | − | 20.2 ± 3.3 | 18.1 ± 3.0 | 850 ± 64 | 1035 ± 65 |
| pClneo | + | 15.2 ± 2.2 | 14.7 ± 2.0 | 963 ± 47 | 851 ± 126 |
| Zf9-pClneo | − | 65.3 ± 7.2* | 48.6 ± 6.5* | 1,029 ± 47* | 1,118 ± 51 |
| Zf9-pClneo | + | 37.4 ± 1.1† | 29.8 ± 1.0† | 1,041 ± 79 | 1,035 ± 53 |
The concentration of active and total TGF-β1 as well as TGF-β2 in each conditioned medium prepared in Figure 6 was determined by subtype-specific ELISAs as described in “Materials and Methods.” Each value represents the average ± SD from triplicate determinations. Each experiment was repeated 3 times with similar results in all, and representative results are shown.
P values obtained by a comparison between pClneo andZf9-pClneo are less than 0.05 except for total TGF-β2 concentration.
P values obtained by a comparison betweenZf9-pClneo and Zf9-pClneo + aprotinin are less than 0.05 for both active TGF-β1 and TGF-β2 concentrations.
Discussion
We have demonstrated a potential role of Zf9 in regulating vascular injury through its marked stimulatory effects on uPA gene expression and TGF-β activity. Cultured BAECs express Zf9 in response to serum as well as PMA (see Figure 1A), which is a model agonist of a hitherto known early response gene, Egr-133; we have not yet identified the responsible serum factor. The kinetic analysis of zf9 mRNA and protein (see Figures 1A-D) suggests that both are labile, and therefore withdrawal of appropriate stimulus culminates in their rapid disappearance. This observation, combined with superinduction by cycloheximide (see Figure 1E), supports the role of Zf9 as an immediate-early gene, as previously observed in hepatic stellate cells in vivo.38 As shown in Figure 1B and D, Zf9 protein is expressed by BAECs as a doublet of 46 kd and 42 kd, consistent with our previous finding in hepatic stellate cells.39 Furthermore, transient transfection of human or rat Zf9 into BAECs also yielded the same doublet in Western blot (data not shown). As we have suggested previously, the 14-kd or 10-kd size differences from 32 kd, which is the predicted size of the unmodified polypeptide, may be explained by posttranslational modifications, but this has not yet been tested directly.39
We have also demonstrated that both the interaction between recombinant Zf9 and the uPA promoter (see Figure 2) and transactivation of theuPA gene by Zf9 (see Figure 3) are dependent on GC boxes within the promoter in vitro. Incubation of Zf9-GST with mutated promoter sequences establishes that the middle GC box is the most essential for Zf9 binding. The substantial increase in endogenous uPA activity after transient transfection of Zf9 (see Figure 4), despite a typical transfection efficiency of only 10% to 20%, underscores the potency of this factor in up-regulating uPA expression. Importantly, vascular injury caused by a balloon catheter induces the expression of Zf9 by endothelial cells in vivo (see Figure 5). Concomitant induction of uPA in this tissue underscores the notion that Zf9 may play an important role in the uPA gene expression induced by vascular injury. This finding supports our in vitro observations and agrees with findings of previous investigators characterizing uPA induction after vascular injury in vivo.12 We acknowledge, however, that the current data do not establish a direct role of Zf9 in uPA expression in vivo.
Zf9 belongs to an enlarging family of zinc finger Krüppel-like transcription factors.42-52 The physiologic roles and distribution of these factors are distinct.52Sp1is widely expressed in various types of cells at a fairly constant level and is involved in basal transactivation of many genes including uPA.36,37Zf9 is also widely expressed but is an early response gene in both hepatic stellate38,39 and vascular endothelial cells (current study) during the wound healing response. In this context, Zf9 resembles Egr-1, a non-Krüppel-like zinc finger transcription factor that has been identified also as an early gene induced by serum, PMA, and tissue injury, and can substitute for Sp1 in promoting transcription of genes including uPA.33 Therefore, it is of great interest to know how Sp1, Egr-1, Zf9, and other Krüppel-like factors interact in regulating uPA expression. In preliminary studies we have documented a synergism and physical interaction between Zf9 and Sp1.61This contrasts with the interplay of Sp1 and Egr-1.33
We previously demonstrated that Zf9 transactivates promoters of collagen α1(I), TGF-β1, and its signaling receptors in hepatic stellate cells.39,53 Activation of latent TGF-β via uPA induction provides an additional mechanism through which TGF-β1 activity is augmented by Zf9 (see Figure 6 and Table 1). This has also been observed in other systems in which uPA is up-regulated, for example, in response to basic fibroblast growth factor or retinoic acid.59,60 Activation of latent TGF-β has been implicated as an important regulator of vascular smooth muscle cell differentiation, endothelial cell quiescence, and extracellular matrix production during vessel morphogenesis. It seems that latent TGF-β is activated through different mechanisms under physiologic and pathologic situations. It is intriguing to determine if Zf9 might also stimulate the expression of other TGF-β activators, such as thrombospondin and integrin, αvβ6.62,63 We speculate that Zf9 may promote different GC box-containing gene promoters in a tissue and cell context-dependent manner. For example, the current (see Figure 3B) and previous39,53 data suggest that in S2 cells Zf9 transactivates the SV40 promoter as well as promoters of TGF-β1 and uPA, but not promoters of collagen α1(I), types I and II TGF-β receptors. Although the percentage of active TGF-β in our system was low (2.4% in Figure 6 and 4.3%-6.3% in Table 1), 40 to 60 pg/mL TGF-β is enough to exert its biologic effects on BAECs as we and others have reported.23,56,59 We cannot explain the reason for the differences in the concentration of active TGF-β determined by the biologic assays (see Figure 6) and ELISAs (see Table 1). We speculate that artificial activation might occur during concentration of the conditioned medium prior to ELISA, resulting in high basal levels. It would be difficult to evaluate if serum-induced Zf9 also generates active TGF-β in BAEC cultures because 10% serum used in the current study already contains 99.5 ± 4.7 pg/mL active TGF-β1 and 11.2 ± 3.8 pg/mL active TGF-β2 as measured by ELISA. This preexisting TGF-β might counteract the ability of Zf9 to enhance uPA expression, inferring that the system is self-regulated, similar to what has been reported previously in other systems.64
In summary, we have shown that both in vitro and in vivo Zf9 is expressed by vascular endothelial cells in response to certain stimuli and can promote transactivation of uPA, which leads to activation of latent TGF-β, suggesting that this system may have an important role in vascular injury response. The present data and previous findings25,38,39 strongly suggest that rapid, early induction of Zf9 may promote fibrogenesis by potentiating the proteolytic activation of latent TGF-β in hepatic stellate cells. It will be important to determine if Zf9 regulates uPA activity and TGF-β1 responses in other in vivo models of tissue injury including atherosclerosis and restenosis,6,20 cutaneous wound healing,2 pulmonary intestinal pneumonia and fibrosis,22,63 and hepatic fibrosis.1 39
Acknowledgments
We gratefully acknowledge F. Blasi for helpful advice and critical reading of the manuscript; H. Tanaka, W.-D. Schleuning, and D. J. Loskutoff for Drosophila S2 cells and constructs; and L. Wong and C. Iijima for their technical assistance.
Supported partly by Grant-in-Aids from the Ministry of Education, Science, Sports and Culture 10780395, Grant for “Biodesign Research Program” and “Multibioprobe Research Program” from RIKEN, The Special Coordination Funds for Promoting Science and Technology from the Science and Technology Agency, and the National Institutes of Health (DK37340).
S.K. and S.H. contributed equally to this work.
Reprints:Soichi Kojima, Laboratory of Molecular Cell Sciences, Tsukuba Life Science Center, The Institute of Physical and Chemical Research (RIKEN), Koyadai, Tsukuba, Ibaraki 305-0074, Japan; e-mail: kojima@rtc.riken.go.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal